WO2003070918A2 - Rna interference by modified short interfering nucleic acid - Google Patents
Rna interference by modified short interfering nucleic acid Download PDFInfo
- Publication number
- WO2003070918A2 WO2003070918A2 PCT/US2003/005346 US0305346W WO03070918A2 WO 2003070918 A2 WO2003070918 A2 WO 2003070918A2 US 0305346 W US0305346 W US 0305346W WO 03070918 A2 WO03070918 A2 WO 03070918A2
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- sina
- sina molecule
- nucleotides
- molecule
- gene
- Prior art date
Links
- 0 B*(*)(*1)C(*)(*)C(*)(*)[C@@]1(*)C(*)(*)* Chemical compound B*(*)(*1)C(*)(*)C(*)(*)[C@@]1(*)C(*)(*)* 0.000 description 1
Classifications
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/63—Introduction of foreign genetic material using vectors; Vectors; Use of hosts therefor; Regulation of expression
- C12N15/79—Vectors or expression systems specially adapted for eukaryotic hosts
- C12N15/82—Vectors or expression systems specially adapted for eukaryotic hosts for plant cells, e.g. plant artificial chromosomes (PACs)
- C12N15/8216—Methods for controlling, regulating or enhancing expression of transgenes in plant cells
- C12N15/8218—Antisense, co-suppression, viral induced gene silencing [VIGS], post-transcriptional induced gene silencing [PTGS]
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K47/00—Medicinal preparations characterised by the non-active ingredients used, e.g. carriers or inert additives; Targeting or modifying agents chemically bound to the active ingredient
- A61K47/50—Medicinal preparations characterised by the non-active ingredients used, e.g. carriers or inert additives; Targeting or modifying agents chemically bound to the active ingredient the non-active ingredient being chemically bound to the active ingredient, e.g. polymer-drug conjugates
- A61K47/51—Medicinal preparations characterised by the non-active ingredients used, e.g. carriers or inert additives; Targeting or modifying agents chemically bound to the active ingredient the non-active ingredient being chemically bound to the active ingredient, e.g. polymer-drug conjugates the non-active ingredient being a modifying agent
- A61K47/54—Medicinal preparations characterised by the non-active ingredients used, e.g. carriers or inert additives; Targeting or modifying agents chemically bound to the active ingredient the non-active ingredient being chemically bound to the active ingredient, e.g. polymer-drug conjugates the non-active ingredient being a modifying agent the modifying agent being an organic compound
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P1/00—Drugs for disorders of the alimentary tract or the digestive system
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P1/00—Drugs for disorders of the alimentary tract or the digestive system
- A61P1/04—Drugs for disorders of the alimentary tract or the digestive system for ulcers, gastritis or reflux esophagitis, e.g. antacids, inhibitors of acid secretion, mucosal protectants
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P1/00—Drugs for disorders of the alimentary tract or the digestive system
- A61P1/16—Drugs for disorders of the alimentary tract or the digestive system for liver or gallbladder disorders, e.g. hepatoprotective agents, cholagogues, litholytics
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P11/00—Drugs for disorders of the respiratory system
- A61P11/06—Antiasthmatics
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P13/00—Drugs for disorders of the urinary system
- A61P13/08—Drugs for disorders of the urinary system of the prostate
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P13/00—Drugs for disorders of the urinary system
- A61P13/10—Drugs for disorders of the urinary system of the bladder
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P13/00—Drugs for disorders of the urinary system
- A61P13/12—Drugs for disorders of the urinary system of the kidneys
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P17/00—Drugs for dermatological disorders
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P17/00—Drugs for dermatological disorders
- A61P17/02—Drugs for dermatological disorders for treating wounds, ulcers, burns, scars, keloids, or the like
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P19/00—Drugs for skeletal disorders
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P19/00—Drugs for skeletal disorders
- A61P19/02—Drugs for skeletal disorders for joint disorders, e.g. arthritis, arthrosis
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P21/00—Drugs for disorders of the muscular or neuromuscular system
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P25/00—Drugs for disorders of the nervous system
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P25/00—Drugs for disorders of the nervous system
- A61P25/02—Drugs for disorders of the nervous system for peripheral neuropathies
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P25/00—Drugs for disorders of the nervous system
- A61P25/28—Drugs for disorders of the nervous system for treating neurodegenerative disorders of the central nervous system, e.g. nootropic agents, cognition enhancers, drugs for treating Alzheimer's disease or other forms of dementia
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P27/00—Drugs for disorders of the senses
- A61P27/02—Ophthalmic agents
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P27/00—Drugs for disorders of the senses
- A61P27/16—Otologicals
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P29/00—Non-central analgesic, antipyretic or antiinflammatory agents, e.g. antirheumatic agents; Non-steroidal antiinflammatory drugs [NSAID]
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P3/00—Drugs for disorders of the metabolism
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P3/00—Drugs for disorders of the metabolism
- A61P3/08—Drugs for disorders of the metabolism for glucose homeostasis
- A61P3/10—Drugs for disorders of the metabolism for glucose homeostasis for hyperglycaemia, e.g. antidiabetics
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P31/00—Antiinfectives, i.e. antibiotics, antiseptics, chemotherapeutics
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P31/00—Antiinfectives, i.e. antibiotics, antiseptics, chemotherapeutics
- A61P31/04—Antibacterial agents
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P31/00—Antiinfectives, i.e. antibiotics, antiseptics, chemotherapeutics
- A61P31/10—Antimycotics
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P31/00—Antiinfectives, i.e. antibiotics, antiseptics, chemotherapeutics
- A61P31/12—Antivirals
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P31/00—Antiinfectives, i.e. antibiotics, antiseptics, chemotherapeutics
- A61P31/12—Antivirals
- A61P31/14—Antivirals for RNA viruses
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P31/00—Antiinfectives, i.e. antibiotics, antiseptics, chemotherapeutics
- A61P31/12—Antivirals
- A61P31/14—Antivirals for RNA viruses
- A61P31/16—Antivirals for RNA viruses for influenza or rhinoviruses
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P31/00—Antiinfectives, i.e. antibiotics, antiseptics, chemotherapeutics
- A61P31/12—Antivirals
- A61P31/14—Antivirals for RNA viruses
- A61P31/18—Antivirals for RNA viruses for HIV
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P31/00—Antiinfectives, i.e. antibiotics, antiseptics, chemotherapeutics
- A61P31/12—Antivirals
- A61P31/20—Antivirals for DNA viruses
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P31/00—Antiinfectives, i.e. antibiotics, antiseptics, chemotherapeutics
- A61P31/12—Antivirals
- A61P31/20—Antivirals for DNA viruses
- A61P31/22—Antivirals for DNA viruses for herpes viruses
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P35/00—Antineoplastic agents
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P35/00—Antineoplastic agents
- A61P35/02—Antineoplastic agents specific for leukemia
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P37/00—Drugs for immunological or allergic disorders
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P37/00—Drugs for immunological or allergic disorders
- A61P37/02—Immunomodulators
- A61P37/04—Immunostimulants
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P37/00—Drugs for immunological or allergic disorders
- A61P37/02—Immunomodulators
- A61P37/06—Immunosuppressants, e.g. drugs for graft rejection
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P37/00—Drugs for immunological or allergic disorders
- A61P37/08—Antiallergic agents
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P43/00—Drugs for specific purposes, not provided for in groups A61P1/00-A61P41/00
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07H—SUGARS; DERIVATIVES THEREOF; NUCLEOSIDES; NUCLEOTIDES; NUCLEIC ACIDS
- C07H21/00—Compounds containing two or more mononucleotide units having separate phosphate or polyphosphate groups linked by saccharide radicals of nucleoside groups, e.g. nucleic acids
- C07H21/02—Compounds containing two or more mononucleotide units having separate phosphate or polyphosphate groups linked by saccharide radicals of nucleoside groups, e.g. nucleic acids with ribosyl as saccharide radical
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/11—DNA or RNA fragments; Modified forms thereof; Non-coding nucleic acids having a biological activity
- C12N15/111—General methods applicable to biologically active non-coding nucleic acids
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/11—DNA or RNA fragments; Modified forms thereof; Non-coding nucleic acids having a biological activity
- C12N15/113—Non-coding nucleic acids modulating the expression of genes, e.g. antisense oligonucleotides; Antisense DNA or RNA; Triplex- forming oligonucleotides; Catalytic nucleic acids, e.g. ribozymes; Nucleic acids used in co-suppression or gene silencing
- C12N15/1131—Non-coding nucleic acids modulating the expression of genes, e.g. antisense oligonucleotides; Antisense DNA or RNA; Triplex- forming oligonucleotides; Catalytic nucleic acids, e.g. ribozymes; Nucleic acids used in co-suppression or gene silencing against viruses
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/11—DNA or RNA fragments; Modified forms thereof; Non-coding nucleic acids having a biological activity
- C12N15/113—Non-coding nucleic acids modulating the expression of genes, e.g. antisense oligonucleotides; Antisense DNA or RNA; Triplex- forming oligonucleotides; Catalytic nucleic acids, e.g. ribozymes; Nucleic acids used in co-suppression or gene silencing
- C12N15/1138—Non-coding nucleic acids modulating the expression of genes, e.g. antisense oligonucleotides; Antisense DNA or RNA; Triplex- forming oligonucleotides; Catalytic nucleic acids, e.g. ribozymes; Nucleic acids used in co-suppression or gene silencing against receptors or cell surface proteins
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61K—PREPARATIONS FOR MEDICAL, DENTAL OR TOILETRY PURPOSES
- A61K38/00—Medicinal preparations containing peptides
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/10—Type of nucleic acid
- C12N2310/11—Antisense
- C12N2310/111—Antisense spanning the whole gene, or a large part of it
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/10—Type of nucleic acid
- C12N2310/14—Type of nucleic acid interfering N.A.
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/30—Chemical structure
- C12N2310/31—Chemical structure of the backbone
- C12N2310/315—Phosphorothioates
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/30—Chemical structure
- C12N2310/31—Chemical structure of the backbone
- C12N2310/317—Chemical structure of the backbone with an inverted bond, e.g. a cap structure
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/30—Chemical structure
- C12N2310/31—Chemical structure of the backbone
- C12N2310/318—Chemical structure of the backbone where the PO2 is completely replaced, e.g. MMI or formacetal
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/30—Chemical structure
- C12N2310/32—Chemical structure of the sugar
- C12N2310/321—2'-O-R Modification
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/30—Chemical structure
- C12N2310/32—Chemical structure of the sugar
- C12N2310/322—2'-R Modification
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/30—Chemical structure
- C12N2310/33—Chemical structure of the base
- C12N2310/332—Abasic residue
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/30—Chemical structure
- C12N2310/34—Spatial arrangement of the modifications
- C12N2310/346—Spatial arrangement of the modifications having a combination of backbone and sugar modifications
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2310/00—Structure or type of the nucleic acid
- C12N2310/50—Physical structure
- C12N2310/53—Physical structure partially self-complementary or closed
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2320/00—Applications; Uses
- C12N2320/50—Methods for regulating/modulating their activity
- C12N2320/51—Methods for regulating/modulating their activity modulating the chemical stability, e.g. nuclease-resistance
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N2330/00—Production
- C12N2330/30—Production chemically synthesised
Definitions
- the present invention concerns methods and reagents useful in modulating gene expression in a variety of applications, including use in therapeutic, diagnostic, target validation, and genomic discovery applications.
- the invention relates to synthetic small nucleic acid molecules, such as short interfering nucleic acid (siNA), short interfering RNA (siRNA), double-stranded RNA (dsRNA), micro-RNA (miRNA), and short hairpin RNA (shRNA) molecules capable of mediating RNA interference (RNAi).
- siNA short interfering nucleic acid
- siRNA short interfering RNA
- dsRNA double-stranded RNA
- miRNA micro-RNA
- shRNA short hairpin RNA
- RNA interference refers to the process of sequence-specific post-transcriptional gene silencing in animals mediated by short interfering RNAs (siRNAs) (Fire et al, 1998, Nature, 391, 806). The corresponding process in plants is commonly referred to as post- transcriptional gene silencing or RNA silencing and is also referred to as quelling in fungi.
- the process of post-transcriptional gene silencing is thought to be an evolutionarily-conserved cellular defense mechanism used to prevent the expression of foreign genes and is commonly shared by diverse flora and phyla (Fire et al, 1999, Trends Genet, 15, 358).
- Such protection from foreign gene expression may have evolved in response to the production of double-stranded RNAs (dsRNAs) derived from viral infection or from the random integration of transposon elements into a host genome via a cellular response that specifically destroys homologous single-stranded RNA or viral genomic RNA.
- dsRNAs double-stranded RNAs
- the presence of dsRNA in cells triggers the RNAi response though a mechanism that has yet to be fully characterized. This mechanism appears to be different from the interferon response that results from dsRNA-mediated activation of protein kinase PKR and 2',5'-oligoadenylate synthetase resulting in non-specific cleavage of mRNA by ribonuclease L.
- dsRNAs ribonuclease III enzyme
- Dicer is involved in the processing of the dsRNA into short pieces of dsRNA known as short interfering RNAs (siRNAs) (Berstein et al, 2001, Nature, 409, 363).
- Short interfering RNAs derived from dicer activity are typically about 21 to about 23 nucleotides in length and comprise about 19 base pair duplexes (Elbashir et al, 2001, Genes Dev., 15, 188).
- Dicer has also been implicated in the excision of 21- and 22-nucleotide small temporal RNAs (stRNAs) from precursor RNA of conserved structure that are implicated in translational control (Hutvagner et al, 2001, Science, 293, 834).
- the RNAi response also features an endonuclease complex, commonly referred to as an RNA-induced silencing complex (RISC), which mediates cleavage of single- stranded RNA having sequence complementary to the antisense strand of the siRNA duplex. Cleavage of the target RNA takes place in the middle of the region complementary to the antisense strand of the siRNA duplex (Elbashir et al, 2001, Genes Dev., 15, 188).
- RISC RNA-induced silencing complex
- RNAi has been studied in a variety of systems. Fire et al, 1998, Nature, 391, 806, were the first to observe RNAi in C. elegans. Wianny and Goetz, 1999, Nature Cell Biol, 2, 70, describe RNAi mediated by dsRNA in mouse embryos. Hammond et al, 2000, Nature, 404, 293, describe RNAi in Drosophila cells transfected with dsRNA. Elbashir et al, 2001, Nature, 411, 494, describe RNAi induced by introduction of duplexes of synthetic 21 -nucleotide RNAs in cultured mammalian cells including human embryonic kidney and HeLa cells.
- siRNA may include modifications to either the phosphate-sugar backbone or the nucleoside to include at least one of a nitrogen or sulfur heteroatom, however, neither application postulates to what extent such modifications would be tolerated in siRNA molecules, nor provides any further guidance or examples of such modified siRNA. Kreutzer et al, Canadian Patent Application No.
- 2,359,180 also describe certain chemical modifications for use in dsRNA constructs in order to counteract activation of double-stranded RNA-dependent protein kinase PKR, specifically 2'-amino or 2'-O- methyl nucleotides, and nucleotides containing a 2'-O or 4'-C methylene bridge.
- W RNA-dependent protein kinase
- the authors describe the introduction of thiophosphate residues into these siRNA transcripts by incorporating thiophosphate nucleotide analogs with T7 and T3 RNA polymerase and observed that RNAs with two phosphorothioate modified bases also had substantial decreases in effectiveness as RNAi.
- Parrish et al. reported that phosphorothioate modification of more than two residues greatly destabilized the RNAs in vitro such that interference activities could not be assayed. Id. at 1081.
- the authors also tested certain modifications at the 2'-position of the nucleotide sugar in the long siRNA transcripts and found that substituting deoxynucleotides for ribonucleotides produced a substantial decrease in interference activity, especially in the case of Uridine to Thymidine and/or Cytidine to deoxy-Cytidine substitutions. Id.
- the authors tested certain base modifications, including substituting, in sense and antisense strands of the siRNA, 4-thiouracil, 5-bromouracil, 5-iodouracil, and 3-(aminoallyl)uracil for uracil, and inosine for guanosine.
- Parrish reported that inosine produced a substantial decrease in interference activity when incorporated in either strand. Parrish also reported that incorporation of 5-iodouracil and 3-(aminoallyl)uracil in the antisense strand resulted in a substantial decrease in RNAi activity as well.
- RNAi can be used to cure genetic diseases or viral infection due to the danger of activating interferon response.
- Li et al, International PCT Publication No. WO 01/68836 describes specific methods for attenuating gene expression using endogenously-derived dsRNA.
- Tuschl et al, International PCT Publication No. WO 01/75164 describe a Drosophila in vitro RNAi system and the use of specific siRNA molecules for certain functional genomic and certain therapeutic applications; although Tuschl, 2001, Chem. Biochem., 2, 239-245, doubts that RNAi can be used to cure genetic diseases or viral infection due to the danger of activating interferon response.
- WO 00/44914 describe the use of specific dsRNAs for attenuating the expression of certain target genes.
- Zernicka-Goetz et al International PCT Publication No. WO 01/36646, describe certain methods for inhibiting the expression of particular genes in mammalian W cells using certain dsRNA molecules.
- Fire et al International PCT Publication No. WO 99/32619, describe particular methods for introducing certain dsRNA molecules into cells for use in inhibiting gene expression.
- Plaetinck et al International PCT Publication No. WO 00/01846, describe certain methods for identifying specific genes responsible for conferring a particular phenotype in a cell using specific dsRNA molecules.
- RNAi and gene-silencing systems have reported on various RNAi and gene-silencing systems. For example, Parrish et al, 2000, Molecular Cell, 6, 1977-1087, describe specific chemically-modified siRNA constructs targeting the unc-22 gene of C. elegans. Grossniklaus, International PCT Publication No. WO 01/38551, describes certain methods for regulating polycomb gene expression in plants using certain dsRNAs. Churikov et al, International PCT Publication No. WO 01/42443, describe certain methods for modifying genetic characteristics of an organism using certain dsRNAs. Cogoni et al, International PCT Publication No. WO 01/53475, describe certain methods for isolating a Neurospora silencing gene and uses thereof.
- Reed et al International PCT Publication No. WO 01/68836, describe certain methods for gene silencing in plants.
- Honer et al International PCT Publication No. WO 01/70944, describe certain methods of drug screening using transgenic nematodes as Parkinson's Disease models using certain dsRNAs.
- Deak et al International PCT Publication No. WO 01/72774, describe certain Drosophila-de ⁇ vQd gene products that may be related to RNAi .in Drosophila.
- Arndt et al, International PCT Publication No. WO 01/92513 describe certain methods for mediating gene suppression by using factors that enhance RNAi. Tuschl et al, International PCT Publication No.
- WO 02/44321 describe certain synthetic siRNA constructs.
- Pachuk et al, International PCT Publication No. WO 00/63364, and Satishchandran et al, International PCT Publication No. WO 01/04313, describe certain methods and compositions for inhibiting the function of certain polynucleotide sequences using certain dsRNAs.
- Echeverri et al, International PCT Publication No. WO 02/38805 describe certain C. elegans genes identified via RNAi.
- Kreutzer et al, International PCT Publications Nos. WO 02/055692, WO 02/055693, and EP 1144623 Bl describes certain methods for inhibiting gene expression using RNAi.
- This invention relates to compounds, compositions, and methods useful for modulating RNA function and/or gene expression in a cell.
- the instant invention features synthetic small nucleic acid molecules, such as short interfering nucleic acid (siNA), short interfering RNA (siRNA), double-stranded RNA (dsRNA), micro- RNA (miRNA), and short hairpin RNA (shRNA) molecules capable of modulating gene expression in cells by RNA inference (RNAi).
- the siNA molecules of the invention can be chemically modified.
- the use of chemically modified siNA can improve various properties of native siRNA molecules through increased resistance to nuclease degradation in vivo and/or improved cellular uptake.
- the chemically modified siNA molecules of the instant invention provide useful reagents and methods for a variety of therapeutic, diagnostic, agricultural, target validation, genomic discovery, genetic engineering and pharmacogenomic applications.
- the introduction of chemically modified nucleotides into nucleic acid molecules provides a powerful tool in overcoming potential limitations of in vivo stability and bioavailabihty inherent to native RNA molecules that are delivered exogenously.
- the use of chemically modified nucleic acid molecules can enable a lower dose of a particular nucleic acid molecule for a given therapeutic effect since chemically modified nucleic acid molecules tend to have a longer half-life in serum.
- certain chemical modifications can improve the bioavailabihty of nucleic acid molecules by targeting particular cells or tissues and/or improving cellular uptake of the nucleic acid molecule.
- chemically modified nucleic acid molecule can be reduced as compared to a native nucleic acid molecule, for example when compared to an all RNA nucleic acid molecule, the overall activity of the modified nucleic acid molecule can be greater than the native molecule due to improved stability and/or delivery of the molecule.
- chemically modified siNA can also minimize the possibility of activating interferon activity in humans.
- the nucleic acid molecules of the invention that act as mediators of the RNA interference gene silencing response are chemically modified double stranded nucleic acid molecules.
- these siNA molecules typically consist of duplexes containing about 19 base pairs between oligonucleotides comprising about 19 to about 25 nucleotides.
- the most active siRNA molecules are thought to have such duplexes with overhanging ends of 1-3 nucleotides, for example 21 nucleotide duplexes with 19 base pairs and 2 nucleotide 3'- overhangs. These overhanging segments are readily hydrolyzed by endonucleases in vivo.
- siRNA may include modifications to either the phosphate-sugar back bone or the nucleoside to include at least one of a nitrogen or sulfur heteroatom, however neither application teaches to what extent these modifications are tolerated in siRNA molecules nor provide any examples of such modified siRNA. Kreutzer and Limmer, Canadian Patent Application No.
- 2,359,180 also describe certain chemical modifications for use in dsRNA constructs in order to counteract activation of double stranded-RNA-dependent protein kinase PKR, specifically 2'-amino or 2'-O-methyl nucleotides, and nucleotides containing a 2'-O or 4'-C methylene bridge.
- PKR double stranded-RNA-dependent protein kinase
- 2'-amino or 2'-O-methyl nucleotides specifically 2'-amino or 2'-O-methyl nucleotides, and nucleotides containing a 2'-O or 4'-C methylene bridge.
- Kreutzer and Limmer similarly fail to show to what extent these modifications are tolerated in siRNA molecules nor provide any examples of such modified siRNA.
- the invention features chemically modified siNA constructs having specificity for target nucleic acid molecules in a cell.
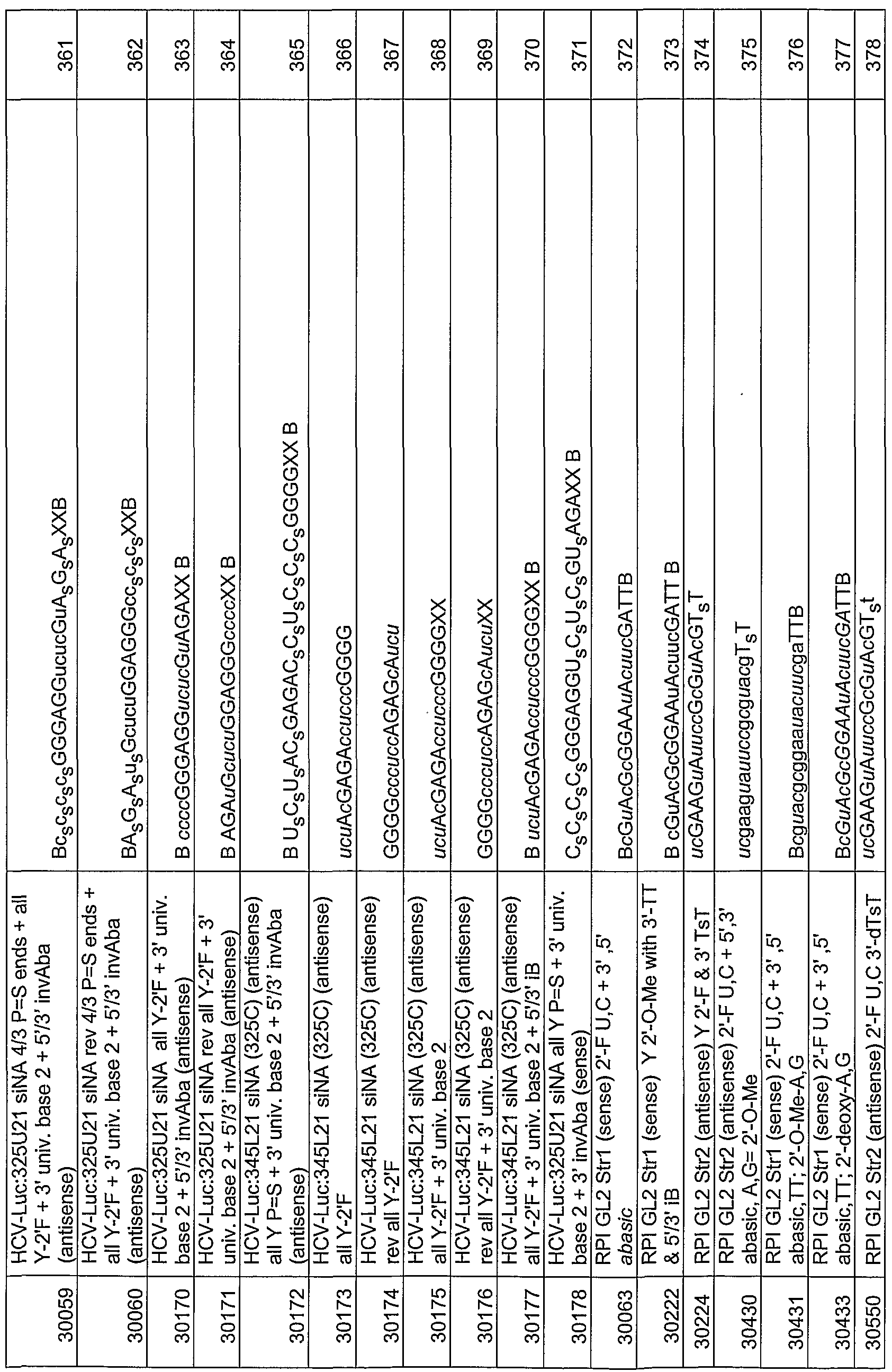
- chemical modifications include without limitation phosphorothioate internucleotide linkages, 2'-O-methyl ribonucleotides, 2 '-deoxy-2' -fluoro ribonucleotides, 2'-deoxy ribonucleotides, "universal base” nucleotides, 5-C-methyl nucleotides, and inverted deoxyabasic residue incorporation.
- the chemically-modified siNA molecules of the invention comprise a duplex having two strands, one or both of which can be chemically-modified, wherein each strand is about 19 to about 29 (e.g., about 19, 20, 21, 22, 23, 24, 25, 26, 27, 28,or 29) nucleotides.
- the chemically-modified siNA molecules of the invention comprise a duplex having two strands, one or both of which can be chemically-modified, wherein each strand is about 19 to about 23 (e.g., about 19, 20, 21, 22, or 23) nucleotides.
- a siNA molecule of the invention comprises modified nucleotides while maintaining the ability to mediate RNAi.
- the modified nucleotides can be used to improve in vitro or in vivo characteristics such as stability, activity, and/or bioavailabihty.
- a siNA molecule of the invention can comprise modified nucleotides as a percentage of the total number of nucleotides present in the siNA molecule.
- a siNA molecule of the invention can generally comprise modified nucleotides from about 5 to about 100% of the nucleotide positions (e.g., 5%, 10%, 15%, 20%, 25%, 30%, 35%, 40%, 45%, 50%, 55%, 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95% or 100% of the nucleotide positions).
- the actual percentage of modified nucleotides present in a given siNA molecule depends on the total number of nucleotides present in the siNA. If the siNA molecule is single stranded, the percent modification can be based upon the total number of nucleotides present in the single stranded siNA molecules.
- the percent modification can be based upon the total number of nucleotides present in the sense strand, antisense strand, or both the sense and antisense strands, hi addition, the actual percentage of modified nucleotides present in a given siNA molecule can also depend on the total number of purine and pyrimidine nucleotides present in the siNA, for example, wherein all pyrimidine nucleotides and/or all purine nucleotides present in the siNA molecule are modified.
- the antisense region of a siNA molecule of the mvention can comprise a phosphorothioate internucleotide linkage at the 3 '-end of said antisense region.
- the antisense region can comprise about one to about five phosphorothioate internucleotide linkages at the 5 '-end of said antisense region.
- the 3'-terminal nucleotide overhangs of a siNA molecule of the invention can comprise ribonucleotides or deoxyribonucleotides that are chemically-modified at a nucleic acid sugar, base, or backbone.
- the 3 '-terminal nucleotide overhangs can comprise one or more universal base ribonucleotides.
- the 3'- terminal nucleotide overhangs can comprise one or more acyclic nucleotides.
- the invention features a double-stranded short interfering nucleic acid (siNA) molecule that down-regulates expression of a target gene, wherein the siNA molecule comprises no ribonucleotides and each strand of the double-stranded siNA comprises about 21 nucleotides.
- siNA short interfering nucleic acid
- one of the strands of a double-stranded siNA molecule of the invention comprises a nucleotide sequence that is complementary to a nucleotide sequence or a portion thereof of a target gene, and wherein the second strand of a double- stranded siNA molecule comprises a nucleotide sequence substantially similar to the nucleotide sequence or a portion thereof of the target gene.
- a siNA molecule of the invention comprises about 19 to about 23 nucleotides, and each strand comprises at least about 19 nucleotides that are complementary to the nucleotides of the other strand.
- a siNA molecule of the invention comprises an antisense region comprising a nucleotide sequence that is complementary to a nucleotide sequence or a portion thereof of a target gene, and the siNA further comprises a sense region, wherein the sense region comprises a nucleotide sequence substantially similar to the nucleotide sequence or a portion thereof of the target gene.
- the antisense region and the sense region each comprise about 19 to about 23 nucleotides, and the antisense region comprises at least about 19 nucleotides that are complementary to nucleotides of the sense region.
- a siNA molecule of the invention comprises a sense region and an antisense region, wherein the antisense region comprises a nucleotide sequence that is complementary to a nucleotide sequence or a portion thereof of RNA encoded by a target gene and the sense region comprises a nucleotide sequence that is complementary to the antisense region.
- a siNA molecule of the invention is assembled from two separate oligonucleotide fragments wherein one fragment comprises the sense region and the second fragment comprises the antisense region of the siNA molecule.
- the sense region is connected to the antisense region via a linker molecule, which can be a polynucleotide linker or a non-nucleotide linker.
- a siNA molecule of the invention comprises a sense region and antisense region, wherein pyrimidine nucleotides in the sense region complies 2'-O- methyl pyrimidine nucleotides and purine nucleotides in the sense region comprise 2'- deoxy purine nucleotides.
- a siNA molecule of the invention comprises a sense region and antisense region, wherein pyrimidine nucleotides present in the sense region comprise 2'- deoxy-2'-fluoro pyrimidine nucleotides and wherein purine nucleotides present in the sense region comprise 2'-deoxy purine nucleotides.
- a siNA molecule of the invention comprises a sense region and antisense region, wherein the sense region includes a terminal cap moiety at the 5'-end, the 3'-end, or both of the 5' and 3' ends of the sense region.
- the terminal cap moiety is an inverted deoxy abasic moiety.
- a siNA molecule of the invention comprises a sense region and antisense region, wherein pyrimidine nucleotides of the antisense region comprise 2'- deoxy-2'-fluoro pyrimidine nucleotides and purine nucleotides of the antisense region comprise 2'-O-methyl purine nucleotides.
- a siNA molecule of the invention comprises a sense region and antisense region, wherein pyrimidine nucleotides present in the antisense region are 2'- deoxy-2'-fluoro pyrimidine nucleotides and wherein purine nucleotides present in the antisense region comprise 2'-deoxy- purine nucleotides.
- a siNA molecule of the invention comprises a sense region and antisense region, wherein the antisense region comprises a phosphorothioate internucleotide linkage at the 3' end of the antisense region.
- a siNA molecule of the invention comprises a sense region and antisense region, wherein the antisense region comprises a glyceryl modification at the 3' end of the antisense region.
- a siNA molecule of the invention is assembled from two separate oligonucleotide fragments, wherein each of the two fragments of the siNA molecule comprise 21 nucleotides.
- a siNA molecule of the invention is assembled from two separate oligonucleotide fragments, wherein about 19 nucleotides of each fragment of the siNA molecule are base-paired to the complementary nucleotides of the other fragment of the siNA molecule and wherein at least two 3' terminal nucleotides of each fragment of the siNA molecule are not base-paired to the nucleotides of the other fragment of the siNA molecule.
- a siNA molecule of the invention is assembled from two separate oligonucleotide fragments, wherein each of the two 3' terminal nucleotides of each fragment of the siNA molecule are 2'-deoxy-pyrimidines, such as 2'-deoxy- thymidine.
- a siNA molecule of the invention is assembled from two separate oligonucleotide fragments, wherein each of the two fragments of the siNA molecule comprise 21 nucleotides and wherein all 21 nucleotides of each fragment of the siNA molecule are base-paired to the complementary nucleotides of the other fragment of the siNA molecule.
- a siNA molecule of the invention comprises a sense region and antisense region, wherein about 19 nucleotides of the antisense region are base-paired to the nucleotide sequence or a portion thereof of the RNA encoded by a target gene.
- a siNA molecule of the invention comprises a sense region and antisense region, wherein the siNA is assembled from two separate oligonucleotide fragments, wherein each of the two fragments of the siNA molecule comprise 21 nucleotides, and wherein 21 nucleotides of the antisense region are base-paired to the nucleotide sequence or a portion thereof of the RNA encoded by a target gene.
- a siNA molecule of the invention is assembled from two separate oligonucleotide fragments, wherein the 5 '-end of a fragment comprising the antisense region of the siNA optionally includes a phosphate group.
- the invention features a double-stranded short interfering nucleic acid (siNA) molecule that inhibits the expression of a target RNA sequence, wherein the siNA molecule comprises no ribonucleotides and wherein each strand of the double-stranded siNA molecule comprises about 21 nucleotides.
- siNA short interfering nucleic acid
- a target RNA sequence contemplated by the invention is encoded by a viral genome, bacterial gene, mammalian gene, human gene, or plant gene.
- the invention features a double-stranded short interfering nucleic acid (siNA) molecule that inhibits the replication of a virus (e.g, as mammalian virus, plant virus, hepatitis C virus, human immunodeficiency virus, hepatitis B virus, herpes simplex virus, cytomegalovirus, human papilloma virus, respiratory syncytial virus, or influenza virus) wherein the siNA molecule comprises no ribonucleotides and each strand of the double-stranded siNA molecule comprises about 21 nucleotides.
- a virus e.g, as mammalian virus, plant virus, hepatitis C virus, human immunodeficiency virus, hepatitis B virus, herpes simplex virus, cytomegalovirus, human papilloma virus, respiratory syncytial virus, or influenza virus
- the invention features a double-stranded short interfering nucleic acid (siNA) molecule that inhibits the expression of a target gene, wherein the siNA molecule does not require the presence of a ribonucleotide within the siNA molecule for the inhibition of expression of a target gene and wherein each strand of the double-stranded siNA molecule comprises about 21 nucleotides .
- siNA short interfering nucleic acid
- the invention features a double-stranded short interfering nucleic acid (siNA) molecule that inhibits the expression of a target gene by mediating RNA interference (RNAi) process, wherein the siNA molecule comprises no ribonucleotides and wherein each strand of the double-stranded siNA molecule comprises about 21 nucleotides.
- siNA short interfering nucleic acid
- the invention features a double-stranded short interfering nucleic acid (siNA) molecule that inhibits the replication of a virus (e.g, as mammalian virus, plant virus, hepatitis C virus, human immunodeficiency virus, hepatitis B virus, herpes simplex virus, cytomegalovirus, human papilloma virus, respiratory syncytial virus, or influenza virus), wherein the siNA molecule does not require the presence of a ribonucleotide within the siNA molecule for the inhibition of replication of the virus and each strand of the double-stranded siNA molecule comprises about 21 nucleotides.
- a virus e.g, as mammalian virus, plant virus, hepatitis C virus, human immunodeficiency virus, hepatitis B virus, herpes simplex virus, cytomegalovirus, human papilloma virus, respiratory syncytial virus, or influenza virus
- a virus e
- the invention features a medicament comprising a siNA molecule of the invention.
- the invention features an active ingredient comprising a siNA molecule of the invention.
- the invention features the use of a double-stranded short interfering nucleic acid (siNA) molecule to down-regulate expression of a target gene, wherein the siNA molecule comprises no ribonucleotides and each strand of the double- stranded siNA comprises about 21 nucleotides.
- siNA short interfering nucleic acid
- the invention features the use of a double-stranded short interfering nucleic acid (siNA) molecule to inhibit the expression of a target RNA sequence, wherein the siNA molecule comprises no ribonucleotides and wherein each strand of the double-stranded siNA molecule comprises about 21 nucleotides.
- siNA short interfering nucleic acid
- the invention features the use of a double-stranded short interfering nucleic acid (siNA) molecule to inhibit the replication of a virus, wherein the siNA molecule comprises no ribonucleotides and each strand of the double-stranded siNA molecule comprises about 21 nucleotides.
- siNA short interfering nucleic acid
- the invention features the use of a double-stranded short interfering nucleic acid (siNA) molecule to inhibit the expression of a target gene, wherein the siNA molecule does not require the presence of a ribonucleotide within the siNA molecule for the inhibition of expression of a target gene and wherein each strand of the double-stranded siNA molecule comprises about 21 nucleotides.
- siNA short interfering nucleic acid
- the invention features the use of a double-stranded short interfering nucleic acid (siNA) molecule to inhibit the expression of a target gene by mediating RNA interference (RNAi) process, wherein the siNA molecule comprises no ribonucleotides and wherein each strand of the double-stranded siNA molecule comprises about 21 nucleotides.
- siNA short interfering nucleic acid
- the invention features the use of a double-stranded short interfering nucleic acid (siNA) molecule to inhibit the replication of a virus, wherein the siNA molecule does not require the presence of a ribonucleotide within the siNA molecule for the inhibition of replication of a virus and each strand of the double-stranded siNA molecule comprises about 21 nucleotides.
- siNA short interfering nucleic acid
- the invention features a chemically-modified short interfering nucleic acid (siNA) molecule capable of mediating RNA interference (RNAi) inside a cell or reconstituted in vitro system, wherein the chemical modification comprises one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) nucleotides comprising a backbone modified internucleotide linkage having Formula I: wherein each Rl and R2 is independently any nucleotide, non-nucleotide, or polynucleotide which can be naturally-occurring or chemically-modified, each X and Y is independently O, S, N, alkyl, or substituted alkyl, each Z and W is independently O, S, N, alkyl, substituted alkyl, O-alkyl, S-alkyl, alkaryl, or aralkyl, and wherein W, X, Y, and Z are optionally not all O.
- siNA short interfering nucleic acid
- the chemically-modified internucleotide linkages having Formula I can be present in one or both oligonucleotide strands of the siNA duplex, for example, in the sense strand, the antisense strand, or both strands.
- the siNA molecules of the invention can comprise one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) chemically- modified internucleotide linkages having Formula I at the 3 '-end, the 5'-end, or both of the 3' and 5'-ends of the sense strand, the antisense strand, or both strands.
- an exemplary siNA molecule of the invention can comprise about 1 to about 5 or more (e.g., about 1, 2, 3, 4, 5, or more) chemically-modified internucleotide linkages having Formula I at the 5'-end of the sense strand, the antisense strand, or both strands.
- an exemplary siNA molecule of the invention can comprise one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) pyrimidine nucleotides with chemically-modified internucleotide linkages having Formula I in the sense strand, the antisense strand, or both strands.
- an exemplary siNA molecule of the invention can comprise one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) purine nucleotides with chemically-modified internucleotide linkages having Formula I in the sense strand, the antisense strand, or both strands.
- a siNA molecule of the invention having internucleotide linkage(s) of Formula I also comprises a chemically-modified nucleotide or non- nucleotide having any of Formulae I-NII.
- the invention features a chemically-modified short interfering nucleic acid (si ⁇ A) molecule capable of mediating R A interference (R ⁇ Ai) inside a cell or reconstituted in vitro system, wherein the chemical modification comprises one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) nucleotides or non-nucleotides having Formula II: wherein each R3, R4, R5, R6, R7, R8, RIO, Rll and R12 is independently H, OH, alkyl, substituted alkyl, alkaryl or aralkyl, F, Cl, Br, CN, CF3, OCF3, OCN, O-alkyl, S-alkyl, N-alkyl, O-alkenyl, S-alkenyl, N-alkenyl, SO-alkyl, alkyl-OSH, alkyl-OH, O-alkyl-OH, O-alkyl-SH, S-alkyl-OH, S
- the chemically-modified nucleotide or non-nucleotide of Formula II can be present in one or both oligonucleotide strands of the siNA duplex, for example in the sense strand, the antisense strand, or both strands.
- the siNA molecules of the invention can comprise one or more chemically-modified nucleotide or non-nucleotide of Formula II at the 3'-end, the 5'-end, or both of the 3' and 5'-ends of the sense strand, the antisense strand, or both strands.
- an exemplary siNA molecule of the invention can comprise about 1 to about 5 or more (e.g., about 1, 2, 3, 4, 5, or more) chemically- modified nucleotides or non-nucleotides of Formula II at the 5'-end of the sense strand, the antisense strand, or both strands.
- an exemplary siNA molecule of the invention can comprise about 1 to about 5 or more (e.g., about 1, 2, 3, 4, 5, or more) chemically-modified nucleotides or non-nucleotides of Formula II at the 3'- end of the sense strand, the antisense strand, or both strands.
- the invention features a chemically-modified short interfering nucleic acid (siNA) molecule capable of mediating RNA interference (RNAi) inside a cell or reconstituted in vitro system, wherein the chemical modification comprises one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) nucleotides or non-nucleotides having Formula III: wherein each R3, R4, R5, R6, R7, R8, RIO, Rll and R12 is independently H, OH, alkyl, substituted alkyl, alkaryl or aralkyl, F, Cl, Br, CN, CF3, OCF3, OCN, O-alkyl, S-alkyl, N-alkyl, O-alkenyl, S-alkenyl, N-alkenyl, SO-alkyl, alkyl-OSH, alkyl-OH, O-alkyl-OH, O-alkyl-SH, S-alkyl-OH, S-alkyl-al
- the chemically-modified nucleotide or non-nucleotide of Formula III can be present in one or both oligonucleotide strands of the siNA duplex, for example, in the sense strand, the antisense strand, or both strands.
- the siNA molecules of the invention can comprise one or more chemically-modified nucleotide or non-nucleotide of Formula III at the 3'-end, the 5'-end, or both of the 3' and 5'-ends of the sense strand, the antisense strand, or both strands.
- an exemplary siNA molecule of the invention can comprise about 1 to about 5 or more (e.g., about 1, 2, 3, 4, 5, or more) chemically- modified nucleotide(s) or non-nucleotide(s) of Formula III at the 5 '-end of the sense strand, the antisense strand, or both strands.
- an exemplary siNA molecule of the invention can comprise about 1 to about 5 or more (e.g., about 1, 2, 3, 4, 5, or more) chemically-modified nucleotide or non-nucleotide of Formula III at the 3'-end of the sense strand, the antisense strand, or both strands.
- a siNA molecule of the invention comprises a nucleotide having Formula II or III, wherein the nucleotide having Formula II or III is in an inverted configuration.
- the nucleotide having Formula II or III is connected to the siNA construct in a 3'-3', 3'-2', 2'-3', or 5'-5' configuration, such as at the 3'-end, the 5'- end, or both of the 3' and 5'-ends of one or both siNA strands.
- the invention features a chemically-modified short interfering nucleic acid (siNA) molecule capable of mediating RNA interference (RNAi) inside a cell or reconstituted in vitro system, wherein the chemical modification comprises a 5'- terminal phosphate group having Formula IN:
- siNA short interfering nucleic acid
- each X and Y is independently O, S, ⁇ , alkyl, substituted alkyl, or alkylhalo; wherein each Z and W is independently O, S, ⁇ , alkyl, substituted alkyl, O-alkyl, S-alkyl, alkaryl, aralkyl, or alkylhalo; and wherein W, X, Y and Z are not all O.
- the invention features a si ⁇ A molecule having a 5 '-terminal phosphate group having Formula IN on the target-complementary strand, for example, a strand complementary to a target R ⁇ A, wherein the si ⁇ A molecule comprises an all R ⁇ A si ⁇ A molecule.
- the invention features a si ⁇ A molecule having a 5 '-terminal phosphate group having Formula IN on the target-complementary strand wherein the si ⁇ A molecule also comprises about 1 to about 3 (e.g., about 1, 2, or 3) nucleotide 3'-terminal nucleotide overhangs having about 1 to about 4 (e.g., about 1, 2, 3, or 4) deoxyribonucleotides on the 3'-end of one or both strands.
- a 5'-terminal phosphate group having Formula IN is present on the target-complementary strand of a siNA molecule of the invention, for example a siNA molecule having chemical modifications having any of Formulae I-VII.
- the invention features a chemically-modified short interfering nucleic acid (siNA) molecule capable of mediating RNA interference (RNAi) inside a cell or reconstituted in vitro system, wherein the chemical modification comprises one or more phosphorothioate internucleotide linkages.
- siNA short interfering nucleic acid
- the invention features a chemically-modified short interfering nucleic acid (siNA) having about 1, 2, 3, 4, 5, 6, 7, 8 or more phosphorothioate internucleotide linkages in one siNA strand.
- the invention features a chemically-modified short interfering nucleic acid (siNA) individually having about 1, 2, 3, 4, 5, 6, 7, 8 or more phosphorothioate internucleotide linkages in both siNA strands.
- the phosphorothioate internucleotide linkages can be present in one or both oligonucleotide strands of the siNA duplex, for example in the sense strand, the antisense strand, or both strands.
- the siNA molecules of the invention can comprise one or more phosphorothioate internucleotide linkages at the 3'-end, the 5'-end, or both of the 3'- and 5'-ends of the sense strand, the antisense strand, or both strands.
- an exemplary siNA molecule of the invention can comprise about 1 to about 5 or more (e.g., about 1, 2, 3, 4, 5, or more) consecutive phosphorothioate internucleotide linkages at the 5 '-end of the sense strand, the antisense strand, or both strands.
- an exemplary siNA molecule of the invention can comprise one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) pyrimidine phosphorothioate internucleotide linkages in the sense strand, the antisense strand, or both strands.
- an exemplary siNA molecule of the invention can comprise one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) purine phosphorothioate internucleotide linkages in the sense strand, the antisense strand, or both strands.
- the invention features a siNA molecule, wherein the sense strand comprises one or more, for example, about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more phosphorothioate internucleotide linkages, and/or one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) 2'-deoxy, 2'-O-methyl, 2'-deoxy-2'-fluoro, and/or about one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) universal base modified nucleotides, and optionally a terminal cap molecule at the 3'-end, the 5'-end, or both of the 3'- and 5'-ends of the sense strand; and wherein the antisense strand comprises about 1 to about 10 or more, specifically about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more phosphorothioate internucleotide linkages, and/or one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9,
- the invention features a siNA molecule, wherein the sense strand comprises about 1 to about 5, specifically about 1, 2, 3, 4, or 5 phosphorothioate internucleotide linkages, and/or one or more (e.g., about 1, 2, 3, 4, 5, or more) 2'-deoxy, 2'-O-methyl, 2'-deoxy-2'-fluoro, and/or one or more (e.g., about 1, 2, 3, 4, 5, or more) universal base modified nucleotides, and optionally a terminal cap molecule at the 3-end, the 5'-end, or both of the 3'- and 5'-ends of the sense strand; and wherein the antisense strand comprises about 1 to about 5 or more, specifically about 1, 2, 3, 4, 5, or more phosphorothioate internucleotide linkages, and/or one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) 2'-deoxy, 2'-O-methyl, 2'-deoxy-2'-
- one or more, for example about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more, pyrimidine nucleotides of the sense and/or antisense siNA strand are chemically-modified with 2'-deoxy, 2'-O-methyl and/or 2'-deoxy-2'-fluoro nucleotides, with or without about 1 to about 5 or more, for example about 1, 2, 3, 4, 5, or more phosphorothioate internucleotide linkages and/or a terminal cap molecule at the 3'- end, the 5 '-end, or both of the 3'- and 5 '-ends, being present in the same or different strand.
- the invention features a siNA molecule, wherein the antisense strand comprises one or more, for example, about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more phosphorothioate internucleotide linkages, and/or about one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) 2'-deoxy, 2'-O-methyl, 2'-deoxy-2'- fluoro, and/or one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) universal base modified nucleotides, and optionally a terminal cap molecule at the 3 '-end, the 5 '-end, or both of the 3'- and 5 '-ends of the sense strand; and wherein the antisense strand comprises about 1 to about 10 or more, specifically about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more phosphorothioate internucleotide linkages, and/or one or more (e.g., about 1, 2, 3, 4,
- one or more, for example about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more pyrimidine nucleotides of the sense and/or antisense siNA strand are chemically- modified with 2'-deoxy, 2'-O-methyl and/or 2'-deoxy-2'-fluoro nucleotides, with or without one or more, for example, about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more phosphorothioate internucleotide linkages and/or a terminal cap molecule at the 3'-end, the 5'-end, or both of the 3' and 5'-ends, being present in the same or different strand.
- the invention features a siNA molecule, wherein the antisense strand comprises about 1 to about 5 or more, specifically about 1, 2, 3, 4, 5 or more phosphorothioate internucleotide linkages, and/or one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) 2'-deoxy, 2'-O-methyl, 2'-deoxy-2'-fluoro, and/or one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) universal base modified nucleotides, and optionally a terminal cap molecule at the 3'-end, the 5'-end, or both of the 3'- and 5'-ends of the sense strand; and wherein the antisense strand comprises about 1 to about 5 or more, specifically about 1, 2, 3, 4, 5 or more phosphorothioate internucleotide linkages, and/or one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more) 2'-
- one or more, for example about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10 or more pyrimidine nucleotides of the sense and/or antisense siNA strand are chemically-modified with 2'-deoxy, 2'-O- methyl and/or 2'-deoxy-2'-fluoro nucleotides, with or without about 1 to about 5, for example about 1, 2, 3, 4, 5 or more phosphorothioate internucleotide linkages and/or a terminal cap molecule at the 3'-end, the 5'-end, or both of the 3'- and 5'-ends, being present in the same or different strand.
- the mvention features a chemically-modified short interfering nucleic acid (siNA) molecule having about 1 to about 5, specifically about 1, 2, 3, 4, 5 or more phosphorothioate internucleotide linkages in each strand of the siNA molecule.
- siNA short interfering nucleic acid
- the invention features a siNA molecule comprising 2'-5' internucleotide linkages.
- the 2'-5' internucleotide linkage(s) can be at the 3'-end, the 5'- end, or both of the 3'- and 5 '-ends of one or both siNA sequence strands.
- the 2'-5' internucleotide linkage(s) can be present at various other positions within one or both siNA sequence strands, for example, about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more including every internucleotide linkage of a pyrimidine nucleotide in one or both strands of the siNA molecule can comprise a 2'-5' internucleotide linkage, or about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more including every internucleotide linkage of a purine nucleotide in one or both strands of the siNA molecule can comprise a 2'-5' internucleotide linkage.
- a chemically-modified siNA molecule of the invention comprises a duplex having two strands, one or both of which can be chemically-modified, wherein each strand is about 18 to about 27 (e.g., about 18, 19, 20, 21, 22, 23, 24, 25, 26, or 27) nucleotides in length, wherein the duplex has about 18 to about 23 (e.g., about 18, 19, 20, 21, 22, or 23) base pairs, and wherein the chemical modification comprises a structure having any of Formulae I-NII.
- an exemplary chemically-modified si ⁇ A molecule of the invention comprises a duplex having two strands, one or both of which can be chemically-modified with a chemical modification having any of Formulae I-NII or any combination thereof, wherein each strand consists of about 21 nucleotides, each having a 2-nucleotide 3 '-terminal nucleotide overhang, and wherein the duplex has about 19 base pairs.
- a si ⁇ A molecule of the invention comprises a single stranded hairpin structure, wherein the si ⁇ A is about 36 to about 70 (e.g., about 36, 40, 45, 50, 55, 60, 65, or 70) nucleotides in length having about 18 to about 23 (e.g., about 18, 19, 20, 21, 22, or 23) base pairs, and wherein the si ⁇ A can include a chemical modification comprising a structure having any of Formulae I-NII or any combination thereof.
- an exemplary chemically-modified si ⁇ A molecule of the invention comprises a linear oligonucleotide having about 42 to about 50 (e.g., about 42, 43, 44, 45, 46, 47, 48, 49, or 50) nucleotides that is chemically-modified with a chemical modification having any of Formulae I-NII or any combination thereof, wherein the linear oligonucleotide forms a hairpin structure having about 19 base pairs and a 2-nucleotide 3 '-terminal nucleotide overhang.
- a linear hairpin si ⁇ A molecule of the invention contains a stem loop motif, wherein the loop portion of the si ⁇ A molecule is biodegradable.
- a linear hairpin si ⁇ A molecule of the mvention is designed such that degradation of the loop portion of the si ⁇ A molecule in vivo can generate a double-stranded si ⁇ A molecule with 3 '-terminal overhangs, such as 3 '-terminal nucleotide overhangs comprising about 2 nucleotides.
- a si ⁇ A molecule of the invention comprises a circular nucleic acid molecule, wherein the si ⁇ A is about 38 to about 70 (e.g., about 38, 40, 45, 50, 55, 60, 65, or 70) nucleotides in length having about 18 to about 23 (e.g., about 18, 19, 20, 21, 22, or 23) base pairs, and wherein the si ⁇ A can include a chemical modification, which comprises a structure having any of Formulae I-NII or any combination thereof.
- an exemplary chemically-modified si ⁇ A molecule of the invention comprises a circular oligonucleotide having about 42 to about 50 (e.g., about 42, 43, 44, 45, 46, 47, 48, 49, or 50) nucleotides that is chemically-modified with a chemical modification having any of Formulae I-NII or any combination thereof, wherein the circular oligonucleotide forms a dumbbell shaped structure having about 19 base pairs and 2 loops.
- a circular si ⁇ A molecule of the invention contains two loop motifs, wherein one or both loop portions of the si ⁇ A molecule is biodegradable.
- a circular si ⁇ A molecule of the mvention is designed such that degradation of the loop portions of the si ⁇ A molecule in vivo can generate a double-stranded si ⁇ A molecule with 3'-terminal overhangs, such as 3'-terminal nucleotide overhangs comprising about 2 nucleotides.
- a si ⁇ A molecule of the invention comprises at least one (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) abasic moiety, for example a compound having Formula N: W wherein each R3, R4, R5, R6, R7, R8, RIO, Rll, R12, and R13 is independently H, OH, alkyl, substituted alkyl, alkaryl or aralkyl, F, Cl, Br, CN, CF3, OCF3, OCN, O-alkyl, S- alkyl, N-alkyl, O-alkenyl, S-alkenyl, N-alkenyl, SO-alkyl, alkyl-OSH, alkyl-OH, O-alkyl- OH, O-alkyl-SH, S-alkyl-OH, S-alkyl-SH, alkyl-S-alkyl, alkyl-O-alkyl, ONO2, NO2, N3, NH2, aminoalkyl, amino
- a siNA molecule of the invention comprises at least one (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) inverted abasic moiety, for example a compound having Formula VI: wherein each R3, R4, R5, R6, R7, R8, R10, Rll, R12, and R13 is independently H, OH, alkyl, substituted alkyl, alkaryl or aralkyl, F, Cl, Br, CN, CF3, OCF3, OCN, O-alkyl, S- alkyl, N-alkyl, O-alkenyl, S-alkenyl, N-alkenyl, SO-alkyl, alkyl-OSH, alkyl-OH, O-alkyl- OH, O-alkyl-SH, S-alkyl-OH, S-alkyl-SH, alkyl-S-alkyl, alkyl-O-alkyl, ONO2, NO2, N3, NH2, aminoalkyl, amino
- a siNA molecule of the invention comprises at least one (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) substituted polyalkyl moieties, for example a compound having Formula Nil: wherein each n is independently an integer from 1 to 12, each Rl, R2 and R3 is independently H, OH, alkyl, substituted alkyl, alkaryl or aralkyl, F, Cl, Br, C ⁇ , CF3, OCF3, OC ⁇ , O-alkyl, S-alkyl, ⁇ -alkyl, O-alkenyl, S-alkenyl, ⁇ -alkenyl, SO-alkyl, alkyl- OSH, alkyl-OH, O-alkyl-OH, O-alkyl-SH, S-alkyl-OH, S-alkyl-SH, alkyl-S-alkyl, alkyl- O-alkyl, O ⁇ O2, NO2, N3, NH2, aminoalkyl, aminoa
- This modification is referred to herein as "glyceryl" (for example modification 6 in Figure 22).
- a moiety having any of Formula N, NI or Nil of the invention is at the 3'-end, the 5'-end, or both of the 3' and 5'-ends of a si ⁇ A molecule of the invention.
- a moiety having Formula N, NI or Nil can be present at the 3'-end, the 5'-end, or both of the 3' and 5'-ends of the antisense strand, the sense strand, or both antisense and sense strands of the si ⁇ A molecule
- a moiety having Formula VII can be present at the 3'-end or the 5'-end of a hairpin si ⁇ A molecule as described herein.
- a siNA molecule of the invention comprises an abasic residue having Formula V or VI, wherein the abasic residue having Formula VI or VI is connected to the siNA construct in a 3'-3', 3'-2', 2'-3', or 5'-5' configuration, such as at the 3 '-end, the 5 '-end, or both of the 3' and 5 '-ends of one or both siNA strands.
- a siNA molecule of the invention comprises one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) locked nucleic acid (LNA) nucleotides, for example at the 5'-end, the 3'-end, both of the 5' and 3'-ends, or any combination thereof, of the siNA molecule.
- LNA locked nucleic acid
- a siNA molecule of the invention comprises one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) acyclic nucleotides, for example at the 5'- end, the 3 '-end, both of the 5' and 3 '-ends, or any combination thereof, of the siNA molecule.
- the invention features a chemically-modified short interfering nucleic acid (siNA) molecule of the invention, wherein the chemically-modified siNA comprises a sense region, where any (e.g., one or more or all) pyrimidine nucleotides present in the sense region are 2'-deoxy-2 '-fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), and where any (e.g., one or more or all) purine nucleotides present in the sense region are 2'- deoxy purine nucleotides (e.g., wherein all purine nucleotides are 2'-deoxy purine nucleotides or alternately a plurality of purine nucle
- the invention features a chemically-modified short interfering nucleic acid (siNA) molecule of the invention, wherein the chemically-modified siNA comprises a sense region, where any (e.g., one or more or all) pyrimidine nucleotides present in the sense region are 2'-deoxy-2'-fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), and where any (e.g., one or more or all) purine nucleotides present in the sense region are 2'- deoxy purine nucleotides (e.g., wherein all purine nucleotides are 2'-deoxy purine nucleotides or alternately a plurality of purine
- the invention features a chemically-modified short interfering nucleic acid (siNA) molecule of the invention, wherein the chemically-modified siNA comprises an antisense region, where any (e.g., one or more or all) pyrimidine nucleotides present in the antisense region are 2'-deoxy-2'-fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), and wherein any (e.g., one or more or all) purine nucleotides present in the antisense region are 2'-O-methyl purine nucleotides (e.g., wherein all purine nucleotides are 2'-O-methyl purine nucleotides or alternate
- the invention features a chemically-modified short interfering nucleic acid (siNA) molecule of the invention, wherein the chemically-modified siNA comprises an antisense region, where any (e.g., one or more or all) pyrimidine nucleotides present in the antisense region are 2'-deoxy-2'- fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'-fiuoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), and wherein any (e.g., one or more or all) purine nucleotides present in the antisense region are 2'-O-methyl purine nucleotides (e.g., wherein all purine nucleotides are 2'-O-methyl purine nucleotides
- the invention features a chemically-modified short interfering nucleic acid (siNA) molecule of the invention, wherein the chemically-modified siNA comprises an antisense region, where any (e.g., one or more or all) pyrimidine nucleotides present in the antisense region are 2'-deoxy-2'-fiuoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), and where any (e.g., one or more or all) purine nucleotides present in the antisense region are 2'-deoxy purine nucleotides (e.g., wherein all purine nucleotides are 2'-deoxy purine nucleotides or alternate
- the invention features a chemically-modified short interfering nucleic acid (siNA) molecule of the invention capable of mediating RNA interference (RNAi) inside a cell or reconstituted in vitro system comprising a sense region and an antisense region.
- siNA short interfering nucleic acid
- the sense region comprises n one or more 2'-deoxy- 2'-fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'- fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'- deoxy-2'-fluoro pyrimidine nucleotides), and one or more 2'-deoxy purine nucleotides (e.g., wherein all purine nucleotides are 2'-deoxy purine nucleotides or alternately a plurality of purine nucleotides are 2'-deoxy purine nucleotides).
- 2'-deoxy- 2'-fluoro pyrimidine nucleotides e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'- fluoro pyrimidine nucleotides or alternately a plurality of pyr
- the sense region can comprise inverted deoxy abasic modifications that are optionally present at the 3 '-end, the 5'-end, or both of the 3' and 5'-ends of the sense region.
- the sense region can optionally further comprise a 3'-terminal overhang having about 1 to about 4 (e.g., about 1, 2, 3, or 4) 2'-deoxyribonucleotides.
- the antisense region comprisesone or more 2'-deoxy-2'- fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'- fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'- deoxy-2'-fluoro pyrimidine nucleotides), and one or more 2'-O-methyl purine nucleotides (e.g., wherein all purine nucleotides are 2'-O-methyl purine nucleotides or alternately a plurality of purine nucleotides are 2'-O-methyl purine nucleotides).
- 2'-deoxy-2'- fluoro pyrimidine nucleotides e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'- fluoro pyrimidine nucleotides or alternately a plurality of
- the antisense region can comprise a terminal cap modification, such as any modification described herein or shown in Figure 22, that is optionally present at the 3 '-end, the 5 '-end, or both of the 3' and 5 '-ends of the antisense sequence.
- the antisense region optionally further comprises a 3'-terminal nucleotide overhang having about 1 to about 4 (e.g., about 1, 2, 3, or 4) 2'- deoxynucleotides, wherein the overhang nucleotides can further comprise one or more (e.g., 1, 2, 3, or 4 ) phosphorothioate internucleotide linkages.
- Non-limiting examples of these chemically-modified siNAs are shown in Figures 18 and 19 and Table IV herein.
- the sense region comprisesone or more 2'-deoxy-2'-fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), and one or more purine ribonucleotides (e.g., wherein all purine nucleotides are purine ribonucleotides or alternately a plurality of purine nucleotides are purine ribonucleotides).
- 2'-deoxy-2'-fluoro pyrimidine nucleotides e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'
- the sense region can also compriseinverted deoxy abasic modifications that are optionally present at the 3'- end, the 5 '-end, or both of the 3' and 5 '-ends of the sense region.
- the sense region optionally further comprises a 3'-terminal overhang having about 1 to about 4 (e.g., about 1, 2, 3, or 4) 2'-deoxyribonucleotides.
- the antisense region comprises one or more 2'- deoxy-2'-fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'- deoxy-2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), and one or more 2'-O-methyl purine nucleotides (e.g., wherein all purine nucleotides are 2'-O-methyl purine nucleotides or alternately a plurality of purine nucleotides are 2'-O-methyl purine nucleotides).
- 2'- deoxy-2'-fluoro pyrimidine nucleotides e.g., wherein all pyrimidine nucleotides are 2'- deoxy-2'-fluoro pyrimidine nucleotides or alternately a plurality
- the antisense region can also comprise a terminal cap modification, such as any modification described herein or shown in Figure 22, that is optionally present at the 3 '-end, the 5 '-end, or both of the 3' and 5'-ends of the antisense sequence.
- the antisense region optionally further comprises a 3'-terminal nucleotide overhang having about 1 to about 4 (e.g., about 1, 2, 3, or 4) 2'-deoxynucleotides, wherein the overhang nucleotides can further comprise one or more (e.g., 1, 2, 3, or 4) phosphorothioate internucleotide linkages.
- Non-limiting examples of these chemically-modified siNAs are shown in Figures 18 and 19 and Table IV herein.
- the sense region comprises one or more 2'-deoxy-2'-fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), and one or more purine nucleotides selected from the group consisting of 2'-deoxy nucleotides, locked nucleic acid (LNA) nucleotides, 2'-methoxyethyl nucleotides, 4'-thionucleotides, and 2'-O- methyl nucleotides (e.g., wherein all purine nucleotides are selected from the group consisting of 2 '-deoxy nucleotides
- the sense region can comprise inverted deoxy abasic modifications that are optionally present at the 3'-end, the 5'-end, or both of the 3' and 5'-ends of the sense region.
- the sense region can optionally further comprise a 3'-terminal overhang having about 1 to about 4 (e.g., about 1, 2, 3, or 4) 2'- deoxyribonucleotides.
- the antisense region comprises one or more 2'-deoxy-2'-fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy- 2'-fluoro pyrimidine nucleotides), and one or more purine nucleotides selected from the group consisting of 2'-deoxy nucleotides, locked nucleic acid (LNA) nucleotides, 2'- methoxyethyl nucleotides, 4'-thionucleotides, and 2'-O-methyl nucleotides (e.g., wherein all purine nucleotides are selected from the group consisting of 2 '-deoxy nucleotides, locked nucleic acid (LNA) nucleotides, 2'-methoxyethyl nucle
- the antisense can also comprise a terminal cap modification, such as any modification described herein or shown in Figure 22, that is optionally present at the 3'- end, the 5'-end, or both of the 3' and 5'-ends of the antisense sequence.
- the antisense region optionally further comprises a 3 '-terminal nucleotide overhang having about 1 to about 4 (e.g., about 1, 2, 3, or 4) 2'-deoxynucleotides, wherein the overhang nucleotides can further comprise one or more (e.g., 1, 2, 3, or 4) phosphorothioate internucleotide linkages.
- any modified nucleotides present in the siNA molecules of the invention preferably in the antisense strand of the siNA molecules of the invention, but also optionally in the sense and/or both antisense and sense strands, comprise modified nucleotides having properties or characteristics similar to naturally occurring ribonucleotides.
- the invention features siNA molecules including modified nucleotides having a Northern conformation (e.g., Northern pseudorotation cycle, see for example Saenger, Principles of Nucleic Acid Structure, Springer- Verlag ed., 1984).
- chemically modified nucleotides present in the siNA molecules of the mvention preferably in the antisense strand of the siNA molecules of the invention, but also optionally in the sense and/or both antisense and sense strands, are resistant to nuclease degradation while at the same time maintaining the capacity to mediate RNAi.
- Non- limiting examples of nucleotides having a northern configuration include locked nucleic acid (LNA) nucleotides (e.g., 2'-O,4'-C-methylene-(D-ribofuranosyl) nucleotides); 2'- methoxyethoxy (MOE) nucleotides; 2'-methyl-thio-ethyl, 2 '-deoxy-2' -fluoro nucleotides, 2 '-deoxy-2' -chloro nucleotides, 2'-azido nucleotides, and 2'-O-methyl nucleotides.
- LNA locked nucleic acid
- MOE methoxyethoxy
- the invention features a chemically-modified short interfering nucleic acid molecule (siNA) capable of mediating RNA interference (RNAi) inside a cell or reconstituted in vitro system, wherein the chemical modification comprises a conjugate attached to the chemically-modified siNA molecule.
- the conjugate can be attached to the chemically-modified siNA molecule via a covalent attachment.
- the conjugate is attached to the chemically-modified siNA molecule via a biodegradable linker.
- the conjugate molecule is attached at the 3 '-end ofeither the sense strand, the antisense strand, or both strands of the chemically-modified siNA molecule.
- the conjugate molecule is attached at the 5 '-end of either the sense strand, the antisense strand, or both strands of the chemically-modified siNA molecule. In yet another embodiment, the conjugate molecule is attached both the 3'-end and 5'-end of either the sense strand, the antisense strand, or both strands of the chemically-modified siNA molecule, or any combination thereof. In one embodiment, the conjugate molecule of the invention comprises a molecule that facilitates delivery of a chemically-modified siNA molecule into a biological system, such as a cell.
- the conjugate molecule attached to the chemically-modified siNA molecule is a poly ethylene glycol, human serum albumin, or a ligand for a cellular receptor that can mediate cellular uptake.
- Examples of specific conjugate molecules contemplated by the instant invention that can be attached to chemically-modified siNA molecules are described in Vargeese et al, U.S. Serial No. 10/201,394, incorporated by reference herein.
- the type of conjugates used and the extent of conjugation of siNA molecules of the invention can be evaluated for improved pharmacokinetic profiles, bioavailabihty, and/or stability of siNA constructs while at the same time maintaining the ability of the siNA to mediate RNAi activity.
- the invention features a short interfering nucleic acid (siNA) molecule of the invention, wherein the siNA further comprises a nucleotide, non- nucleotide, or mixed nucleotide/non-nucleotide linker that joins the sense region of the siNA to the antisense region of the siNA.
- siNA short interfering nucleic acid
- a nucleotide linker of the invention can be a linker of > 2 nucleotides in length, for example 3, 4, 5, 6, 7, 8, 9, or 10 nucleotides in length.
- the nucleotide linker can be a nucleic acid aptamer.
- aptamer or “nucleic acid aptamer” as used herein is meant a nucleic acid molecule that binds specifically to a target molecule wherein the nucleic acid molecule has sequence that comprises a sequence recognized by the target molecule in its natural setting.
- an aptamer can be a nucleic acid molecule that binds to a target molecule where the target molecule does not naturally bind to a nucleic acid.
- the target molecule can be any molecule of interest.
- the aptamer can be used to bind to a ligand-binding domain of a protein, thereby preventing interaction of the naturally occurring ligand with the protein. This is a non-limiting example and those in the art will recognize that other embodiments can be readily generated using techniques generally known in the art. (See, for example, Gold et al, 1995, Annu. Rev. Biochem., 64, 763; Brody and Gold, 2000, J. Biotechnol, 74, 5; Sun, 2000, Curr.
- a non-nucleotide linker of the invention can comprise abasic nucleotide, polyether, polyamine, polyamide, peptide, carbohydrate, lipid, polyhydrocarbon, or other polymeric compounds (e.g. polyethylene glycols such as those having between 2 and 100 ethylene glycol units).
- polyethylene glycols such as those having between 2 and 100 ethylene glycol units.
- Specific examples include those described by Seela and Kaiser, Nucleic Acids Res. 1990, 18:6353 and Nucleic Acids Res. 1987, 75:3113; Cload and Schepartz, J. Am. Chem. Soc. 1991, 113:632A; Richardson and Schepartz, J. Am. Chem. Soc. 1991, 773:5109; Ma et al, Nucleic Acids Res.
- non-nucleotide further means any group or compound that can be incorporated into a nucleic acid chain in the place of one or more nucleotide units, including either sugar and/or phosphate substitutions, and allows the remaining bases to exhibit their enzymatic activity.
- the group or compound can be abasic in that it does not contain a commonly recognized nucleotide base, such as adenosine, guanine, cytosine, uracil or thymine, for example at the Cl position of the sugar.
- the invention features a short interfering nucleic acid (siNA) molecule capable of mediating RNA interference (RNAi) inside a cell or reconstituted in vitro system, wherein one or both strands of the siNA molecule that are assembled from two separate oligonucleotides do not comprise any ribonucleotides.
- a siNA molecule can be assembled from a single oligonculeotide where the sense and antisense regions of the siNA comprise separate oligonucleotides not having any ribonucleotides (e.g., nucleotides having a 2'-OH group) present in the oligonucleotides.
- a siNA molecule in another embodiment, can be assembled from a single oligonculeotide where the sense and antisense regions of the siNA are linked or circularized by a nucleotide or non-nucleotide linker as desrcibed herein, wherein the oligonucleotide does not have any ribonucleotides (e.g., nucleotides having a 2' -OH group) present in the oligonucleotide.
- ribonucleotides e.g., nucleotides having a 2' -OH group
- all positions within the siNA can include chemically modified nucleotides and/or non-nucleotides, such as nucleotides and or non-nucleotides having Formula I, II, III, IV, V, NI, or Nil or any combination thereof to the extent that the ability of the si ⁇ A molecule to support R ⁇ Ai activity in a cell is maintained.
- a si ⁇ A molecule of the invention is a single stranded si ⁇ A molecule that mediates R ⁇ Ai activity in a cell or reconstituted in vitro system, wherein the si ⁇ A molecule comprises a single stranded polynucleotide having complementarity to a target nucleic acid sequence.
- the single stranded si ⁇ A molecule of the invention can comprise about 19 to about 29 nucleotides.
- the single stranded si ⁇ A molecule of the invention comprises a 5 '-terminal phosphate group.
- the single stranded si ⁇ A molecule of the invention comprises a 5 '-terminal phosphate group and a 3 '-terminal phosphate group (e.g., a 2', 3 '-cyclic phosphate).
- the single stranded siNA molecule of the invention comprises one or more chemically modified nucleotides or non-nucleotides described herein. For example, all the positions within the siNA molecule can include chemically-modified nucleotides such as nucleotides having any of Formulae I-NII, or any combination thereof to the extent that the ability of the si ⁇ A molecule to support R ⁇ Ai activity in a cell is maintained.
- the single stranded si ⁇ A molecule having complementarity to a target nucleic acid sequence comprises one or more 2'-deoxy-2'-fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), and one or more 2'-O-methyl purine nucleotides (e.g., wherein all purine nucleotides are 2'-O-methyl purine nucleotides or alternately a plurality of purine nucleotides are 2'-O-methyl purine nucleotides.
- the single stranded si ⁇ A molecule comprises one or more 2'-deoxy-2'-fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nu
- the single stranded si ⁇ A molecule comprises one or more 2'-deoxy-2'-fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'-deoxy-2'-fluoro pyrimidine nucleotides), wherein any purine nucleotides present in the antisense region are locked nucleic acid (L ⁇ A) nucleotides (e.g., wherein all purine nucleotides are L ⁇ A nucleotides or alternately a plurality of purine nucleotides are L ⁇ A nucleotides).
- L ⁇ A locked nucleic acid
- the single stranded si ⁇ A molecule comprises one or more 2'-deoxy-2'- fluoro pyrimidine nucleotides (e.g., wherein all pyrimidine nucleotides are 2'-deoxy-2'- fluoro pyrimidine nucleotides or alternately a plurality of pyrimidine nucleotides are 2'- deoxy-2'-fluoro pyrimidine nucleotides), and one or more 2'-methoxyethyl purine nucleotides (e.g., wherein all purine nucleotides are 2'-methoxyethyl purine nucleotides or alternately a plurality of purine nucleotides are 2'-methoxyethyl purine nucleotides), the single stranded si ⁇ A can comprise a terminal cap modification, such as any modification described herein or shown in Figure 22, that is optionally present at the 3'- end, the 5'-end, or both
- the single stranded siNA optionally further comprises about 1 to about 4 (e.g., about 1, 2, 3, or 4) terminal 2'-deoxynucleotides at the 3 '-end of the siNA molecule, wherein the te ⁇ ninal nucleotides can further comprise one or more (e.g., 1, 2, 3, or 4) phosphorothioate internucleotide linkages.
- the single stranded siNA optionally further comprises a terminal phosphate group, such as a 5'-terminal phosphate group.
- any modified nucleotides present in the single stranded siNA molecules of the invention comprise modified nucleotides having properties or characteristics similar to naturally occurring ribonucleotides.
- the invention features siNA molecules including modified nucleotides having a Northern conformation (e.g., Northern pseudorotation cycle, see for example Saenger, Principles of Nucleic Acid Structure, Springer-Nerlag ed., 1984).
- chemically modified nucleotides present in the single stranded siNA molecules of the invention are preferably resistant to nuclease degradation while at the same time maintaining the capacity to mediate RNAi.
- the invention features a method for modulating the expression of a gene within a cell comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically-modified, wherein one of the siNA strands comprises a sequence complementary to RNA of the gene; and (b) introducing the siNA molecule into a cell under conditions suitable to modulate the expression of the gene in the cell.
- the invention features a method for modulating the expression of a gene within a cell comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically-modified, wherein one of the siNA strands comprises a sequence complementary to RNA of the gene and wherein the sense strand sequence of the siNA comprises a sequence substantially similar to the sequence of the target RNA; and (b) introducing the siNA molecule into a cell under conditions suitable to modulate the expression of the gene in the cell.
- the invention features a method for modulating the expression of more than one gene within a cell comprising: (a) synthesizing siNA molecules of the invention, which can be chemically-modified, wherein one of the siNA strands comprises a sequence complementary to RNA of the genes; and (b) introducing the siNA molecules into a cell under conditions suitable to modulate the expression of the genes in the cell.
- the invention features a method for modulating the expression of more than one gene within a cell comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically-modified, wherein one of the siNA strands comprises a sequence complementary to RNA of the gene and wherein the sense strand sequence of the siNA comprises a sequence substantially similar to the sequence of the target RNA; and (b) introducing the siNA molecules into a cell under conditions suitable to modulate the expression of the genes in the cell.
- siNA molecules of the invention are used as reagents in ex vivo applications.
- siNA reagents are intoduced into tissue or cells that are transplanted into a subject for therapeutic effect.
- the cells and/or tissue can be derived from an organism or subject that later receives the explant, or can be derived from another organism or subject prior to transplantation.
- the siNA molecules can be used to modulate the expression of one or more genes in the cells or tissue, such that the cells or tissue obtain a desired phenotype or are able to perform a function when transplanted in vivo.
- certain target cells from a patient are extracted.
- These extracted cells are contacted with siNAs targeteing a specific nucleotide sequence within the cells under conditions suitable for uptake of the siNAs by these cells (e.g. using delivery reagents such as cationic lipids, liposomes and the like or using techniques such as electroporation to facilitate the delivery of siNAs into cells).
- the cells are then reintroduced back into the same patient or other patients.
- Non-limiting examples of ex vivo applications include use in organ/tissue transplant, tissue grafting, or treatment of pulmonary disease (e.g., restenosis) or prevent neointimal hyperplasia and atherosclerosis in vein grafts.
- Such ex vivo applications may also used to treat conditions associated with coronary and peripheral bypass graft failure, for example, such methods can be used in conjunction with peripheral vascular bypass graft surgery and coronary artery bypass graft surgery.
- Additional applications include transplants to treat CNS lesions or injury, including use in treatment of neurodegenerative conditions such as Alzheimer's disease, Parkinson's Disease, Epilepsy, Dementia, Huntington's disease, or amyotrophic lateral sclerosis (ALS).
- ALS amyotrophic lateral sclerosis
- the invention features a method of modulating the expression of a gene in a tissue explant comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically-modified, wherein one of the siNA strands comprises a sequence complementary to RNA of the gene; and (b) introducing the siNA molecule into a cell of the tissue explant derived from a particular organism under conditions suitable to modulate the expression of the gene in the tissue explant.
- the method further comprises introducing the tissue explant back into the organism the tissue was derived from or into another organism under conditions suitable to modulate the expression of the gene in that organism.
- the invention features a method of modulating the expression of a gene in a tissue explant comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically-modified, wherein one of the siNA strands comprises a sequence complementary to RNA of the gene and wherein the sense strand sequence of the siNA comprises a sequence substantially similar to the sequence of the target RNA; and (b) introducing the siNA molecule into a cell of the tissue explant derived from a particular organism under conditions suitable to modulate the expression of the gene in the tissue explant.
- the method further comprises introducing the tissue explant back into the organism the tissue was derived from or into another organism under conditions suitable to modulate the expression of the gene in that organism.
- the invention features a method of modulating the expression of more than one gene in a tissue explant comprising: (a) synthesizing siNA molecules of the invention, which can be chemically-modified, wherein one of the siNA strands comprises a sequence complementary to RNA of the genes; and (b) introducing the siNA molecules into a cell of the tissue explant derived from a particular organism under conditions suitable to modulate the expression of the genes in the tissue explant.
- the method further comprises introducing the tissue explant back into the organism the tissue was derived from or into another organism under conditions suitable to modulate the expression of the genes in that organism.
- the invention features a method of modulating the expression of a gene in an organism comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically-modified, wherein one of the siNA strands comprises a sequence complementary to RNA of the gene; and (b) introducing the siNA molecule into the organism under conditions suitable to modulate the expression of the gene in the organism.
- the invention features a method of modulating the expression of more than one gene in an organism comprising: (a) synthesizing siNA molecules of the invention, which can be chemically-modified, wherein one of the siNA strands comprises a sequence complementary to RNA of the genes; and (b) introducing the siNA molecules into the organism under conditions suitable to modulate the expression of the genes in the organism.
- the invention features a method for modulating the expression of a gene within a cell comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically-modified, wherein the siNA comprises a single stranded sequence having complementarity to RNA of the gene; and (b) introducing the siNA molecule into a cell under conditions suitable to modulate the expression of the gene in the cell.
- the invention features a method for modulating the expression of more than one gene within a cell comprising: (a) synthesizing siNA molecules of the invention, which can be chemically-modified, wherein the siNA comprises a single stranded sequence having complementarity to RNA of the gene; and (b) contacting the siNA molecule with a cell in vitro or in vivo under conditions suitable to modulate the expression of the genes in the cell.
- the invention features a method of modulating the expression of a gene in a tissue explant comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically-modified, wherein the siNA comprises a single stranded sequence having complementarity to RNA of the gene; and (b) contacting the siNA molecule with a cell of the tissue explant derived from a particular organism under conditions suitable to modulate the expression of the gene in the tissue explant.
- the method further comprises introducing the tissue explant back into the organism the tissue was derived from or into another organism under conditions suitable to modulate the expression of the gene in that organism.
- the invention features a method of modulating the expression of more than one gene in a tissue explant comprising: (a) synthesizing siNA molecules of the invention, which can be chemically-modified, wherein the siNA comprises a single stranded sequence having complementarity to RNA of the gene; and (b) introducing the siNA molecules into a cell of the tissue explant derived from a particular organism under conditions suitable to modulate the expression of the genes in the tissue explant.
- the method further comprises introducing the tissue explant back into the organism the tissue was derived from or into another organism under conditions suitable to modulate the expression of the genes in that organism.
- the invention features a method of modulating the expression of a gene in an organism comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically-modified, wherein the siNA comprises a single stranded sequence having complementarity to RNA of the gene; and (b) introducing the siNA molecule into the organism under conditions suitable to modulate the expression of the gene in the organism.
- the invention features a method of modulating the expression of more than one gene in an organism comprising: (a) synthesizing siNA molecules of the invention, which can be chemically-modified, wherein the siNA comprises a single stranded sequence having complementarity to RNA of the gene; and (b) introducing the siNA molecules into the organism under conditions suitable to modulate the expression of the genes in the organism.
- the invention features a method of modulating the expression of a gene in an organism comprising contacting the organism with a siNA molecule of the invention under conditions suitable to modulate the expression of the gene in the organism.
- the invention features a method of modulating the expression of more than one gene in an organism comprising contacting the organism with one or more siNA molecules of the invention under conditions suitable to modulate the expression of the genes in the organism.
- the siNA molecules of the invention can be designed to inhibit target gene expression through RNAi targeting of a variety of RNA molecules.
- the siNA molecules of the invention are used to target various RNAs co ⁇ esponding to a target gene.
- Non-limiting examples of such RNAs include messenger RNA (mRNA), alternate RNA splice variants of target gene(s), post-transcriptionally modified RNA of target gene(s), pre-mRNA of target gene(s), and/or RNA templates.
- the instant invention can be used to inhibit gene expression through the appropriate exons to specifically inhibit or to distinguish among the functions of gene family members.
- a protein that contains an alternatively spliced transmembrane domain can be expressed in both membrane bound and secreted forms.
- Use of the invention to target the exon containing the transmembrane domain can be used to determine the functional consequences of pharmaceutical targeting of membrane bound as opposed to the secreted form of the protein.
- Non-limiting examples of applications of the invention relating to targeting these RNA molecules include therapeutic pharmaceutical applications, pharmaceutical discovery applications, molecular diagnostic and gene function applications, and gene mapping, for example using single nucleotide polymorphism mapping with siNA molecules of the invention.
- Such applications can be implemented using known gene sequences or from partial sequences available from an expressed sequence tag (EST).
- EST expressed sequence tag
- the siNA molecules of the invention are used to target conserved sequences corresponding to a gene family or gene families. As such, siNA molecules targeting multiple gene targets can provide increased therapeutic effect.
- siNA can be used to characterize pathways of gene function in a variety of applications.
- the present invention can be used to inhibit the activity of target gene(s) in a pathway to determine the function of uncharacterized gene(s) in gene function analysis, mRNA function analysis, or translational analysis.
- the invention can be used to determine potential target gene pathways involved in various diseases and conditions toward pharmaceutical development.
- the invention can be used to understand pathways of gene expression involved in, for example, in development, such as prenatal development and postnatal development, and/or the progression and/or maintenance of cancer, infectious disease, autoimmunity, inflammation, endocrine disorders, renal disease, pulmonary disease, cardiovascular disease, birth defects, ageing, any other disease or condition related to gene expression.
- the invention features a method comprising: (a) generating a library of siNA constructs having a predetermined complexity; and (b) assaying the siNA constructs of (a) above, under conditions suitable to determine RNAi target sites within the target RNA sequence.
- the siNA molecules of (a) have strands of a fixed length, for example, about 23 nucleotides in length.
- the siNA molecules of (a) are of differing length, for example having strands of about 19 to about 25 (e.g., about 19, 20, 21, 22, 23, 24, or 25) nucleotides in length.
- the assay can comprise a reconstituted in vitro siNA assay as described herein.
- the assay can comprise a cell culture system in which target RNA is expressed, hi another embodiment, fragments of target RNA are analyzed for detectable levels of cleavage, for example by gel electrophoresis, northern blot analysis, or RNAse protection assays, to determine the most suitable target site(s) within the target RNA sequence.
- the target RNA sequence can be obtained as is known in the art, for example, by cloning and/or transcription for in vitro systems, and by cellular expression in in vivo systems.
- the invention features a method comprising: (a) generating a randomized library of siNA constructs having a predetermined complexity, such as of 4 N , where N represents the number of base paired nucleotides in each of the siNA construct strands (eg. for a siNA construct having 21 nucleotide sense and antisense strands with 19 base pairs, the complexity would be 4 19 ); and (b) assaying the siNA constructs of (a) above, under conditions suitable to determine RNAi target sites within the target RNA sequence.
- the siNA molecules of (a) have strands of a fixed length, for example about 23 nucleotides in length.
- the siNA molecules of (a) are of differing length, for example having strands of about 19 to about 25 (e.g., about 19, 20, 21, 22, 23, 24, or 25) nucleotides in length.
- the assay can comprise a reconstituted in vitro siNA assay as described in Example 7 herein.
- the assay can comprise a cell culture system in which target RNA is expressed.
- fragments of target RNA are analyzed for detectable levels of cleavage, for example by gel electrophoresis, northern blot analysis, or RNAse protection assays, to determine the most suitable target site(s) within the target RNA sequence.
- the target RNA sequence can be obtained as is known in the art, for example, by cloning and/or transcription for in vitro systems, and by cellular expression in in vivo systems.
- the invention features a method comprising: (a) analyzing the sequence of a RNA target encoded by a target gene; (b) synthesizing one or more sets of siNA molecules having sequence complementary to one or more regions of the RNA of (a); and (c) assaying the siNA molecules of (b) under conditions suitable to determine RNAi targets within the target RNA sequence.
- the siNA molecules of (b) have strands of a fixed length, for example about 23 nucleotides in length.
- the siNA molecules of (b) are of differing length, for example having strands of about 19 to about 25 (e.g., about 19, 20, 21, 22, 23, 24, or 25) nucleotides in length,
- the assay can comprise a reconstituted in vitro siNA assay as described herein.
- the assay can comprise a cell culture system in which target RNA is expressed. Fragments of target RNA are analyzed for detectable levels of cleavage, for example by gel electrophoresis, northern blot analysis, or RNAse protection assays, to determine the most suitable target site(s) within the target RNA sequence.
- the target RNA sequence can be obtained as is known in the art, for example, by cloning and/or transcription for in vitro systems, and by expression in in vivo systems.
- target site is meant a sequence within a target RNA that is “targeted” for cleavage mediated by a siNA construct which contains sequences within its antisense region that are complementary to the target sequence.
- detecttable level of cleavage is meant cleavage of target RNA (and fo ⁇ nation of cleaved product RNAs) to an extent sufficient to discern cleavage products above the background of RNAs produced by random degradation of the target RNA. Production of cleavage products from 1-5% of the target RNA is sufficient to detect above the background for most methods of detection.
- the invention features a composition comprising a siNA molecule of the invention, which can be chemically-modified, in a pharmaceutically acceptable carrier or diluent.
- the invention features a pharmaceutical composition comprising siNA molecules of the invention, which can be chemically-modified, targeting one or more genes in a pharmaceutically acceptable carrier or diluent.
- the invention features a method for treating or preventing a disease or condition in a subject comprising administering to the subject a composition of the invention under conditions suitable for the treatment or prevention of the disease or condition in the subject, alone or in conjunction with one or more other therapeutic compounds.
- the invention features a method for reducing or preventing tissue rejection in a subject comprising administering to the subject a composition of the mvention under conditions suitable for the reduction or prevention of tissue rejection in the subject.
- the invention features a method for validating a gene target, comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically- modified, wherein one of the siNA strands includes a sequence complementary to RNA of a target gene; (b) introducing the siNA molecule into a cell, tissue, or organism under conditions suitable for modulating expression of the target gene in the cell, tissue, or organism; and (c) determining the function of the gene by assaying for any phenotypic change in the cell, tissue, or organism.
- the invention features a method for validating a target gene comprising: (a) synthesizing a siNA molecule of the invention, which can be chemically- modified, wherein one of the siNA strands includes a sequence complementary to RNA of a target gene; (b) introducing the siNA molecule into a biological system under conditions suitable for modulating expression of the target gene in the biological system; and (c) determining the function of the gene by assaying for any phenotypic change in the biological system.
- biological system is meant, material, in a purified or unpurified form, from biological sources, including but not limited to human, animal, plant, insect, bacterial, viral or other sources, wherein the system comprises the components required for RNAi acitivity.
- biological system includes, for example, a cell, tissue, or organism, or extract thereof.
- biological system also includes reconstituted RNAi systems that can be used in an in vitro setting.
- phenotypic change is meant any detectable change to a cell that occurs in response to contact or treatment with a nucleic acid molecule of the invention (e.g., siNA).
- detectable changes include, but are not limited to, changes in shape, size, proliferation, motility, protein expression or RNA expression or other physical or chemical changes as can be assayed by methods known in the art.
- the detectable change can also include expression of reporter genes/molecules such as Green Florescent Protein (GFP) or various tags that are used to identify an expressed protein or any other cellular component that can be assayed.
- GFP Green Florescent Protein
- the invention features a kit containing a siNA molecule of the invention, which can be chemically-modified, that can be used to modulate the expression of a target gene in a cell, tissue, or organism.
- the invention features a kit containing more than one siNA molecule of the invention, which can be chemically-modified, that can be used to modulate the expression of more than one target gene in a cell, tissue, or organism.
- the invention features a kit containing a siNA molecule of the invention, which can be chemically-modified, that can be used to modulate the expression of a target gene in a biological system.
- the invention features a kit containing more than one siNA molecule of the invention, which can be chemically- modified, that can be used to modulate the expression of more than one target gene in a biological system.
- the invention features a cell containing one or more siNA molecules of the invention, which can be chemically-modified.
- the cell containing a siNA molecule of the invention is a mammalian cell.
- the cell containing a siNA molecule of the invention is a human cell.
- the synthesis of a siNA molecule of the invention comprises: (a) synthesis of two complementary strands of the siNA molecule; (b) annealing the two complementary strands together under conditions suitable to obtain a double-stranded siNA molecule.
- synthesis of the two complementary strands of the siNA molecule is by solid phase oligonucleotide synthesis.
- synthesis of the two complementary strands of the siNA molecule is by solid phase tandem oligonucleotide synthesis.
- the invention features a method for synthesizing a siNA duplex molecule comprising: (a) synthesizing a first oligonucleotide sequence strand of the siNA molecule, wherein the first oligonucleotide sequence strand comprises a cleavable linker molecule that can be used as a scaffold for the synthesis of the second oligonucleotide sequence strand of the siNA; (b) synthesizing the second oligonucleotide sequence strand of siNA on the scaffold of the first oligonucleotide sequence strand, wherein the second oligonucleotide sequence strand further comprises a chemical moiety than can be used to purify the siNA duplex; (c) cleaving the linker molecule of (a) under conditions suitable for the two siNA oligonucleotide strands to hybridize and form a stable duplex; and (d) purifying the siNA duplex utilizing the chemical moiety of the second
- cleavage of the linker molecule in (c) above takes place during deprotection of the oligonucleotide, for example under hydrolysis conditions using an alkylamine base such as methylamine.
- the method of synthesis comprises solid phase synthesis on a solid support such as controlled pore glass (CPG) or polystyrene, wherein the first sequence of (a) is synthesized on a cleavable linker, such as a succinyl linker, using the solid support as a scaffold.
- CPG controlled pore glass
- a cleavable linker such as a succinyl linker
- the cleavable linker in (a) used as a scaffold for synthesizing the second strand can comprise similar reactivity as the solid support derivatized linker, such that cleavage of the solid support derivatized linker and the cleavable linker of (a) takes place concomitantly.
- the chemical moiety of (b) that can be used to isolate the attached oligonucleotide sequence comprises a trityl group, for example a dimethoxytrityl group, which can be employed in a trityl-on synthesis strategy as described herein.
- the chemical moiety, such as a dimethoxytrityl group is removed during purification, for example, using acidic conditions.
- the method for siNA synthesis is a solution phase synthesis or hybrid phase synthesis wherein both strands of the siNA duplex are synthesized in tandem using a cleavable linker attached to the first sequence which acts a scaffold for synthesis of the second sequence. Cleavage of the linker under conditions suitable for hybridization of the separate siNA sequence strands results in formation of the double-stranded siNA molecule.
- the invention features a method for synthesizing a siNA duplex molecule comprising: (a) synthesizing one oligonucleotide sequence strand of the siNA molecule, wherein the sequence comprises a cleavable linker molecule that can be used as a scaffold for the synthesis of another oligonucleotide sequence; (b) synthesizing a second oligonucleotide sequence having complementarity to the first sequence strand on the scaffold of (a), wherein the second sequence comprises the other strand of the double- stranded siNA molecule and wherein the second sequence further comprises a chemical moiety than can be used to isolate the attached oligonucleotide sequence; (c) purifying the product of (b) utilizing the chemical moiety of the second oligonucleotide sequence strand under conditions suitable for isolating the full-length sequence comprising both siNA oligonucleotide strands connected by the cleavable linker and under conditions suitable
- cleavage of the linker molecule in (c) above takes place during deprotection of the oligonucleotide, for example under hydrolysis conditions. In another embodiment, cleavage of the linker molecule in (c) above takes place after deprotection of the oligonucleotide.
- the method of synthesis comprises solid phase synthesis on a solid support such as controlled pore glass (CPG) or polystyrene, wherein the first sequence of (a) is synthesized on a cleavable linker, such as a succinyl linker, using the solid support as a scaffold.
- CPG controlled pore glass
- cleavable linker such as a succinyl linker
- the cleavable linker in (a) used as a scaffold for synthesizing the second strand can comprise similar reactivity or differing reactivity as the solid support derivatized linker, such that cleavage of the solid support derivatized linker and the cleavable linker of (a) takes place either concomitantly or sequentially.
- the chemical moiety of (b) that can be used to isolate the attached oligonucleotide sequence comprises a trityl group, for example a dimethoxytrityl group.
- the invention features a method for making a double- stranded siNA molecule in a single synthetic process comprising: (a) synthesizing an oligonucleotide having a first and a second sequence, wherein the first sequence is complementary to the second sequence, and the first oligonucleotide sequence is linked to the second sequence via a cleavable linker, and wherein a terminal 5 '-protecting group, for example, a 5'-O-dimethoxytrityl group (5'-O-DMT) remains on the oligonucleotide having the second sequence; (b) deprotecting the oligonucleotide whereby the deprotection results in the cleavage of the linker joining the two oligonucleotide sequences; and (c) purifying the product of (b) under conditions suitable for isolating the double-stranded siNA molecule, for example using a trityl-on synthesis strategy as described herein.
- the method of synthesis of siNA molecules of the invention comprises the teachings of Scaringe et al, US Patent Nos. 5,889,136; 6,008,400; and 6,111 ,086, incorporated by reference herein in their entirety.
- the invention features siNA constructs that mediate RNAi in a cell or reconstituted system, wherein the siNA construct comprises one or more chemical modifications, for example, one or more chemical modifications having any of Formulae
- the invention features a method for generating si ⁇ A molecules with increased nuclease resistance comprising (a) introducing nucleotides having any of Formula I-NII or any combination thereof into a si ⁇ A molecule, and (b) assaying the si ⁇ A molecule of step (a) under conditions suitable for isolating si ⁇ A molecules having increased nuclease resistance.
- the invention features si ⁇ A constructs that mediate R ⁇ Ai against a target gene, wherein the si ⁇ A construct comprises one or more chemical modifications described herein that modulates the binding affinity between the sense and antisense strands of the si ⁇ A construct.
- the invention features a method for generating si ⁇ A molecules with increased binding affinity between the sense and antisense strands of the si ⁇ A molecule comprising (a) introducing nucleotides having any of Formula I-NII or any combination thereof into a si ⁇ A molecule, and (b) assaying the si ⁇ A molecule of step (a) under conditions suitable for isolating si ⁇ A molecules having increased binding affinity between the sense and antisense strands of the si ⁇ A molecule.
- the invention features si ⁇ A constructs that mediate R ⁇ Ai in a cell or reconstituted system, wherein the si ⁇ A construct comprises one or more chemical modifications described herein that modulates the binding affinity between the antisense strand of the si ⁇ A construct and a complementary target R ⁇ A sequence within a cell.
- the invention features siNA constructs that mediate RNAi in a cell or reconstituted system, wherein the siNA construct comprises one or more chemical modifications described herein that modulates the binding affinity between the antisense strand of the siNA construct and a complementary target DNA sequence within a cell.
- the mvention features a method for generating siNA molecules with increased binding affinity between the antisense strand of the siNA molecule and a complementary target RNA sequence comprising (a) introducing nucleotides having any of Fo ⁇ nula I-NII or any combination thereof into a si ⁇ A molecule, and (b) assaying the si ⁇ A molecule of step (a) under conditions suitable for isolating si ⁇ A molecules having increased binding affinity between the antisense strand of the si ⁇ A molecule and a complementary target R ⁇ A sequence.
- the invention features a method for generating si ⁇ A molecules with increased binding affinity between the antisense strand of the si ⁇ A molecule and a complementary target D ⁇ A sequence comprising (a) introducing nucleotides having any of Formula I-NII or any combination thereof into a si ⁇ A molecule, and (b) assaying the si ⁇ A molecule of step (a) under conditions suitable for isolating si ⁇ A molecules having increased binding affinity between the antisense strand of the si ⁇ A molecule and a complementary target D ⁇ A sequence.
- the invention features si ⁇ A constructs that mediate R ⁇ Ai in a cell or reconstituted system, wherein the si ⁇ A construct comprises one or more chemical modifications described herein that modulate the polymerase activity of a cellular polymerase capable of generating additional endogenous si ⁇ A molecules having sequence homology to the chemically-modified si ⁇ A construct.
- the invention features a method for generating si ⁇ A molecules capable of mediating increased polymerase activity of a cellular polymerase capable of generating additional endogenous si ⁇ A molecules having sequence homology to a chemically-modified si ⁇ A molecule comprising (a) introducing nucleotides having any of Formula I-NII or any combination thereof into a si ⁇ A molecule, and (b) assaying the si ⁇ A molecule of step (a) under conditions suitable for isolating si ⁇ A molecules capable of mediating increased polymerase activity of a cellular polymerase capable of generating additional endogenous si ⁇ A molecules having sequence homology to the chemically-modified siNA molecule, h one embodiment, the invention features chemically-modified siNA constructs that mediate RNAi in a cell or reconstituted system, wherein the chemical modifications do not significantly effect the interaction of siNA with a target RNA molecule, DNA molecule and/or proteins or other factors that are essential for RNAi in a manner that would decrease the effic
- the invention features a method for generating siNA molecules with improved RNAi activity comprising (a) introducing nucleotides having any of Formula I-NII or any combination thereof into a si ⁇ A molecule, and (b) assaying the si ⁇ A molecule of step (a) under conditions suitable for isolating si ⁇ A molecules having improved R ⁇ Ai activity.
- the invention features a method for generating si ⁇ A molecules with improved R ⁇ Ai activity against a target R ⁇ A comprising (a) introducing nucleotides having any of Formula I-NII or any combination thereof into a si ⁇ A molecule, and (b) assaying the si ⁇ A molecule of step (a) under conditions suitable for isolating si ⁇ A molecules having improved R ⁇ Ai activity against the target R ⁇ A.
- the invention features a method for generating si ⁇ A molecules with improved R ⁇ Ai activity against a D ⁇ A target comprising (a) introducing nucleotides having any of Formula I-NII or any combination thereof into a si ⁇ A molecule, and (b) assaying the si ⁇ A molecule of step (a) under conditions suitable for isolating si ⁇ A molecules having improved R ⁇ Ai activity against the D ⁇ A target, such as a gene, chromosome, or portion thereof.
- the invention features si ⁇ A constmcts that mediate R ⁇ Ai in a cell or reconstituted system, wherein the si ⁇ A constmct comprises one or more chemical modifications described herein that modulates the cellular uptake of the si ⁇ A constmct.
- the invention features a method for generating si ⁇ A molecules against a target gene with improved cellular uptake comprising (a) introducing nucleotides having any of Formula I-NII or any combination thereof into a si ⁇ A molecule, and (b) assaying the si ⁇ A molecule of step (a) under conditions suitable for isolating si ⁇ A molecules having improved cellular uptake.
- the invention features siNA constmcts that mediate RNAi against a target gene, wherein the siNA constmct comprises one or more chemical modifications described herein that increases the bioavailabihty of the siNA constmct, for example, by attaching polymeric conjugates such as polyethyleneglycol or equivalent conjugates that improve the pharmacokinetics of the siNA constmct, or by attaching conjugates that target specific tissue types or cell types in vivo.
- polymeric conjugates such as polyethyleneglycol or equivalent conjugates that improve the pharmacokinetics of the siNA constmct
- conjugates that target specific tissue types or cell types in vivo.
- Non-limiting examples of such conjugates are described in Nargeese et al, U.S. Serial No. 10/201,394 incorporated by reference herein.
- the invention features a method for generating siNA molecules of the invention with improved bioavailabihty, comprising (a) introducing a conjugate into the structure of a siNA molecule, and (b) assaying the siNA molecule of step (a) under conditions suitable for isolating siNA molecules having improved bioavailabihty.
- Such conjugates can include ligands for cellular receptors, such as peptides derived from naturally occurring protein ligands; protein localization sequences, including cellular ZIP code sequences; antibodies; nucleic acid aptamers; vitamins and other co-factors, such as folate and N-acetylgalactosamine; polymers, such as polyethyleneglycol (PEG); phospholipids; polyamines, such as spermine or spermidine; and others.
- ligands for cellular receptors such as peptides derived from naturally occurring protein ligands; protein localization sequences, including cellular ZIP code sequences; antibodies; nucleic acid aptamers; vitamins and other co-factors, such as folate and N-acetylgalactosamine; polymers, such as polyethyleneglycol (PEG); phospholipids; polyamines, such as spermine or spermidine; and others.
- the invention features a method for generating siNA molecules of the invention with improved bioavailabihty comprising (a) introducing an excipient formulation to a siNA molecule, and (b) assaying the siNA molecule of step (a) under conditions suitable for isolating siNA molecules having improved bioavailabihty.
- excipients include polymers such as cyclodextrins, lipids, cationic lipids, polyamines, phospholipids, and others.
- the invention features a method for generating siNA molecules of the invention with improved bioavailabihty comprising (a) introducing nucleotides having any of Formulae I-NII or any combination thereof into a si ⁇ A molecule, and (b) assaying the si ⁇ A molecule of step (a) under conditions suitable for isolating si ⁇ A molecules having improved bioavailabihty.
- polyethylene glycol can be covalently attached to si ⁇ A compoxmds of the present invention.
- the attached PEG can be any molecular weight, preferably from about 2,000 to about 50,000 daltons (Da).
- the present invention can be used alone or as a component of a kit having at least one of the reagents necessary to carry out the in vitro or in vivo introduction of RNA to test samples and/or subjects.
- preferred components of the kit include a siNA molecule of the invention and a vehicle that promotes introduction of the siNA into cells of interest as described herein (e.g., using lipids and other methods of transfection known in the art, see for example Beigelman et al, US 6,395,713).
- the kit can be used for target validation, such as in determining gene function and/or activity, or in dmg optimization, and in drag discovery (see for example Usman et al., USSN 60/402,996).
- target validation such as in determining gene function and/or activity, or in dmg optimization, and in drag discovery (see for example Usman et al., USSN 60/402,996).
- kit can also include instructions to allow a user of the kit to practice the invention.
- short interfering nucleic acid short interfering nucleic acid
- siNA short interfering nucleic acid
- siRNA short interfering RNA
- RNA short interfering nucleic acid molecule
- short interfering oligonucleotide molecule or “chemically-modified short interfering nucleic acid molecule” as used herein refers to any nucleic acid molecule capable of inhibiting or down regulating gene expression or viral replication, for example by mediating RNA interference "RNAi” or gene silencing in a sequence-specific manner; see for example Bass, 2001, Nat -e, 411, 428-429; Elbashir et al, 2001, Nature, Al l, 494-498; and Kreutzer et al, International PCT Publication No.
- RNAi RNA interference
- the si ⁇ A can be a double-stranded polynucleotide molecule comprising self-complementary sense and antisense regions, wherein the antisense region comprises nucleotide sequence that is complementary to nucleotide sequence in a target nucleic acid molecule or a portion thereof and the sense region having nucleotide sequence co ⁇ esponding to the target nucleic acid sequence or a portion thereof.
- the si ⁇ A can be assembled from two separate oligonucleotides, where one strand is the sense strand and the other is the antisense strand, wherein the antisense and sense strands are self-complementary (i.e.
- each strand comprises nucleotide sequence that is complementary to nucleotide sequence in the other strand; such as where the antisense strand and sense strand form a duplex or double stranded structure, for example wherein the double stranded region is about 19 base pairs); the antisense strand comprises nucleotide sequence that is complementary to nucleotide sequence in a target nucleic acid molecule or a portion thereof and the sense strand comprises nucleotide sequence corresponding to the target nucleic acid sequence or a portion thereof.
- the siNA is assembled from a single oligonucleotide, where the self-complementary sense and antisense regions of the siNA are linked by means of a nucleic acid based or non-nucleic acid-based linker(s).
- the siNA can be a polynucleotide with a hairpin secondary structure, having self-complementary sense and antisense regions, wherein the antisense region comprises nucleotide sequence that is complementary to nucleotide sequence in a separate target nucleic acid molecule or a portion thereof and the sense region having nucleotide sequence corresponding to the target nucleic acid sequence or a portion thereof.
- the siNA can be a circular single- stranded polynucleotide having two or more loop structures and a stem comprising self- complementary sense and antisense regions, wherein the antisense region comprises nucleotide sequence that is complementary to nucleotide sequence in a target nucleic acid molecule or a portion thereof and the sense region having nucleotide sequence co ⁇ esponding to the target nucleic acid sequence or a portion thereof, and wherein the circular polynucleotide can be processed either in vivo or in vitro to generate an active siNA molecule capable of mediating RNAi.
- the siNA can also comprise a single stranded polynucleotide having nucleotide sequence complementary to nucleotide sequence in a target nucleic acid molecule or a portion thereof (for example, where such siNA molecule does not require the presence within the siNA molecule of nucleotide sequence corresponding to the target nucleic acid sequence or a portion thereof), wherein the single stranded polynucleotide can further comprise a terminal phosphate group, such as a 5'-phosphate (see for example Martinez et al, 2002, Cell, 110, 563-574 and Schwarz et al, 2002, Molecular Cell, 10, 537-568), or 5',3'-diphosphate.
- a 5'-phosphate see for example Martinez et al, 2002, Cell, 110, 563-574 and Schwarz et al, 2002, Molecular Cell, 10, 537-568
- the siNA molecule of the invention comprises separate sense and antisense sequences or regions, wherein the sense and antisense regions are covalently linked by nucleotide or non-nucleotide linkers molecules as is known in the art, or are alternately non-covalently linked by ionic interactions, hydrogen bonding, van der waals interactions, hydrophobic intercations, and/or stacking interactions.
- the siNA molecules of the invention comprise nucleotide sequence that is complementary to nucleotide sequence of a target gene.
- the siNA molecule of the invention interacts with nucleotide sequence of a target gene in a manner that causes inhibition of expression of the target gene.
- siNA molecules need not be limited to those molecules containing only RNA, but further encompasses chemically-modified nucleotides and non-nucleotides.
- the short interfering nucleic acid molecules of the invention lack 2'- hydroxy (2'-OH) containing nucleotides.
- Applicant describes in certain embodiments short interfering nucleic acids that do not require the presence of nucleotides having a 2'- hydroxy group for mediating RNAi and as such, short interfering nucleic acid molecules of the mvention optionally do not include any ribonucleotides (e.g., nucleotides having a 2'-OH group).
- siNA molecules that do not require the presence of ribonucleotides within the siNA molecule to support RNAi can however have an attached linker or linkers or other attached or associated groups, moieties, or chains containing one or more nucleotides with 2'-OH groups.
- siNA molecules can comprise ribonucleotides at about 5, 10, 20, 30, 40, or 50% of the nucleotide positions.
- modified short interfering nucleic acid molecules of the invention can also be refe ⁇ ed to as short interfering modified oligonucleotides "siMON.”
- siNA is meant to be equivalent to other terms used to describe nucleic acid molecules that are capable of mediating sequence specific RNAi, for example short interfering RNA (siRNA), double- stranded RNA (dsRNA), micro-RNA (miRNA), short hairpin RNA (shRNA), short interfering oligonucleotide, short interfering nucleic acid, short interfering modified oligonucleotide, chemically-modified siRNA, post-transcriptional gene silencing RNA (ptgsRNA), and others.
- siRNA short interfering RNA
- dsRNA double- stranded RNA
- miRNA micro-RNA
- shRNA short hairpin RNA
- ptgsRNA post-transcriptional gene silencing RNA
- RNAi is meant to be equivalent to other terms used to describe sequence specific RNA interference, such as post transcriptional gene silencing, or epigenetics.
- siNA molecules of the invention can be used to epigenetically silence genes at both the post-transcriptional level or the pre-transcriptional level.
- epigenetic regulation of gene expression by siNA molecules of the invention can result from siNA mediated modification of chromatin structure to alter gene expression (see, for example, Allshire, 2002, Science, 297, 1818-1819; Volpe et al, 2002, Science, 297, 1833-1837; Jenuwein, 2002, Science, 297, 2215-2218; and Hall et al, 2002, Science, 297, 2232-2237).
- modulate is meant that the expression of the gene, or level of RNA molecule or equivalent RNA molecules encoding one or more proteins or protein subunits, or activity of one or more proteins or protein subunits is up regulated or down regulated, such that expression, level, or activity is greater than or less than that observed in the absence of the modulator.
- modulate can mean “inhibit,” but the use of the word “modulate” is not limited to this definition.
- inhibitor By “inhibit”, “down-regulate”, or “reduce”, it is meant that the expression of the gene, or level of RNA molecules or equivalent RNA molecules encoding one or more proteins or protein subunits, or activity of one or more proteins or protein subunits, is reduced below that observed in the absence of the nucleic acid molecules (e.g., siNA) of the invention.
- nucleic acid molecules e.g., siNA
- inhibition, down-regulation or reduction with an siNA molecule is below that level observed in the presence of an inactive or attenuated molecule.
- inhibition, down-regulation, or reduction with siNA molecules is below that level observed in the presence of, for example, an siNA molecule with scrambled sequence or with mismatches.
- inhibition, down- regulation, or reduction of gene expression with a nucleic acid molecule of the instant invention is greater in the presence of the nucleic acid molecule than in its absence.
- RNA nucleic acid that encodes an RNA
- the target gene can be a gene derived from a cell, an endogenous gene, a transgene, or exogenous genes such as genes of a pathogen, for example a vims, which is present in the cell after infection thereof.
- the cell containing the target gene can be derived from or contained in any organism, for example a plant, animal, protozoan, vims, bacterium, or fungus.
- Non-limiting examples of plants include monocots, dicots, or gymnosperms.
- Non-limiting examples of animals include vertebrates or invertebrates.
- fungi include molds or yeasts.
- highly conserved sequence region a nucleotide sequence of one or more regions in a target gene does not vary significantly from one generation to the other or from one biological system to the other.
- cancer is meant a group of diseases characterized by uncontrolled growth and spread of abnormal cells.
- sense region is meant a nucleotide sequence of a siNA molecule having complementarity to an antisense region of the siNA molecule.
- the sense region of a siNA molecule can comprise a nucleic acid sequence having homology with a target nucleic acid sequence.
- antisense region is meant a nucleotide sequence of a siNA molecule having complementarity to a target nucleic acid sequence.
- the antisense region of a siNA molecule can optionally comprise a nucleic acid sequence having complementarity to a sense region of the siNA molecule.
- target nucleic acid is meant any nucleic acid sequence whose expression or activity is to be modulated.
- the target nucleic acid can be DNA or RNA.
- nucleic acid can form hydrogen bond(s) with another nucleic acid sequence by either traditional Watson-Crick or other non-traditional types.
- the binding free energy for a nucleic acid molecule with its complementary sequence is sufficient to allow the relevant function of the nucleic acid to proceed, e.g., RNAi activity. Determination of binding free energies for nucleic acid molecules is well known in the art (see, e.g., Turner et al, 1987, CSHSymp. Quant. Biol. Ill pp.123-133; Frier et al, 1986, Proc. Nat. Acad. Sci.
- a percent complementarity indicates the percentage of contiguous residues in a nucleic acid molecule that can fo ⁇ n hydrogen bonds (e.g., Watson-Crick base pairing) with a second nucleic acid sequence (e.g., 5, 6, 7, 8, 9, 10 out of 10 being 50%, 60%, 70%, 80%, 90%, and 100% complementary).
- Perfectly complementary means that all the contiguous residues of a nucleic acid sequence will hydrogen bond with the same number of contiguous residues in a second nucleic acid sequence.
- the siNA molecules of the invention represent a novel therapeutic approach to a broad spectrum of diseases and conditions, including cancer or cancerous disease, infectious disease, cardiovascular disease, neurological disease, prion disease, inflammatory disease, autoimmune disease, pulmonary disease, renal disease, liver disease, mitochondrial disease, endocrine disease, reproduction related diseases and conditions, and any other indications that can respond to the level of an expressed gene product in a cell or organsim.
- each sequence of a siNA molecule of the invention is independently about 18 to about 24 nucleotides in length, in specific embodiments about 18, 19, 20, 21, 22, 23, or 24 nucleotides in length.
- the siNA duplexes of the invention independently comprise about 17 to about 23 base pairs (e.g., about 17, 18, 19, 20, 21, 22 or 23).
- siNA molecules of the invention comprising hairpin or circular stmctures are about 35 to about 55 (e.g., about 35, 40, 45, 50 or 55) nucleotides in length, or about 38 to about 44 (e.g., 38, 39, 40, 41, 42, 43 or 44) nucleotides in length and comprising about 16 to about 22 (e.g., about 16, 17, 18, 19, 20, 21 or 22) base pairs.
- Exemplary siNA molecules of the invention are shown in Table II.
- Exemplary synthetic siNA molecules of the invention are shown in Table I and/or Figures 18-19.
- cell is used in its usual biological sense, and does not refer to an entire multicellular organism, e.g., specifically does not refer to a human.
- the cell can be present in an organism, e.g., birds, plants and mammals such as humans, cows, sheep, apes, monkeys, swine, dogs, and cats.
- the cell can be prokaryotic or eukaryotic (e.g., mammalian or plant cell).
- the cell can be of somatic or germ line origin, totipotent or pluripotent, dividing or non-dividing.
- the cell can also be derived from or can comprise a gamete or embryo, a stem cell, or a fully differentiated cell.
- the siNA molecules of the invention are added directly, or can be complexed with cationic lipids, packaged within liposomes, or otherwise delivered to target cells or tissues.
- the nucleic acid or nucleic acid complexes can be locally administered to relevant tissues ex vivo, or in vivo through injection, infusion pump or stent, with or without their incorporation in biopolymers.
- the nucleic acid molecules of the invention comprise sequences shown in Tables I-II and/or Figures 18- 19. Examples of such nucleic acid molecules consist essentially of sequences defined in these tables and figures.
- the chemically modified constmcts described in Table IV can be applied to any siNA sequence of the invention.
- the invention provides mammalian cells containing one or more siNA molecules of this mvention.
- the one or more siNA molecules can independently be targeted to the same or different sites.
- RNA is meant a molecule comprising at least one ribonucleotide residue.
- ribonucleotide is meant a nucleotide with a hydroxyl group at the 2' position of a ⁇ -D- ribo-furanose moiety.
- the terms include double-stranded RNA, single-stranded RNA, isolated RNA such as partially purified RNA, essentially pure RNA, synthetic RNA, recombinantly produced RNA, as well as altered RNA that differs from naturally occurring RNA by the addition, deletion, substitution and/or alteration of one or more nucleotides.
- Such alterations can include addition of non-nucleotide material, such as to the end(s) of the siNA or internally, for example at one or more nucleotides of the RNA.
- Nucleotides in the RNA molecules of the instant invention can also comprise non- standard nucleotides, such as non-naturally occurring nucleotides or chemically synthesized nucleotides or deoxynucleotides. These altered RNAs can be referred to as analogs or analogs of naturally-occurring RNA.
- subject is meant an organism, which is a donor or recipient of explanted cells or the cells themselves. “Subject” also refers to an organism to which the nucleic acid molecules of the invention can be administered. In one embodiment, a subject is a mammal or mammalian cells. In another embodiment, a subject is a human or human cells.
- phosphorothioate refers to an internucleotide linkage having Fo ⁇ nula I, wherein Z and/or W comprise a sulfur atom. Hence, the term phosphorothioate refers to both phosphorothioate and phosphorodithioate internucleotide linkages.
- universal base refers to nucleotide base analogs that form base pairs with each of the natural DNA/RNA bases with little discrimination between them.
- Non-limiting examples of universal bases include C-phenyl, C-naphthyl and other aromatic derivatives, inosine, azole carboxamides, and nitroazole derivatives such as 3- nitropyrrole, 4-nitroindole, 5-nitroindole, and 6-nitroindole as known in the art (see for example Loakes, 2001 , Nucleic Acids Research, 29, 2437-2447).
- acyclic nucleotide refers to any nucleotide having an acyclic ribose sugar, for example where any of the ribose carbons (Cl, C2, C3, C4, or C5), are independently or in combination absent from the nucleotide.
- the nucleic acid molecules of the instant invention can be used to treat diseases or conditions discussed herein.
- the siNA molecules can be administered to a subject or can be administered to other appropriate cells evident to those skilled in the art, individually or in combination with one or more drags under conditions suitable for the treatment.
- the siNA molecules can be used in combination with other known treatments to treat conditions or diseases discussed above.
- the described molecules could be used in combination with one or more known therapeutic agents to treat a disease or condition.
- Non-limiting examples of other therapeutic agents that can be readily combined with a siNA molecule of the invention are enzymatic nucleic acid molecules, allosteric nucleic acid molecules, antisense, decoy, or aptamer nucleic acid molecules, antibodies such as monoclonal antibodies, small molecules, and other organic and/or inorganic compounds including metals, salts and ions.
- Figure 1 shows a non-limiting example of a scheme for the synthesis of siNA molecules.
- the complementary siNA sequence strands, strand 1 and strand 2 are synthesized in tandem and are connected by a cleavable linkage, such as a nucleotide succinate or abasic succinate, which can be the same or different from the cleavable linker used for solid phase synthesis on a solid support.
- the synthesis can be either solid phase or solution phase, in the example shown, the synthesis is a solid phase synthesis.
- the synthesis is performed such that a protecting group, such as a dimethoxytrityl group, remains intact on the terminal nucleotide of the tandem oligonucleotide.
- the two siNA strands spontaneously hybridize to form a siNA duplex, which allows the purification of the duplex by utilizing the properties of the terminal protecting group, for example by applying a trityl on purification method wherein only duplexes/oligonucleotides with the terminal protecting group are isolated.
- Figure 2 shows a MALDI-TOV mass spectrum of a purified siNA duplex synthesized by a method of the invention.
- the two peaks shown correspond to the predicted mass of the separate siNA sequence strands. This result demonstrates that the siNA duplex generated from tandem synthesis can be purified as a single entity using a simple trityl-on purification methodology.
- Figure 3 shows the results of a stability assay used to determine the serum stability of chemically modified siNA constructs compared to a siNA control consisting of all RNA with 3'-TT termini. T l A values are shown for duplex stability.
- Figure 4 shows the results of an RNAi activity screen of several phosphorothioate modified siNA constmcts using a luciferase reporter system.
- Figure 5 shows the results of an RNAi activity screen of several phosphorothioate and universal base modified siNA constmcts using a luciferase reporter system.
- Figure 6 shows the results of an RNAi activity screen of several 2'-O-methyl modified siNA constmcts using a luciferase reporter system.
- Figure 7 shows the results of an RNAi activity screen of several 2'-O-methyl and 2 '-deoxy-2' -fluoro modified siNA constmcts using a luciferase reporter system.
- Figure 8 shows the results of an RNAi activity screen of a phosphorothioate modified siNA construct using a luciferase reporter system.
- Figure 9 shows the results of an RNAi activity screen of an inverted deoxyabasic modified siNA construct generated via tandem synthesis using a luciferase reporter system.
- Figure 10 shows the results of an RNAi activity screen of chemically modifed siNA constmcts including 3'-glyceryl modified siNA constmcts compared to an all RNA control siNA construct using a luciferase reporter system.
- These chemically modified siNAs were compared in the luciferase assay described herein at 1 nM and lOnM concentration using an all RNA siNA control (siGL2) having 3 '-terminal dithymidine (TT) and its corresponding inverted control (Inv siGL2).
- the background level of luciferase expression in the HeLa cells is designated by the "cells" column.
- Sense and antisense strands of chemically modified siNA constmcts are shown by RPI number (sense strand/antisense strand). Sequences correspoding to these RPI numbers are shown in Table I.
- Figure 11 shows the results of an RNAi activity screen of chemically modifed siNA constmcts.
- the screen compared various combinations of sense strand chemical modifications and antisense strand chemical modifications.
- These chemically modified siNAs were compared in the luciferase assay described herein at 1 nM and lOnM concentration using an all RNA siNA control (siGL2) having 3 '-terminal dithymidine (TT) and its corresponding inverted control (Inv siGL2).
- the background level of luciferase expression in the HeLa cells is designated by the "cells" column.
- Sense and antisense strands of chemically modified siNA constmcts are shown by RPI number (sense strand/antisense strand). Sequences co ⁇ espoding to these RPI numbers are shown in Table I.
- Figure 12 shows the results of an RNAi activity screen of chemically modifed siNA constmcts.
- the screen compared various combinations of sense strand chemical modifications and antisense strand chemical modifications.
- These chemically modified siNAs were compared in the luciferase assay described herein at 1 nM and lOnM concentration using an all RNA siNA control (siGL2) having 3 '-terminal dithymidine (TT) and its co ⁇ esponding inverted control (Inv siGL2).
- the background level of luciferase expression in the HeLa cells is designated by the "cells" column.
- Sense and antisense strands of chemically modified siNA constmcts are shown by RPI number (sense strand/antisense strand). Sequences correspoding to these RPI numbers are shown in Table I.
- the antisense strand alone (RPI 30430) and an inverted control (RPI 30227/30229, having matched chemistry to RPI (30063/30224) was compared to the siNA duplexes described above.
- Figure 13 shows the results of an RNAi activity screen of chemically modifed siNA constmcts.
- the screen compared various combinations of sense strand chemical modifications and antisense strand chemical modifications.
- These chemically modified siNAs were compared in the luciferase assay described herein at 1 nM and lOnM concentration using an all RNA siNA control (siGL2) having 3 '-terminal dithymidine (TT) and its co ⁇ esponding inverted control (Inv siGL2).
- the background level of luciferase expression in the HeLa cells is designated by the "cells" column.
- Sense and antisense strands of chemically modified siNA constmcts are shown by RPI number (sense strand/antisense strand). Sequences correspoding to these RPI numbers are shown in Table I. hi addition, an inverted control (RPI 30226/30229), having matched chemistry to RPI (30222/30224) was compared to the siNA duplexes described above.
- Figure 14 shows the results of an RNAi activity screen of chemically modifed siNA constmcts including various 3 '-terminal modified siNA constructs compared to an all RNA control siNA constmct using a luciferase reporter system.
- These chemically modified siNAs were compared in the luciferase assay described herein at 1 nM and lOnM concentration using an all RNA siNA control (siGL2) having 3 '-terminal dithymidine (TT) and its co ⁇ esponding inverted control (Inv siGL2).
- the background level of luciferase expression in the HeLa cells is designated by the "cells" column.
- Sense and antisense strands of chemically modified siNA constructs are shown by RPI number (sense strand/antisense strand). Sequences correspoding to these RPI numbers are shown in Table I.
- Figure 15 shows the results of an RNAi activity screen of chemically modifed siNA constructs.
- the screen compared various combinations of sense strand chemistries compared to a fixed antisense strand chemistry.
- These chemically modified siNAs were compared in the luciferase assay described herein at 1 nM and lOnM concentration using an all RNA siNA control (siGL2) having 3 '-terminal dithymidine (TT) and its co ⁇ esponding inverted control (Inv siGL2).
- the background level of luciferase expression in the HeLa cells is designated by the "cells" column.
- Sense and antisense strands of chemically modified siNA constmcts are shown by RPI number (sense strand/antisense strand). Sequences co ⁇ espoding to these RPI numbers are shown in Table I.
- Figure 16 shows the results of a siNA titration study using a luciferase reporter system, wherein the RNAi activity of a phosphorothioate modified siNA constmct is compared to that of a siNA construct consisting of all ribonucleotides except for two terminal thymidine residues.
- Figure 17 shows a non-limiting proposed mechanistic representation of target RNA degradation involved in RNAi.
- Double-stranded RNA dsRNA
- RdRP RNA-dependent RNA polymerase
- siNA duplexes RNA-dependent RNA polymerase
- synthetic or expressed siNA can be introduced directely into a cell by appropriate means.
- An active siNA complex forms which recognizes a target RNA, resulting in degradation of the target RNA by the RISC endonuclease complex or in the synthesis of additional RNA by RNA-dependent RNA polymerase (RdRP), which can activate DICER and result in additional siNA molecules, thereby amplifying the RNAi response.
- RdRP RNA-dependent RNA polymerase
- Figure 18A-F shows non-limiting examples of chemically-modified siNA constructs of the present invention.
- N stands for any nucleotide (adenosine, guanosine, cytosine, uridine, or optionally thymidine, for example thymidine can be substituted in the overhanging regions designated by parenthesis (N N).
- Various modifications are shown for the sense and antisense strands of the siNA constmcts.
- the sense strand comprises 21 nucleotides having four phosphorothioate 5'- and 3'-terminal internucleotide linkages, wherein the two terminal 3'- nucleotides are optionally base paired and wherein all pyrimidine nucleotides that may be present are 2'-O-methyl or 2'-deoxy-2'-fluoro modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein.
- the antisense strand comprises 21 nucleotides, optionally having a 3 '-terminal glyceryl moiety and wherein the two terminal 3 '-nucleotides are optionally complementary to the target RNA sequence, and having one 3 '-terminal phosphorothioate internucleotide linkage and four 5 '-terminal phosphorothioate internucleotide linkages and wherein all pyrimidine nucleotides that may be present are 2'-deoxy-2'-fluoro modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein.
- the sense strand comprises 21 nucleotides wherein the two terminal 3 '-nucleotides are optionally base paired and wherein all pyrimidine nucleotides that may be present are 2'-O-methyl or 2'-deoxy-2'-fluoro modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein.
- the antisense strand comprises 21 nucleotides, optionally having a 3 '-terminal glyceryl moiety and wherein the two terminal 3 '-nucleotides are optionally complementary to the target RNA sequence, and wherein all pyrimidine nucleotides that may be present are 2'-deoxy-2'-fluoro modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein.
- the sense strand comprises 21 nucleotides having 5'- and 3'- terminal cap moieties wherein the two terminal 3 '-nucleotides are optionally base paired and wherein all pyrimidine nucleotides that may be present are 2'-O-methyl or 2'-deoxy-2'- fluoro modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein.
- the antisense strand comprises 21 nucleotides, optionally having a 3'- terminal glyceryl moiety and wherein the two terminal 3 '-nucleotides are optionally complementary to the target RNA sequence, and having one 3 '-terminal phosphorothioate internucleotide linkage and wherein all pyrimidine nucleotides that may be present are 2'- deoxy-2'-fluoro modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein.
- the sense strand comprises 21 nucleotides having 5'- and 3'- terminal cap moieties wherein the two terminal 3 '-nucleotides are optionally base paired and wherein all pyrimidine nucleotides that may be present are 2'-deoxy-2'-fluoro modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein and wherein and all purine nucleotides that may be present are 2'-deoxy nucleotides.
- the antisense strand comprises 21 nucleotides, optionally having a 3 '-terminal glyceryl moiety and wherein the two terminal 3 '-nucleotides are optionally complementary to the target RNA sequence, and having one 3 '-terminal phosphorothioate internucleotide linkage and wherein all pyrimidine nucleotides that may be present are 2'-deoxy-2'-fluoro modified nucleotides and all purine nucleotides that may be present are 2'-O-methyl modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein.
- the sense strand comprises 21 nucleotides having 5'- and 3'- terminal cap moieties wherein the two terminal 3 '-nucleotides are optionally base paired and wherein all pyrimidine nucleotides that may be present are 2'-deoxy-2'-fluoro modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein.
- the antisense strand comprises 21 nucleotides, optionally having a 3 '-terminal glyceryl moiety and wherein the two terminal 3'-nucleotides are optionally complementary to the target RNA sequence, and wherein all pyrimidine nucleotides that may be present are 2'-deoxy- 2'-fluoro modified nucleotides and all purine nucleotides that may be present are 2'-O- methyl modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein.
- the sense strand comprises 21 nucleotides having 5'- and 3'- terminal cap moieties wherein the two terminal 3 '-nucleotides are optionally base paired and wherein all pyrimidine nucleotides that may be present are 2'-deoxy-2'-fluoro modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein.
- the antisense strand comprises 21 nucleotides, optionally having a 3 '-terminal glyceryl moiety and wherein the two terminal 3 '-nucleotides are optionally complementary to the target RNA sequence, and having one 3 '-terminal phosphorothioate internucleotide linkage and wherein all pyrimidine nucleotides that may be present are 2'-deoxy-2'-fluoro modified nucleotides and all purine nucleotides that may be present are 2'-deoxy modified nucleotides except for (N N) nucleotides, which can comprise ribonucleotides, deoxynucleotides, universal bases, or other chemical modifications described herein.
- the antisense strand of constmcts A-F comprise sequence complementary to target RNA sequence of the invention.
- Figure 19 shows non-limiting examples of specific chemically modified siNA sequences of the invention.
- A-F applies the chemical modifications described in Figure 18A-F to a representative siNA sequence targeting the hepatitis C vims (HCV).
- Figure 20 shows non-limiting examples of different siNA constructs of the mvention.
- the examples shown (constmcts 1, 2, and 3) have 19 representative base pairs, however, different embodiments of the invention include any number of base pairs described herein.
- Bracketed regions represent nucleotide overhangs, for example comprising about 1, 2, 3, or 4 nucleotides in length, preferably about 2 nucleotides.
- Constructs 1 and 2 can be used independently for RNAi activity.
- Constmct 2 can comprise a polynucleotide or non-nucleotide linker, which can optionally be designed as a biodegradable linker.
- the loop structure shown in constmct 2 can comprise a biodegradable linker that results in the formation of constmct 1 in vivo and/or in vitro
- construct 3 can be used to generate constmct 2 under the same principle wherein a linker is used to generate the active siNA constmct 2 in vivo and/or in vitro, which can optionally utilize another biodegradable linker to generate the active siNA constmct 1 in vivo and/or in vitro.
- the stability and/or activity of the siNA constmcts can be modulated based on the design of the siNA constmct for use in vivo or in vitro and/or in vitro.
- Figure 21 is a diagrammatic representation of a method used to determine target sites for siNA mediated RNAi within a particular target nucleic acid sequence, such as messenger RNA.
- a pool of siNA oligonucleotides are synthesized wherein the antisense region of the siNA constmcts has complementarity to target sites across the target nucleic acid sequence, and wherein the sense region comprises sequence complementary to the antisense region of the siNA.
- the sequences are transfected into cells.
- Cells are selected based on phenotypic change that is associated with modulation of the target nucleic acid sequence.
- D The siNA is isolated from the selected cells and is sequenced to identify efficacious target sites within the target nucleic acid sequence.
- Figure 22 shows non-limiting examples of different stabilization chemistries (1-10) that can be used, for example, to stabilize the 3'-end of siNA sequences of the invention, including (1) [3 -3'] -inverted deoxyribose; (2) deoxyribonucleotide; (3) [5'-3']-3'- deoxyribonucleotide; (4) [5'-3']-ribonucleotide; (5) [5'-3']-3'-O-methyl ribonucleotide; (6) 3'-glyceryl; (7) [3'-5']-3'-deoxyribonucleotide; (8) [3 '-3'] -deoxyribonucleotide; (9) [5'-2']- deoxyribonucleotide; and (10) [5-3']-dideoxyribonucleotide.
- stabilization chemistries (1-10) that can be used, for example, to stabilize the 3'-end of siNA sequences
- modified and unmodified backbone chemistries indicated in the figure can be combined with different backbone modifications as described herein, for example, backbone modifications having Formula I.
- the 2'-deoxy nucleotide shown 5' to the terminal modifications shown can be another modified or unmodified nucleotide or non-nucleotide described herein, for example modifications having any of Formulae I- VII or any combination thereof.
- Figure 23 shows a non-limiting example of siNA mediated inhibition of VEGF- induced angiogenesis using the rat comeal model of angiogenesis.
- siNA targeting site 2340 of VEGFR1 RNA shown as RPI No. 29695/29699
- inverted controls shown as RPI No. 29983/29984
- Figure 24 is a non-limiting example of a HBsAg screen of stabilized siNA constmcts ("stab 4/5", see Table IV) targeting HBV pregenomic RNA in HepG2 cells at 25 nM compared to untreated and matched chemistry inverted sequence controls.
- the siNA sense and antisense strands are shown by RPI number (sense/antisense).
- Figure 25 is a non-limiting example of a dose response HBsAg screen of stabilized siNA constmcts ("stab 4/5", see Table IV) targeting sites 262 and 1580 of the HBV pregenomic RNA in HepG2 cells at 0.5, 5, 10 and 25 nM compared to untreated and matched chemistry inverted sequence controls.
- the siNA sense and antisense strands are shown by RPI number (sense/antisense).
- Figure 26 shows a dose response comparison of two different stabilization chemistries ("stab 7/8" and "stab 7/11", see Table IV) targeting site 1580 of the HBV pregenomic RNA in HepG2 cells at 5, 10, 25, 50 and 100 nM compared to untreated and matched chemistry inverted sequence controls.
- the siNA sense and antisense strands are shown by RPI number (sense/antisense).
- Figure 27 shows a non-limiting example of a strategy used to identify chemically modified siNA constracts of the invention that are nuclease resistance while preserving the ability to mediate RNAi activity.
- Chemical modifications are introduced into the siNA constmct based on educated design parameters (e.g. introducing 2'-mofications, base modifications, backbone modifications, terminal cap modifications etc).
- the modified constract in tested in an appropriate system (e.g human serum for nuclease resistance, shown, or an animal model for PK delivery parameters).
- the siNA constract is tested for RNAi activity, for example in a cell culture system such as a luciferase reporter assay).
- siNA constracts are then identified which possess a particular characteristic while maintaining RNAi activity, and can be further modified and assayed once again. This same approach can be used to identify siNA-conjugate molecules with improved pharmacokinetic profiles, delivery, and RNAi activity.
- Figure 28 shows representative data of a chemically modified siNA constmct (Stab
- HBV site 1580 RNA compared to an unstabilized siRNA constmct in a dose response time course HBsAg assay.
- the constmcts were compared at different concentrations (5nM, 10 nM, 25 nM, 50 nM, and 100 nM) over the course of nine days.
- Activity based on HBsAg levels was determined at day 3, day 6, and day 9.
- Figure 29 shows representative data of a chemically modified siNA constract (Stab 7/8, Table IV) targeting HBV site 1580 RNA compared to an unstabilized siRNA constract in a dose response time course HBsAg assay.
- the constracts were compared at different concentrations (5nM, 10 nM, 25 nM, 50 nM, and 100 nM) over the course of nine days.
- SiNA activity based on HBsAg levels was determined at day 3, day 6, and day 9.
- Figure 30 shows representative data of a chemically modified siNA construct (Stab 7/11, Table IV) targeting HBV site 1580 RNA compared to an unstabilized siRNA constract in a dose response time course HBsAg assay.
- the constmcts were compared at different concentrations (5nM, 10 nM, 25 nM, 50 nM, and 100 nM) over the course of nine days.
- SiNA activity based on HBsAg levels was determined at day 3, day 6, and day 9.
- Figure 31 shows representative data of a chemically modified siNA constmct (Stab
- Figure 32 shows non-limiting examples of inhibition of viral replication of a HCV/poliovirus chimera by siNA constracts targeted to HCV chimera (29579/29586; 29578/29585) compared to control (29593/29600).
- Figure 33 shows a non-limiting example of a dose response study demonstrating the inhibition of viral replication of a HCV/poliovims chimera by siNA constract (29579/29586) at various concentrations (lnM, 5nM, lOnM, and 25nM) compared to control (29593/29600).
- Figure 34 shows a non-limiting example demonstrating the inhibition of viral replication of a HCV/poliovirus chimera by a chemically modified siRNA constract (30051/30053) compared to control constract (30052/30054).
- Figure 35 shows a non-limiting example demonstrating the inhibition of viral replication of a HCV/poliovims chimera by a chemically modified siRNA constract (30055/30057) compared to control construct (30056/30058).
- Figure 36 shows a non- limiting example of several chemically modified siRNA constructs targeting viral replication of an HCV/poliovirus chimera at 10 nM treatment in comparison to a lipid control and an inverse siNA control constract 29593/ 29600.
- Figure 37 shows a non-limiting example of several chemically modified siRNA constracts targeting viral replication of a HCV/poliovirus chimera at 25 nM treatment in comparison to a lipid control and an inverse siNA control construct 29593/ 29600.
- Figure 38 shows a non-limiting example of several chemically modified siRNA constructs targeting viral replication of a Huh7 HCV replicon system at 25 nM treatment in comparison to untreated cells (“cells”), cells transfected with lipofectamine (“LFA2K”) and inverse siNA control constmcts.
- RNAi activity measured in vitro and/or in vivo where the RNAi activity is a reflection of both the ability of the siNA to mediate RNAi and the stability of the siNAs of the invention.
- the product of these activities can be increased in vitro and/or in vivo compared to an all RNA siRNA or a siNA containing a plurality of ribonucleotides.
- the activity or stability of the siNA molecule can be decreased (i.e., less than ten-fold), but the overall activity of the siNA molecule is enhanced in vitro and/or in vivo.
- RNA interference refers to the process of sequence specific post-transcriptional gene silencing in animals mediated by short interfering RNAs (siRNAs) (Fire et al, 1998, Nature, 391, 806).
- siRNAs short interfering RNAs
- the co ⁇ esponding process in plants is commonly refe ⁇ ed to as post- transcriptional gene silencing or RNA silencing and is also refe ⁇ ed to as quelling in fungi.
- the process of post-transcriptional gene silencing is thought to be an evolutionarily-conserved cellular defense mechanism used to prevent the expression of foreign genes which is commonly shared by diverse flora and phyla (Fire et al, 1999, Trends Genet., 15, 358).
- Such protection from foreign gene expression may have evolved in response to the production of double-stranded RNAs (dsRNAs) derived from viral infection or the random integration of transposon elements into a host genome via a cellular response that specifically destroys homologous single-stranded RNA or viral genomic RNA.
- dsRNAs double-stranded RNAs
- the presence of dsRNA in cells triggers the RNAi response though a mechanism that has yet to be fully characterized. This mechanism appears to be different from the interferon response that results from dsRNA-mediated activation of protein kinase PKR and 2', 5'-oligoadenylate synthetase resulting in non-specific cleavage of mRNA by ribonuclease L.
- Dicer The presence of long dsRNAs in cells stimulates the activity of a ribonuclease III enzyme refe ⁇ ed to as Dicer.
- Dicer is involved in the processing of the dsRNA into short pieces of dsRNA known as short interfering RNAs (siRNAs) (Berstein et al, 2001, Nature, 409, 363).
- Short interfering RNAs derived from Dicer activity are typically about 21 to about 23 nucleotides in length and comprise about 19 base pair duplexes.
- Dicer has also been implicated in the excision of 21- and 22-nucleotide small temporal RNAs (stRNAs) from precursor RNA of conserved structure that are implicated in translational control (Hutvagner et al, 2001, Science, 293, 834).
- the RNAi response also features an endonuclease complex containing a siRNA, commonly refened to as an RNA-induced silencing complex (RISC), which mediates cleavage of single-stranded RNA having sequence homologous to the siRNA. Cleavage of the target RNA takes place in the middle of the region complementary to the guide sequence of the siRNA duplex (Elbashir et al, 2001, Genes Dev., 15, 188).
- RISC RNA-induced silencing complex
- RNA interference can also involve small RNA (e.g., micro-RNA or miRNA) mediated gene silencing, presumably though cellular mechanisms that regulate chromatin structure and thereby prevent transcription of target gene sequences (see for example Allshire, 2002, Science, 297, 1818-1819; Volpe et al, 2002, Science, 297, 1833-1837; Jenuwein, 2002, Science, 297, 2215-2218; and Hall et al, 2002, Science, 297, 2232-2237).
- siNA molecules of the invention can be used to mediate gene silencing via interaction with RNA transcripts or alternately by interaction with particular gene sequences, wherein such interaction results in gene silencing either at the transcriptional level or post-transcriptional level.
- RNAi has been studied in a variety of systems. Fire et al, 1998, Nature, 391, 806, were the first to observe RNAi in C. elegans. Wianny and Goetz, 1999, Nature Cell Biol, 2, 70, describe RNAi mediated by dsRNA in mouse embryos. Hammond et al, 2000, Nature, 404, 293, describe RNAi in Drosophila cells transfected with dsRNA. Elbashir et al, 2001, Nature, 411, 494, describe RNAi induced by introduction of duplexes of synthetic 21 -nucleotide RNAs in cultured mammalian cells including human embryonic kidney and HeLa cells.
- small nucleic acid motifs refers to nucleic acid motifs no more than 100 nucleotides in length, preferably no more than 80 nucleotides in length, and most preferably no more than 50 nucleotides in length; e.g., individual siNA oligonucleotide sequences or siNA sequences synthesized in tandem) are preferably used for exogenous delivery.
- the simple stmcture of these molecules increases the ability of the nucleic acid to invade targeted regions of protein and/or RNA structure.
- Exemplary molecules of the instant invention are chemically synthesized, and others can similarly be synthesized.
- Oligonucleotides are synthesized using protocols known in the art, for example as described in Caruthers et al, 1992, Methods in Enzymology 211, 3-19, Thompson et al, hitemational PCT Publication No. WO 99/54459, Wincott et al, 1995, Nucleic Acids Res. 23, 2677-2684, Wincott et al, 1997, Methods Mol. Bio., 74, 59, Brennan et al, 1998, Biotechnol Bioeng., 61, 33-45, and Brennan, U.S. Pat. No.
- oligonucleotides make use of common nucleic acid protecting and coupling groups, such as dimethoxytrityl at the 5'-end, and phosphoramidites at the 3'-end.
- small scale syntheses are conducted on a 394 Applied Biosystems, Inc. synthesizer using a 0.2 ⁇ mol scale protocol with a 2.5 min coupling step for 2'-O- methylated nucleotides and a 45 sec coupling step for 2'-deoxy nucleotides or 2'-deoxy-2'- fluoro nucleotides.
- Table II outlines the amounts and the contact times of the reagents used in the synthesis cycle.
- syntheses at the 0.2 ⁇ mol scale can be performed on a 96-well plate synthesizer, such as the instrument produced by Protogene (Palo Alto, CA) with minimal modification to the cycle.
- synthesizer include the following: detritylation solution is 3% TCA in methylene chloride (ABI); capping is performed with 16% N-methyl imidazole in THF (ABI) and 10% acetic anhydride/10%) 2,6-lutidine in THF (ABI); and oxidation solution is 16.9 mM I , 49 mM pyridine, 9% water in THF (PERSEPTIVETM). Burdick & Jackson Synthesis Grade acetonitrile is used directly from the reagent bottle. S-Ethyltetrazole solution (0.25 M in acetonitrile) is made up from the solid obtained from American International Chemical, Inc. Alternately, for the introduction of phosphorothioate linkages, Beaucage reagent (3H-l,2-Benzodithiol-3-one 1,1-dioxide, 0.05 M in acetonitrile) is used.
- Deprotection of the DNA-based oligonucleotides is performed as follows: the polymer-bound trityl-on oligoribonucleotide is transfe ⁇ ed to a 4 mL glass screw top vial and suspended in a solution of 40% aq. methylamine (1 mL) at 65 °C for 10 min. After cooling to -20 °C, the supernatant is removed from the polymer support. The support is washed three times with 1.0 mL of EtOH:MeCN:H2O/3 : 1 : 1 , vortexed and the supematant is then added to the first supematant. The combined supernatants, containing the oligoribonucleotide, are dried to a white powder.
- RNA including certain siNA molecules of the invention follows the procedure as described in Usman et al, 1987, J. Am. Chem. Soc, 109, 7845; Scaringe et al, 1990, Nucleic Acids Res., 18, 5433; and Wincott et al, 1995,
- small scale syntheses are conducted on a 394 Applied Biosystems, Inc. synthesizer using a 0.2 ⁇ mol scale protocol with a 7.5 min coupling step for alkylsilyl protected nucleotides and a 2.5 min coupling step for 2'-O-methylated nucleotides.
- Table II outlines the amounts and the contact times of the reagents used in the synthesis cycle.
- syntheses at the 0.2 ⁇ mol scale can be done on a 96-well plate synthesizer, such as the instrument produced by Protogene (Palo Alto, CA) with minimal modification to the cycle.
- Average coupling yields on the 394 Applied Biosystems, h e. synthesizer, determined by colorimetric quantitation of the trityl fractions, are typically 97.5-99%.
- synthesizer include the following: detritylation solution is 3% TCA in methylene chloride (ABI); capping is performed with 16% N-methyl imidazole in THF (ABI) and 10% acetic anhydride/10% 2,6-lutidine in THF (ABI); oxidation solution is 16.9 mM I2, 49 mM pyridine, 9% water in THF (PERSEPTINETM). Burdick & Jackson Synthesis Grade acetonitrile is used directly from the reagent bottle. S-Ethyltetrazole solution (0.25 M in acetonitrile) is made up from the solid obtained from American International Chemical, Inc. Alternately, for the introduction of phosphorothioate linkages, Beaucage reagent (3H-l,2-Benzodithiol-3-one l,l-dioxide0.05 M in acetonitrile) is used.
- Deprotection of the R ⁇ A is performed using either a two-pot or one-pot protocol.
- the polymer-bound trityl-on oligoribonucleotide is transferred to a 4 mL glass screw top vial and suspended in a solution of 40% aq. methylamine (1 mL) at 65 °C for 10 min. After cooling to -20 °C, the supematant is removed from the polymer support. The support is washed three times with 1.0 mL of EtOH:MeC ⁇ :H2O/3:l:l, vortexed and the supematant is then added to the first supematant. The combined supematants, containing the oligoribonucleotide, are dried to a white powder.
- the base deprotected oligoribonucleotide is resuspended in anhydrous TEA/HF/NMP solution (300 ⁇ L of a solution of 1.5 mL N-methylpy ⁇ olidinone, 750 ⁇ L TEA and 1 mL TEA-3HF to provide a 1.4 M HF concentration) and heated to 65 °C. After 1.5 h, the oligomer is quenched with 1.5 M NH4HCO3.
- the polymer-bound trityl-on oligoribonucleotide is transfe ⁇ ed to a 4 mL glass screw top vial and suspended in a solution of 33% ethanolic methylamine/DMSO: 1/1 (0.8 mL) at 65 °C for 15 min.
- the vial is brought to rt.
- TEA-3HF 0.1 mL is added and the vial is heated at 65 °C for 15 min.
- the sample is cooled at -20 °C and then quenched with 1.5 M NH4HCO3.
- the quenched NH4HCO3 solution is loaded onto a C-18 containing cartridge that had been prewashed with acetonitrile followed by 50 mM TEAA. After washing the loaded cartridge with water, the RNA is detritylated with 0.5% TFA for 13 min. The cartridge is then washed again with water, salt exchanged with 1 M NaCl and washed with water again. The oligonucleotide is then eluted with 30% acetonitrile.
- the average stepwise coupling yields are typically >98% (Wincott et al, 1995
- nucleic acid molecules of the present invention can be synthesized separately and joined together post-synthetically, for example, by ligation (Moore et al, 1992, Science 256, 9923; Draper et al, International PCT publication No. WO 93/23569; Shabarova et al, 1991, Nucleic Acids Research 19, 4247; Bellon et al, 1997, Nucleosides & Nucleotides, 16, 951; Bellon et al, 1997, Bioconjugate Chem. 8, 204), or by hybridization following synthesis and/or deprotection.
- siNA molecules of the invention can also be synthesized via a tandem synthesis methodology as described in Example 1 herein, wherein both siNA strands are synthesized as a single contiguous oligonucleotide fragment or strand separated by a cleavable linker which is subsequently cleaved to provide separate siNA fragments or strands that hybridize and permit purification of the siNA duplex.
- the linker can be a polynucleotide linker or a non-nucleotide linker.
- the tandem synthesis of siNA as described herein can be readily adapted to both multiwell/multiplate synthesis platforms such as 96 well or similarly larger multi-well platforms.
- the tandem synthesis of siNA as described herein can also be readily adapted to large scale synthesis platforms employing batch reactors, synthesis columns and the like.
- a siNA molecule can also be assembled from two distinct nucleic acid strands or fragments wherein one fragment includes the sense region and the second fragment includes the antisense region of the RNA molecule.
- nucleic acid molecules of the present invention can be modified extensively to enhance stability by modification with nuclease resistant groups, for example, 2'-amino, 2'-C-allyl, 2'-fluoro, 2'-O-methyl, 2'-H (for a review see Usman and Cedergren, 1992, TIBS 17, 34; Usman et al, 1994, Nucleic Acids Symp. Ser. 31, 163).
- siNA constmcts can be purified by gel electrophoresis using general methods or can be purified by high pressure liquid chromatography (HPLC; see Wincott et al, supra, the totality of which is hereby incorporated herein by reference) and re-suspended in water.
- siNA molecules of the invention are expressed from transcription units inserted into DNA or RNA vectors.
- the recombinant vectors can be DNA plasmids or viral vectors.
- siNA expressing viral vectors can be constmcted based on, but not limited to, adeno-associated virus, retrovirus, adenovims, or alphaviras.
- the recombmant vectors capable of expressing the siNA molecules can be delivered as described herein, and persist in target cells. Altematively, viral vectors can be used that provide for transient expression of siNA molecules.
- nucleic acid molecules with modifications can prevent their degradation by serum ribonucleases, which can increase their potency (see e.g., Eckstein et al, hitemational Publication No. WO 92/07065; Pe ⁇ ault et al, 1990 Nature 344, 565; Pieken et al, 1991, Science 253, 314; Usman and Cedergren, 1992, T ⁇ -ends in Biochem. Sci. 17, 334; Usman et al, hitemational Publication No. WO 93/15187; and Rossi et al, hitemational Publication No. WO 91/03162; Sproat, U.S. Pat. No.
- oligonucleotides are modified to enhance stability and/or enhance biological activity by modification with nuclease resistant groups, for example, 2'-amino, 2'-C-allyl, 2'-fluoro, 2'-O-methyl, 2'-O- allyl, 2'-H, nucleotide base modifications (for a review see Usman and Cedergren, 1992, TIBS. 17, 34; Usman et al, 1994, Nucleic Acids Symp. Ser. 31, 163; Burgin et al, 1996, Biochemistry, 35, 14090).
- nuclease resistant groups for example, 2'-amino, 2'-C-allyl, 2'-fluoro, 2'-O-methyl, 2'-O- allyl, 2'-H, nucleotide base modifications (for a review see Usman and Cedergren, 1992, TIBS. 17, 34; Usman et al, 1994, Nucleic Acids Symp. Ser. 31, 163; Burgin
- Short interfering nucleic acid (siNA) molecules having chemical modifications that maintain or enhance activity are provided.
- Such a nucleic acid is also generally more resistant to nucleases than an unmodified nucleic acid. Accordingly, the in vitro and/or in vivo activity should not be significantly lowered.
- therapeutic nucleic acid molecules delivered exogenously should optimally be stable within cells until translation of the target RNA has been modulated long enough to reduce the levels of the undesirable protein. This period of time varies between hours to days depending upon the disease state. Improvements in the chemical synthesis of RNA and DNA (Wincott et al, 1995, Nucleic Acids Res.
- nucleic acid molecules of the invention include one or more
- G-clamp nucleotides e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more G-clamp nucleotides.
- a G-clamp nucleotide is a modified cytosine analog wherein the modifications confer the ability to hydrogen bond both Watson-Crick and Hoogsteen faces of a complementary guanine within a duplex, see for example Lin and Matteucci, 1998, J. Am. Chem. Soc, 120, 8531- 8532.
- a single G-clamp analog substitution within an oligonucleotide can result in substantially enhanced helical thermal stability and mismatch discrimination when hybridized to complementary oligonucleotides.
- nucleic acid molecules of the invention include one or more (e.g., about 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, or more) LNA "locked nucleic acid" nucleotides such as a 2', 4'-C methylene bicyclo nucleotide (see for example Wengel et al, International PCT Publication No. WO 00/66604 and WO 99/14226).
- the invention features conjugates and/or complexes of siNA molecules of the invention.
- conjugates and/or complexes can be used to facilitate delivery of siNA molecules into a biological system, such as a cell.
- the conjugates and complexes provided by the instant invention can impart therapeutic activity by transferring therapeutic compounds across cellular membranes, altering the pharmacokinetics, and/or modulating the localization of nucleic acid molecules of the invention.
- the present invention encompasses the design and synthesis of novel conjugates and complexes for the delivery of molecules, including, but not limited to, small molecules, lipids, phospholipids, nucleosides, nucleotides, nucleic acids, antibodies, toxins, negatively charged polymers and other polymers, for example proteins, peptides, hormones, carbohydrates, polyethylene glycols, or polyamines, across cellular membranes.
- molecules including, but not limited to, small molecules, lipids, phospholipids, nucleosides, nucleotides, nucleic acids, antibodies, toxins, negatively charged polymers and other polymers, for example proteins, peptides, hormones, carbohydrates, polyethylene glycols, or polyamines, across cellular membranes.
- the transporters described are designed to be used either individually or as part of a multi-component system, with or without degradable linkers.
- Conjugates of the molecules described herein can be attached to biologically active molecules via linkers that are biodegradable, such as biodegradable nucleic acid linker molecules.
- biodegradable linker refers to a nucleic acid or non- nucleic acid linker molecule that is designed as a biodegradable linker to connect one molecule to another molecule, for example, a biologically active molecule to a siNA molecule of the invention or the sense and antisense strands of a siNA molecule of the invention.
- the biodegradable linker is designed such that its stability can be modulated for a particular purpose, such as delivery to a particular tissue or cell type.
- the stability of a nucleic acid-based biodegradable linker molecule can be modulated by using various chemistries, for example combinations of ribonucleotides, deoxyribonucleotides, and chemically-modified nucleotides, such as 2'-O-methyl, 2'-fluoro, 2'-amino, 2'-O-amino, 2'-C-allyl, 2'-O-allyl, and other 2'-modified or base modified nucleotides.
- the biodegradable nucleic acid linker molecule can be a dimer, trimer, tetramer or longer nucleic acid molecule, for example, an oligonucleotide of about 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, or 20 nucleotides in length, or can comprise a single nucleotide with a phosphorus-based linkage, for example, a phosphoramidate or phosphodiester linkage.
- the biodegradable nucleic acid linker molecule can also comprise nucleic acid backbone, nucleic acid sugar, or nucleic acid base modifications.
- biodegradable refers to degradation in a biological system, for example enzymatic degradation or chemical degradation.
- biologically active molecule refers to compounds or molecules that are capable of eliciting or modifying a biological response in a system.
- biologically active siNA molecules either alone or in combination with other molecules contemplated by the instant invention include therapeutically active molecules such as antibodies, hormones, antivirals, peptides, proteins, chemotherapeutics, small molecules, vitamins, co-factors, nucleosides, nucleotides, oligonucleotides, enzymatic nucleic acids, antisense nucleic acids, triplex forming oligonucleotides, 2,5-A chimeras, siNA, dsRNA, allozymes, aptamers, decoys and analogs thereof.
- Biologically active molecules of the invention also include molecules capable of modulating the pharmacokinetics and/or pha ⁇ nacodynamics of other biologically active molecules, for example, lipids and polymers such as polyamines, polyamides, polyethylene glycol and other polyethers.
- phospholipid refers to a hydrophobic molecule comprising at least one phosphorus group.
- a phospholipid can comprise a phosphorus-containing group and saturated or unsaturated alkyl group, optionally substituted with OH, COOH, oxo, amine, or substituted or unsubstituted aryl groups.
- nucleic acid molecules delivered exogenously optimally are stable within cells until reverse transcription of the RNA has been modulated long enough to reduce the levels of the RNA transcript.
- the nucleic acid molecules are resistant to nucleases in order to function as effective intracellular therapeutic agents. Improvements in the chemical synthesis of nucleic acid molecules described in the instant invention and in the art have expanded the ability to modify nucleic acid molecules by introducing nucleotide modifications to enhance their nuclease stability as described above.
- siNA molecules having chemical modifications that maintain or enhance enzymatic activity of proteins involved in RNAi are provided.
- Such nucleic acids are also generally more resistant to nucleases than unmodified nucleic acids. Thus, in vitro and/or in vivo the activity should not be significantly lowered.
- nucleic acid-based molecules of the mvention will lead to better treatment of the disease progression by affording the possibility of combination therapies (e.g., multiple siNA molecules targeted to different genes; nucleic acid molecules coupled with known small molecule modulators; or intermittent treatment with combinations of molecules, including different motifs and/or other chemical or biological molecules).
- combination therapies e.g., multiple siNA molecules targeted to different genes; nucleic acid molecules coupled with known small molecule modulators; or intermittent treatment with combinations of molecules, including different motifs and/or other chemical or biological molecules.
- the treatment of subjects with siNA molecules can also include combinations of different types of nucleic acid molecules, such as enzymatic nucleic acid molecules (ribozymes), allozymes, antisense, 2,5-A oligoadenylate, decoys, and aptamers.
- ribozymes enzymatic nucleic acid molecules
- allozymes antisense
- 2,5-A oligoadenylate 2,5-A oligoadeny
- a siNA molecule of the invention comprises one or more 5' and/or a 3'- cap stmcture, for example on only the sense siNA strand, the antisense siNA strand, or both siNA strands.
- cap structure is meant chemical modifications, which have been incorporated at either terminus of the oligonucleotide (see, for example, Adamic et al, U.S. Pat. No. 5,998,203, incorporated by reference herein). These terminal modifications protect the nucleic acid molecule from exonuclease degradation, and may help in delivery and/or localization within a cell.
- the cap may be present at the 5 '-terminus (5 '-cap) or at the 3'- terminal (3'-cap) or may be present on both termini.
- the 5'-cap is selected from the group consisting of glyceryl, inverted deoxy abasic residue (moiety); 4', 5 '-methylene nucleotide; l-(beta-D-erythrofuranosyl) nucleotide, 4'-thio nucleotide; carbocyclic nucleotide; 1,5-anhydrohexitol nucleotide; L-nucleotides; alpha-nucleotides; modified base nucleotide; phosphorodithioate linkage; t/jreo-pentofuranosyl nucleotide; acyclic 3',4'-seco nucleotide; acyclic 3,4-dihydroxybutyl nucleotide; acyclic 3,5- dihydroxypentyl nucleotide, 3 '-3 '-inverted nucleotide moiety; 3 '-3 '-inverted a
- the 3 '-cap is selected from the group consisting of glyceryl, inverted deoxy abasic residue (moiety), 4', 5 '-methylene nucleotide; l-(beta-D- erythrofuranosyl) nucleotide; 4'-thio nucleotide, carbocyclic nucleotide; 5'-amino-alkyl phosphate; l,3-diamino-2-propyl phosphate; 3-aminopropyl phosphate; 6-aminohexyl phosphate; 1,2-aminododecyl phosphate; hydroxypropyl phosphate; 1,5-anhydrohexitol nucleotide; L-nucleotide; alpha-nucleotide; modified base nucleotide; phosphorodithioate; t/jreo-pentofuranosyl nucleotide; acycl
- non-nucleotide any group or compound which can be incorporated into a nucleic acid chain in the place of one or more nucleotide units, including either sugar and/or phosphate substitutions, and allows the remaining bases to exhibit their enzymatic activity.
- the group or compound is abasic in that it does not contain a commonly recognized nucleotide base, such as adenosine, guanine, cytosine, uracil or thymine and therefore lacks a base at the 1 '-position.
- alkyl refers to a saturated aliphatic hydrocarbon, including straight- chain, branched-chain, and cyclic alkyl groups.
- the alkyl group has 1 to 12 carbons. More preferably, it is a lower alkyl of from 1 to 7 carbons, more preferably 1 to 4 carbons.
- the term also includes alkenyl groups that are unsaturated hydrocarbon groups containing at least one carbon-carbon double bond, including straight-chain, branched-chain, and cyclic groups.
- the alkenyl group has 1 to 12 carbons. More preferably, it is a lower alkenyl of from 1 to 7 carbons, more preferably 1 to 4 carbons.
- alkyl also includes alkynyl groups that have an unsaturated hydrocarbon group containing at least one carbon-carbon triple bond, including straight-chain, branched-chain, and cyclic groups. Preferably, the alkynyl group has 1 to 12 carbons.
- the alkynyl group may be substituted or unsubstituted.
- alkyl groups can also include aryl, alkylaryl, carbocyclic aryl, heterocychc aryl, amide and ester groups.
- An "aryl” group refers to an aromatic group that has at least one ring having a conjugated pi electron system and includes carbocyclic aryl, heterocychc aryl and biaryl groups, all of which may be optionally substituted.
- the prefe ⁇ ed substituent(s) of aryl groups are halogen, trihalomethyl, hydroxyl, SH, OH, cyano, alkoxy, alkyl, alkenyl, alkynyl, and amino groups.
- alkylaryl refers to an alkyl group (as described above) covalently joined to an aryl group (as described above).
- Carbocyclic aryl groups are groups wherein the ring atoms on the aromatic ring are all carbon atoms. The carbon atoms are optionally substituted.
- Heterocychc aryl groups are groups having from 1 to 3 heteroatoms as ring atoms in the aromatic ring and the remainder of the ring atoms are carbon atoms.
- Suitable heteroatoms include oxygen, sulfur, and nitrogen, and include furanyl, thienyl, pyridyl, pyrrolyl, N-lower alkyl py ⁇ olo, pyrimidyl, pyrazinyl, imidazolyl and the like, all optionally substituted.
- An "amide” refers to an -C(O)-NH-R, where R is either alkyl, aryl, alkylaryl or hydrogen.
- An “ester” refers to an -C(O)-OR', where R is either alkyl, aryl, alkylaryl or hydrogen.
- nucleotide as used herein is as recognized in the art to include natural bases (standard), and modified bases well known in the art. Such bases are generally located at the 1' position of a nucleotide sugar moiety. Nucleotides generally comprise a base, sugar and a phosphate group. The nucleotides can be unmodified or modified at the sugar, phosphate and/or base moiety, (also refe ⁇ ed to interchangeably as nucleotide analogs, modified nucleotides, non-natural nucleotides, non-standard nucleotides and other; see, for example, Usman and McSwiggen, supra; Eckstein et al, International PCT Publication No.
- base modifications that can be introduced into nucleic acid molecules include, inosine, purine, pyridin-4-one, pyridin-2-one, phenyl, pseudouracil, 2, 4, 6-trimethoxy benzene, 3-methyl uracil, dihydrouridine, naphthyl, aminophenyl, 5-alkylcytidines (e.g., 5-methylcytidine), 5-alkyluridines (e.g., ribothymidine), 5-halouridine (e.g., 5-bromouridine) or 6-azapyrimidines or 6-alkylpyrimidines (e.g.
- modified bases in this aspect is meant nucleotide bases other than adenine, guanine, cytosine and uracil at 1' position or their equivalents.
- the invention features modified siNA molecules, with phosphate backbone modifications comprising one or more phosphorothioate, phosphorodithioate, methylphosphonate, phosphotriester, morpholino, amidate carbamate, carboxymethyl, acetamidate, polyamide, sulfonate, sulfonamide, sulfamate, formacetal, thioformacetal, and/or alkylsilyl, substitutions.
- phosphate backbone modifications comprising one or more phosphorothioate, phosphorodithioate, methylphosphonate, phosphotriester, morpholino, amidate carbamate, carboxymethyl, acetamidate, polyamide, sulfonate, sulfonamide, sulfamate, formacetal, thioformacetal, and/or alkylsilyl, substitutions.
- abasic sugar moieties lacking a base or having other chemical groups in place of a base at the 1' position, see for example Adamic et al, U.S. Pat. No. 5,998,203.
- unmodified nucleoside is meant one of the bases adenine, cytosine, guanine, thymine, or uracil joined to the 1' carbon of ⁇ -D-ribo-furanose.
- modified nucleoside is meant any nucleotide base which contains a modification in the chemical structure of an unmodified nucleotide base, sugar and/or phosphate.
- modified nucleotides are shown by Formulae I-NII and/or other modifications described herein.
- amino 2'- ⁇ H 2 or 2'-O- NH 2 , which can be modified or unmodified.
- modified groups are described, for example, in Eckstein et al, U.S. Pat. No. 5,672,695 and Matulic-Adamic et al, U.S. Pat. No. 6,248,878, which are both incorporated by reference in their entireties.
- nucleic acid siNA structure can be made to enhance the utility of these molecules. Such modifications will enhance shelf-life, half-life in vitro, stability, and ease of introduction of such oligonucleotides to the target site, e.g., to enhance penetration of cellular membranes, and confer the ability to recognize and bind to targeted cells.
- a siNA molecule of the invention can be adapted for use to treat any disease, infection or condition associated with gene expression, and other indications that can respond to the level of gene product in a cell or tissue, alone or in combination with other therapies.
- a siNA molecule can comprise a delivery vehicle, including liposomes, for administration to a subject, carriers and diluents and their salts, and/or can be present in pharmaceutically acceptable formulations.
- Methods for the delivery of nucleic acid molecules are described in Akhtar et al, 1992, Trends Cell Bio., 2, 139; Delivery Strategies for Antisense Oligonucleotide Tlierapeutics, ed. Akhtar, 1995, Maurer et al, 1999, Mol Membr.
- Nucleic acid molecules can be administered to cells by a variety of methods known to those of skill in the art, including, but not restricted to, encapsulation in liposomes, by iontophoresis, or by incorporation into other vehicles, such as hydrogels, cyclodextrins (see for example Gonzalez et al, 1999, Bioconjugate Chem., 10, 1068-1074), biodegradable nanocapsules, and bioadhesive microspheres, or by proteinaceous vectors (O'Hare and Normand, International PCT Publication No. WO 00/53722). Altematively, the nucleic acid/vehicle combination is locally delivered by direct injection or by use of an infusion pump.
- phrases of the instant invention can be used as pharmaceutical agents.
- Pha ⁇ naceutical agents prevent, modulate the occu ⁇ ence, or treat (alleviate a symptom to some extent, preferably all of the symptoms) of a disease state in a subject.
- the invention features the use of methods to deliver the nucleic acid molecules of the instant invention to hematopoietic cells, including monocytes and lymphocytes. These methods are described in detail by Hartmann et al, 1998, J Phamacol Exp. Ther., 285(2), 920-928; Kronenwett et al, 1998, Blood, 91(3), 852-862; Filion and Phillips, 1997, Biochim. Biophys. Ada., 1329(2), 345-356; Ma and Wei, 1996, Leuk Res., 20(11/12), 925-930; and Bongartz et al, 1994, Nucleic Acids Research, 22(22), 4681-8.
- Such methods include the use of free oligonucleitide, cationic lipid formulations, liposome formulations including pH sensitive liposomes and immunoliposomes, and bioconjugates including oligonucleotides conjugated to fusogenic peptides, for the transfection of hematopoietic cells with oligonucleotides.
- the invention features a pharmaceutical composition
- a pharmaceutical composition comprising one or more nucleic acid(s) of the invention in an acceptable carrier, such as a stabilizer, buffer, and the like.
- the polynucleotides of the invention can be administered (e.g., RNA, DNA or protein) and introduced into a subject by any standard means, with or without stabilizers, buffers, and the hke, to form a pharmaceutical composition.
- standard protocols for formation of liposomes can be followed.
- the compositions of the present invention can also be formulated and used as tablets, capsules or elixirs for oral administration, suppositories for rectal administration, sterile solutions, suspensions for injectable administration, and the other compositions known in the art.
- the present invention also includes pharmaceutically acceptable formulations of the compounds described.
- formulations include salts of the above compounds, e.g., acid addition salts, for example, salts of hydrochloric, hydrobromic, acetic acid, and benzene sulfonic acid.
- a pharmacological composition or formulation refers to a composition or formulation in a form suitable for administration, e.g., systemic administration, into a cell or subject, including for example a human. Suitable forms, in part, depend upon the use or the route of entry, for example oral, transdermal, or by injection. Such forms should not prevent the composition or formulation from reaching a target cell (i.e., a cell to which the negatively charged nucleic acid is desirable for delivery). For example, pharmacological compositions injected into the blood stream should be soluble. Other factors are known in the art, and include considerations such as toxicity and forms that prevent the composition or formulation from exerting its effect.
- systemic administration in vivo systemic absorption or accumulation of drugs in the blood stream followed by distribution throughout the entire body.
- Administration routes that lead to systemic absorption include, without limitation: intravenous, subcutaneous, intraperitoneal, inhalation, oral, intrapulmonary and intramuscular.
- Each of these administration routes exposes the siNA molecules of the invention to an accessible diseased tissue.
- the rate of entry of a drag into the circulation has been shown to be a function of molecular weight or size.
- the use of a liposome or other drag carrier comprising the compounds of the instant invention can potentially localize the drug, for example, in certain tissue types, such as the tissues of the reticular endothelial system (RES).
- RES reticular endothelial system
- a liposome formulation that can facilitate the association of drug with the surface of cells, such as, lymphocytes and macrophages is also useful. This approach can provide enhanced delivery of the drag to target cells by taking advantage of the specificity of macrophage and lymphocyte immune recognition of abnormal cells, such as cancer cells.
- pharmaceutically acceptable formulation is meant, a composition or formulation that allows for the effective distribution of the nucleic acid molecules of the instant invention in the physical location most suitable for their desired activity.
- agents suitable for formulation with the nucleic acid molecules of the instant invention include: P-glycoprotein inhibitors (such as Pluronic P85), which can enhance entry of drags into the CNS (Jolliet-Riant and Tillement, 1999, Fundam. Clin.
- biodegradable polymers such as poly (DL-lactide-coglycolide) microspheres for sustained release delivery after intracerebral implantation (Emerich, DF et al, 1999, Cell Transplant, 8, 47-58) (Alkermes, Inc. Cambridge, MA); and loaded nanoparticles, such as those made of polybutylcyanoacrylate, which can deliver drugs across the blood brain barrier and can alter neuronal uptake mechanisms (Prog Neuropsychopharmacol Biol Psychiati ⁇ , 23, 941-949, 1999).
- Other non-limiting examples of delivery strategies for the nucleic acid molecules of the instant invention include material described in Boado et al, 1998, J. Pharm.
- the mvention also features the use of the composition comprising surface-modified liposomes containing poly (ethylene glycol) lipids (PEG-modified, or long-circulating liposomes or stealth liposomes).
- PEG-modified, or long-circulating liposomes or stealth liposomes These formulations offer a method for increasing the accumulation of drags in target tissues.
- This class of drag carriers resists opsonization and elimination by the mononuclear phagocytic system (MPS or RES), thereby enabling longer blood circulation times and enhanced tissue exposure for the encapsulated drag (Lasic et al. Chem. Rev. 1995, 95, 2601-2627; lshiwata et al, Chem. Pharm. Bull 1995, 43, 1005-1011).
- liposomes have been shown to accumulate selectively in tumors, presumably by extravasation and capture in the neovascularized target tissues (Lasic et al, Science 1995, 267, 1275-1276; Oku et /., 1995, Biochim. Biophys. Ada, 1238, 86- 90).
- the long-circulating liposomes enhance the pharmacokinetics and pharmacodynamics of DNA and RNA, particularly compared to conventional cationic liposomes which are known to accumulate in tissues of the MPS (Liu et al, J. Biol Chem. 1995, 42, 24864-24870; Choi et al, hitemational PCT Pubhcation No.
- WO 96/10391 Ansell et al, International PCT Publication No. WO 96/10390; Holland et al, International PCT Publication No. WO 96/10392).
- Long-circulating liposomes are also likely to protect drags from nuclease degradation to a greater extent compared to cationic liposomes, based on their ability to avoid accumulation in metabolically aggressive MPS tissues such as the liver and spleen.
- compositions prepared for storage or administration that include a pharmaceutically effective amount of the desired compounds in a pharmaceutically acceptable carrier or diluent.
- Acceptable carriers or diluents for therapeutic use are well known in the pha ⁇ naceutical art, and are described, for example, in Remington's Pharmaceutical Sciences, Mack Publishing Co. (A.R. Gennaro edit. 1985), hereby incorporated by reference herein.
- preservatives, stabilizers, dyes and flavoring agents can be provided. These include sodium benzoate, sorbic acid and esters of y ⁇ -hydroxybenzoic acid.
- antioxidants and suspending agents can be used.
- a pharmaceutically effective dose is that dose required to prevent, inhibit the occurrence, or treat (alleviate a symptom to some extent, preferably all of the symptoms) of a disease state.
- the pharmaceutically effective dose depends on the type of disease, the composition used, the route of administration, the type of mammal being treated, the physical characteristics of the specific mammal under consideration, concu ⁇ ent medication, and other factors that those skilled in the medical arts will recognize. Generally, an amount between 0.1 mg/kg and 100 mg/kg body weight/day of active ingredients is administered dependent upon potency of the negatively charged polymer.
- nucleic acid molecules of the mvention and formulations thereof can be administered orally, topically, parenterally, by inhalation or spray, or rectally in dosage unit formulations containing conventional non-toxic pharmaceutically acceptable carriers, adjuvants and/or vehicles.
- parenteral as used herein includes percutaneous, subcutaneous, intravascular (e.g., intravenous), intramuscular, or intrathecal injection or infusion techniques and the like.
- a pha ⁇ naceutical formulation comprising a nucleic acid molecule of the invention and a pharmaceutically acceptable carrier.
- nucleic acid molecules of the invention can be present in association with one or more non-toxic pharmaceutically acceptable carriers and/or diluents and/or adjuvants, and if desired other active ingredients.
- the pharmaceutical compositions containing nucleic acid molecules of the invention can be in a form suitable for oral use, for example, as tablets, troches, lozenges, aqueous or oily suspensions, dispersible powders or granules, emulsion, hard or soft capsules, or syrups or elixirs.
- compositions intended for oral use can be prepared according to any method known to the art for the manufacture of pharmaceutical compositions and such compositions can contain one or more such sweetening agents, flavoring agents, coloring agents or preservative agents in order to provide pharmaceutically elegant and palatable preparations.
- Tablets contain the active ingredient in admixture with non-toxic pharmaceutically acceptable excipients that are suitable for the manufacture of tablets.
- excipients can be, for example, inert diluents; such as calcium carbonate, sodium carbonate, lactose, calcium phosphate or sodium phosphate; granulating and disintegrating agents, for example, corn starch, or alginic acid; binding agents, for example starch, gelatin or acacia; and lubricating agents, for example magnesium stearate, stearic acid or talc.
- the tablets can be uncoated or they can be coated by known techniques. In some cases such coatings can be prepared by known techniques to delay disintegration and absorption in the gastrointestinal tract and thereby provide a sustained action over a longer period.
- a time delay material such as glyceryl monosterate or glyceryl distearate can be employed.
- Formulations for oral use can also be presented as hard gelatin capsules wherein the active ingredient is mixed with an inert solid diluent, for example, calcium carbonate, calcium phosphate or kaolin, or as soft gelatin capsules wherein the active ingredient is mixed with water or an oil medium, for example peanut oil, liquid paraffin or olive oil.
- an inert solid diluent for example, calcium carbonate, calcium phosphate or kaolin
- water or an oil medium for example peanut oil, liquid paraffin or olive oil.
- Aqueous suspensions contain the active materials in a mixture with excipients suitable for the manufacture of aqueous suspensions.
- excipients are suspending agents, for example sodium carboxymethylcellulose, methylcellulose, hydropropyl- methylcellulose, sodium alginate, polyvinylpy ⁇ olidone, gum tragacanth and gum acacia; dispersing or wetting agents can be a naturally-occurring phosphatide, for example, lecithin, or condensation products of an alkylene oxide with fatty acids, for example polyoxyethylene stearate, or condensation products of ethylene oxide with long chain aliphatic alcohols, for example heptadecaethyleneoxycetanol, or condensation products of ethylene oxide with partial esters derived from fatty acids and a hexitol such as polyoxyethylene sorbitol monooleate, or condensation products of ethylene oxide with partial esters derived from fatty acids and hexitol anhydrides, for example polyethylene sorbitan monoo
- the aqueous suspensions can also contain one or more preservatives, for example ethyl, or n-propyl p-hydroxybenzoate, one or more coloring agents, one or more flavoring agents, and one or more sweetening agents, such as sucrose or saccharin.
- preservatives for example ethyl, or n-propyl p-hydroxybenzoate
- coloring agents for example ethyl, or n-propyl p-hydroxybenzoate
- flavoring agents for example ethyl, or n-propyl p-hydroxybenzoate
- sweetening agents such as sucrose or saccharin.
- Oily suspensions can be formulated by suspending the active ingredients in a vegetable oil, for example arachis oil, olive oil, sesame oil or coconut oil, or in a mineral oil such as liquid paraffin.
- the oily suspensions can contain a thickening agent, for example beeswax, hard paraffin or cetyl alcohol.
- Sweetening agents and flavoring agents can be added to provide palatable oral preparations.
- These compositions can be preserved by the addition of an anti-oxidant such as ascorbic acid
- Dispersible powders and granules suitable for preparation of an aqueous suspension by the addition of water provide the active ingredient in admixture with a dispersing or wetting agent, suspending agent and one or more preservatives.
- a dispersing or wetting agent e.g., glycerol, glycerol, glycerol, glycerol, glycerol, glycerol, glycerin, glycerin, glycerin, glycerin, glycerin, sorbitol, sorbitol, sorbitol, sorbitol, sorbitol, sorbitol, sorbitol, sorbitol, sorbitol, sorbitol, glycerol, glycerol, glycerol, glycerol, glycerol, glycerol, glycerol, glycerol, glycerol
- compositions of the invention can also be in the form of oil-in- water emulsions.
- the oily phase can be a vegetable oil or a mineral oil or mixtures of these.
- Suitable emulsifying agents can be naturally-occurring gums, for example gum acacia or gum tragacanth, naturally-occurring phosphatides, for example soy bean, lecithin, and esters or partial esters derived from fatty acids and hexitol, anhydrides, for example sorbitan monooleate, and condensation products of the said partial esters with ethylene oxide, for example polyoxyethylene sorbitan monooleate.
- the emulsions can also contain sweetening and flavoring agents.
- Syrups and elixirs can be formulated with sweetening agents, for example glycerol, propylene glycol, sorbitol, glucose or sucrose. Such fonnulations can also contain a demulcent, a preservative and flavoring and coloring agents.
- the pharmaceutical compositions can be in the form of a sterile injectable aqueous or oleaginous suspension.
- the sterile injectable preparation can also be a sterile injectable solution or suspension in a non-toxic parentally acceptable diluent or solvent, for example as a solution in 1,3- butanediol.
- a non-toxic parentally acceptable diluent or solvent for example as a solution in 1,3- butanediol.
- acceptable vehicles and solvents that can be employed are water, Ringer's solution and isotonic sodium chloride solution, hi addition, sterile, fixed oils are conventionally employed as a solvent or suspending medium.
- any bland fixed oil can be employed including synthetic mono-or diglycerides.
- fatty acids such as oleic acid find use n the preparation of injectables.
- the nucleic acid molecules of the invention can also be administered in the form of suppositories, e.g., for rectal administration of the drag.
- suppositories e.g., for rectal administration of the drag.
- These compositions can be prepared by mixing the drag with a suitable non-irritating excipient that is solid at ordinary temperatures but liquid at the rectal temperature and will therefore melt in the rectum to release the drug.
- suitable non-irritating excipient that is solid at ordinary temperatures but liquid at the rectal temperature and will therefore melt in the rectum to release the drug.
- Such materials include cocoa butter and polyethylene glycols.
- Nucleic acid molecules of the invention can be administered parenterally in a sterile medium.
- the drag depending on the vehicle and concentration used, can either be suspended or dissolved in the vehicle.
- adjuvants such as local anesthetics, preservatives and buffering agents can be dissolved in the vehicle.
- Dosage levels of the order of from about 0.1 mg to about 140 mg per kilogram of body weight per day are useful in the treatment of the above-indicated conditions (about 0.5 mg to about 7 g per subject per day).
- the amount of active ingredient that can be combined with the carrier materials to produce a single dosage form varies depending upon the host treated and the particular mode of administration.
- Dosage unit forms generally contain between from about 1 mg to about 500 mg of an active ingredient.
- the specific dose level for any particular subject depends upon a variety of factors including the activity of the specific compound employed, the age, body weight, general health, sex, diet, time of administration, route of administration, and rate of excretion, drag combination and the severity of the particular disease undergoing therapy.
- the composition can also be added to the animal feed or drinking water. It can be convenient to formulate the animal feed and drinking water compositions so that the animal takes in a therapeutically appropriate quantity of the composition along with its diet. It can also be convenient to present the composition as a premix for addition to the feed or drinking water.
- nucleic acid molecules of the present invention can also be administered to a subject in combination with other therapeutic compounds to increase the overall therapeutic effect.
- the use of multiple compounds to treat an indication can increase the beneficial effects while reducing the presence of side effects.
- the invention comprises compositions suitable for administering nucleic acid molecules of the invention to specific cell types.
- ASGPr asialoglycoprotein receptor
- ASOR asialoorosomucoid
- the folate receptor is overexpressed in many cancer cells.
- Binding of such glycoproteins, synthetic glycoconjugates, or folates to the receptor takes place with an affinity that strongly depends on the degree of branching of the oligosaccharide chain, for example, triatennary stractures are bound with greater affinity than biatena ⁇ y or monoatennary chains (Baenziger and Fiete, 1980, Cell, 22, 611-620; Connolly et al, 1982, J. Biol. Chem., 257, 939-945).
- bioconjugates can also provide a reduction in the required dose of therapeutic compounds required for treatment. Furthermore, therapeutic bioavialability, pharmacodynamics, and pharmacokinetic parameters can be modulated through the use of nucleic acid bioconjugates of the invention. Non-limiting examples of such bioconjugates are described in Vargeese et al, USSN 10/201,394, filed August 13, 2001; and Matulic-Adamic et al, USSN 60/362,016, filed March 6, 2002.
- siNA molecules of the invention are synthesized in tandem using a cleavable linker, for example, a succinyl-based linker. Tandem synthesis as described herein is followed by a one-step purification process that provides RNAi molecules in high yield. This approach is highly amenable to siNA synthesis in support of high throughput RNAi screening, and can be readily adapted to multi-column or multi-well synthesis platforms.
- a cleavable linker for example, a succinyl-based linker.
- the oligonucleotides are deprotected as described above. Following deprotection, the siNA sequence strands are allowed to spontaneously hybridize. This hybridization yields a duplex in which one strand has retained the 5'-O-DMT group while the complementary strand comprises a terminal 5'-hydroxyl. The newly formed duplex behaves as a single molecule during routine solid-phase extraction purification (Trityl-On purification) even though only one molecule has a dimethoxytrityl group.
- this dimethoxytrityl group (or an equivalent group, such as other trityl groups or other hydrophobic moieties) is all that is required to purify the pair of oligos, for example, by using a C 18 cartridge.
- Standard phosphoramidite synthesis chemistry is used up to the point of introducing a tandem linker, such as an inverted deoxy abasic succinate or glyceryl succinate linker (see Figure 1) or an equivalent cleavable linker.
- a tandem linker such as an inverted deoxy abasic succinate or glyceryl succinate linker (see Figure 1) or an equivalent cleavable linker.
- linker coupling conditions includes a hindered base such as diisopropylethylamme (DIP A) and/or DMAP in the presence of an activator reagent such as Bromotripynolidinophosphoniumhexaflurorophosphate (PyBrOP).
- DIP A diisopropylethylamme
- PyBrOP Bromotripynolidinophosphoniumhexaflurorophosphate
- the resulting oligonucleotide is deprotected according to the procedures described herein and quenched with a suitable buffer, for example with 50mM NaOAc or 1.5M NH4H2CO3.
- a suitable buffer for example with 50mM NaOAc or 1.5M NH4H2CO3.
- Purification of the siNA duplex can be readily accomplished using solid phase extraction, for example using a Waters C18 SepPak lg cartridge conditioned with 1 column volume (CV) of acetonitrile, 2 CV H2O, and 2 CV 50mM NaOAc. The sample is loaded and then washed with 1 CV H2O or 50mM NaOAc. Failure sequences are eluted with 1 CV 14% ACN (Aqueous with 50mM NaOAc and 50mM NaCl).
- the column is then washed, for example with 1 CV H2O followed by on-column detritylation, for example by passing 1 CV of 1% aqueous trifluoroacetic acid (TFA) over the column, then adding a second CV of 1% aqueous TFA to the column and allowing to stand for approximately 10 minutes.
- TFA trifluoroacetic acid
- the remaining TFA solution is removed and the column washed with H20 followed by 1 CV 1M NaCl and additional H2O.
- the siNA duplex product is then eluted, for example, using 1 CV 20%> aqueous CAN.
- Figure 2 provides an example of MALDI-TOV mass spectrometry analysis of a purified siNA constract in which each peak corresponds to the calculated mass of an individual siNA strand of the siNA duplex.
- the same purified siNA provides three peaks when analyzed by capillary gel electrophoresis (CGE), one peak presumably corresponding to the duplex siNA, and two peaks presumably co ⁇ esponding to the separate siNA sequence strands.
- Ion exchange HPLC analysis of the same siNA contract only shows a single peak.
- Testing of the purified siNA constract using a luciferase reporter assay described below demonstrated the same RNAi activity compared to siNA constracts generated from separately synthesized oligonucleotide sequence strands.
- siNA constracts Chemical modifications were introduced into siNA constracts to determine the stability of these constracts compared to native siNA oligonucleotides (containing two thymidine nucleotide overhangs) in human serum.
- An investigation of the serum stability of RNA duplexes revealed that siNA constmcts consisting of all RNA nucleotides containing two thymidine nucleotide overhangs have a half-life in serum of 15 seconds, whereas chemically modified siNA constmcts remained stable in serum for 1 to 3 days depending on the extent of modification (see Figure 3).
- RNAi stability tests were performed by internally labeling one strand (strand 1) of siNA and duplexing with 1.5 X the concentration of the complementary siNA strand (strand 2) (to insure all labeled material was in duplex form).
- Duplexed siNA constmcts were then tested for stability by incubating at a final concentration of 2 ⁇ M siNA (strand 2 concentration) in 90% mouse or human serum for time-points of 30sec, lmin, 5min, 30min, 90min, 4hrs lOmin, 16hrs 24min, and 49hrs. Time points were run on a 15% denaturing polyacrylamide gels and analyzed on a phosphoimager.
- RNAi was done by adding appropriate concentrations of the siNA oligonucleotides and heating to 95° C for 5minutes followed by slow cooling to room temperature. Reactions were performed by adding 100% serum to the siNA duplexes and incubating at 37° C, then removing aliquots at desired time-points. Results of this study are summarized in Figure 3.
- siNA molecules e.g., SEQ ID NOs: 412/413, 412/414, 412/415, 412/416, and 412/4108
- TT 3 '-terminal dithymidine
- RNA target of interest such as a viral or human mRNA transcript
- sequence of a gene or RNA gene transcript derived from a database is used to generate siNA targets having complementarity to the target.
- a database such as Genbank
- siNA targets having complementarity to the target.
- Such sequences can be obtained from a database, or can be determined experimentally as known in the art.
- Target sites that are known, for example, those target sites determined to be effective target sites based on studies with other nucleic acid molecules, for example ribozymes or antisense, or those targets known to be associated with a disease or condition such as those sites containing mutations or deletions, can be used to design siNA molecules targeting those sites.
- RNA transcripts can be chosen to screen siNA molecules for efficacy, for example by using in vitro RNA cleavage assays, cell culture, or animal models. In a non- limiting example, anywhere from 1 to 1000 target sites are chosen within the transcript based on the size of the siNA constract to be used.
- High throughput screening assays can be developed for screening siNA molecules using methods known in the art, such as with multi-well or multi-plate assays or combinatorial/siNA library screening assays to determine efficient reduction in target gene expression.
- the following non-limiting steps can be used to cany out the selection of siNAs targeting a given gene sequence or transcript.
- the target sequence is parsed in silico into a list of all fragments or subsequences of a particular length, for example 23 nucleotide fragments, contained within the target sequence. This step is typically carried out using a custom Perl script, but commercial sequence analysis programs such as Oligo, Mac Vector, or the GCG Wisconsin Package can be employed as well.
- the siNAs co ⁇ espond to more than one target sequence; such would be the case for example in targeting different transcripts of the same gene, targeting different transcripts of more than one gene, or for targeting both the human gene and an animal homolog.
- a subsequence list of a particular length is generated for each of the targets, and then the lists are compared to find matching sequences in each list.
- the subsequences are then ranked according to the number of target sequences that contain the given subsequence; the goal is to find subsequences that are present in most or all of the target sequences. Alternately, the ranking can identify subsequences that are unique to a target sequence, such as a mutant target sequence. Such an approach would enable the use of siNA to target specifically the mutant sequence and not effect the expression of the normal sequence.
- the siNA subsequences are absent in one or more sequences while present in the desired target sequence; such would be the case if the siNA targets a gene with a paralogous family member that is to remain untargeted.
- a subsequence list of a particular length is generated for each of the targets, and then the lists are compared to find sequences that are present in the target gene but are absent in the untargeted paralog.
- the ranked siNA subsequences can be further analyzed and ranked according to GC content.
- a preference can be given to sites containing 30-70%) GC, with a further preference to sites containing 40-60% GC.
- the ranked siNA subsequences can be further analyzed and ranked according to self-folding and internal hairpins. Weaker internal folds are prefe ⁇ ed; strong hairpin stractures are to be avoided.
- the ranked siNA subsequences can be further analyzed and ranked according to whether they have runs of GGG or CCC in the sequence.
- GGG (or even more Gs) in either strand can make oligonucleotide synthesis problematic and can potentially interfere with RNAi activity, so it is avoided other appropriately suitable sequences are available.
- CCC is searched in the target strand because that will place GGG in the antisense strand.
- the ranked siNA subsequences can be further analyzed and ranked according to whether they have the dinucleotide UU (uridine dinucleotide) on the 3 '-end of the sequence, and/or AA on the 5'-end of the sequence (to yield 3' UU on the antisense sequence). These sequences allow one to design siNA molecules with terminal TT thymidine dinucleotides.
- UU uridine dinucleotide
- target sites are chosen from the ranked list of subsequences as described above. For example, in subsequences having 23 nucleotides, the right 21 nucleotides of each chosen 23-mer subsequence are then designed and synthesized for the upper (sense) strand of the siNA duplex, while the reverse complement of the left 21 nucleotides of each chosen 23-mer subsequence are then designed and synthesized for the lower (antisense) strand of the siNA duplex (see Tables I).
- siNA molecules are screened in an in vitro, cell culture or animal model system to identify the most active siNA molecule or the most preferred target site within the target RNA sequence.
- a pool of siNA constracts specific to a target sequence is used to screen for target sites in cells expressing target RNA, such as human HeLa cells.
- target RNA such as human HeLa cells.
- the general strategy used in this approach is shown in Figure 21.
- a non-limiting example of such a pool is a pool comprising sequences having antisense sequences complementary to the target RNA sequence and sense sequences complementary to the antisense sequences.
- Cells e.g., HeLa cells
- the pool of siNA constracts can be chemically modified as described herein and synthesized, for example, in a high throughput manner.
- the siNA from cells demonstrating a positive phenotypic change are identified, for example by positional analysis within the assay, and are used to detennine the most suitable target site(s) within the target RNA sequence based upon the complementary sequence to the co ⁇ esponding siNA antisense strand identified in the assay.
- siNA Short interfering nucleic acid
- All-ribose siNA duplexes activate the RNAi pathway but have limited utility as therapeutic compounds due to their nuclease sensitivity and short half-life in serum, as shown in Example 2 above.
- siNAs that target luciferase mRNA and contain stabilizing chemical modifications were tested for activity in HeLa cells.
- the sequences for the siNA oligonucleotide sequences used in this study are shown in Table I.
- 2'-O-methyl nucleotides or 2'-fluoro (F) nucleotides in one or both siNA strands
- various 3 '-end stabilization chemistries including 3 '-glyceryl, 3 '-inverted abasic, 3 '-inverted Thymidine, and/or Thymidine.
- RNAi activity of chemically stabilized siNA constracts was compared with the RNAi activity of control siNA constracts consisting of all ribonucleotides at every position except the 3 '-terminus which comprised two thymidine nucleotide overhangs.
- Active siNA molecules containing stabilizing modifications such as described herein should prove useful for in vivo applications, given their enhanced nuclease-resistance.
- luciferase reporter system was utilized to test RNAi activity of chemically modified siNA constracts compared to siNA constracts consisting of all RNA nucleotides containing two thymidine nucleotide overhangs.
- Sense and antisense siNA strands (20 uM each) were annealed by incubation in buffer (100 mM potassium acetate, 30 mM HEPES-KOH, pH 7.4, 2 mM magnesium acetate) for 1 min. at 90°C followed by 1 hour at 37°C.
- Plasmids encoding firefly luciferase (pGL2) and renilla luciferase (pRLSV40) were purchased from Promega Biotech.
- HeLa S3 cells were grown at 37°C in DMEM with 5% FBS and seeded at 15,300 cells in 100 ul media per well of a 96-well plate 24 hours prior to transfection.
- 4 ul Lipofectamine 2000 (Life Technologies) was added to 96 ul OPTI- MEM, vortexed and incubated at room temperature for 5 minutes.
- the 100 ul diluted lipid was then added to a microtiter tube containing 5 ul pGL2 (200ng/ul), 5 ul pRLSV40 (8 ng/ul) 6 ul siNA (25 nM or 10 nM final), and 84 ul OPTI-MEM, vortexed briefly and incubated at room temperature for 20 minutes.
- the transfection mix was then mixed briefly and 50 ul was added to each of three wells that contained HeLa S3 cells in 100 ul media. Cells were incubated for 20 hours after transfection and analyzed for luciferase expression using the Dual luciferase assay according to the manufacturer's instructions (Promega Biotech). The results of this study are summarized in Figures 4-16.
- the sequences of the siNA strands used in this study are shown in Table I and are refe ⁇ ed to by RPI# in the figures. Normalized luciferase activity is reported as the ratio of firefly luciferase activity to renilla luciferase activity in the same sample. Error bars represent standard deviation of triplicate transfections.
- the RNAi activity of chemically modified constracts is often comparable to that of unmodified control siNA constructs, which consist of all ribonucleotides at every position except the 3 '-terminus which comprises two thymidine nucleotide overhangs.
- the RNAi activity of the chemically modified constmcts is greater than the unmodified control siNA constmct consisting of all ribonucleotides.
- Figure 4 shows results obtained from a screen using phosphorothioate modified siNA constmcts.
- the RPI 27654/27659 constmct contains phosphorothioate substitutions for every pyrimidine nucleotide in both sequences
- the RPI 27657/27662 constract contains 5 terminal 3 '-phosphorothioate substitutions in each strand
- the RPI 27649/27658 constract contains all phosphorothioate substitutions only in the antisense strand
- the RPI 27649/27660 and RPI 27649/27661 constructs have unmodified sense strands and varying degrees of phosphorothioate substitutions in the antisense strand. All of these constracts show significant RNAi activity when compared to a scrambled siNA conrol constract (27651/27652).
- FIG. 5 shows results obtained from a screen using phosphorothioate (RPI
- Figure 6 shows results obtained from a screen using 2'-O-methyl modified siNA constracts in which the sense strand contains either 10 (RPI 28244/27650) or 5 (RPI 28245/27650) 2'-O-methyl substitutions, both with comparable activity to the unmodified control siNA construct.
- Figure 7 shows results obtained from a screen using 2'-O-methyl or 2'-deoxy-2'- fluoro modified siNA constmcts compared to a control constract consisting of all ribonucleotides at every position except the 3 '-terminus which comprises two thymidine nucleotide overhangs.
- Figure 8 compares a siNA constmct containing six phosphorothioate substitutions in each strand (RPI 28460/28461), where 5 phosphorothioates are present at the 3' end and a single phosphorothioate is present at the 5' end of each strand.
- This motif shows very similar activity to the control siNA construct consisting of all ribonucleotides at every position except the 3 '-terminus, which comprises two thymidine nucleotide overhangs.
- Figure 9 compares a siNA constract synthesized by the method of the invention described in Example 1, wherein an inverted deoxyabasic succinate linker was used to generate a siNA having a 3 '-inverted deoxyabasic cap on the antisense strand of the siNA.
- This constract shows improved activity compared to the control siNA constract consisting of all ribonucleotides at every position except the 3 '-terminus which comprises two thymidine nucleotide overhangs.
- Figure 10 shows the results of an RNAi activity screen of chemically modifed siNA constracts including 3 '-glyceryl modified siNA constracts compared to an all RNA control siNA constract using a luciferase reporter system.
- These chemically modified siNAs were compared in the luciferase assay described herein at 1 nM and lOnM concentration using an all RNA siNA control (siGL2) having 3 '-terminal dithymidine (TT) and its co ⁇ esponding inverted control (Inv siGL2).
- the background level of luciferase expression in the HeLa cells is designated by the "cells" column.
- Sense and antisense strands of chemically modified siNA constructs are shown by RPI number (sense strand/antisense strand). Sequences conespoding to these RPI numbers are shown in Table I. As shown in the Figure, the 3 '-terminal modified siNA constracts retain significant RNAi activity compared to the unmodified control siNA (siGL2) construct.
- Figure 11 shows the results of an RNAi activity screen of chemically modifed siNA constracts.
- the screen compared various combinations of sense strand chemical modifications and antisense strand chemical modifications.
- These chemically modified siNAs were compared in the luciferase assay described herein at 1 nM and lOnM concentration using an all RNA siNA control (siGL2) having 3 '-terminal dithymidine (TT) and its co ⁇ esponding inverted control (Inv siGL2).
- the background level of luciferase expression in the HeLa cells is designated by the "cells" column.
- Sense and antisense strands of chemically modified siNA constructs are shown by RPI number (sense strand/antisense strand).
- RPI 30063/30430 As shown in the figure, the chemically modified RPI 30063/30430, RPI 30433/30430, and RPI 30063/30224 constracts retain significant RNAi activity compared to the unmodified control siNA constract. It should be noted that RPI 30433/30430 is a siNA constract having no ribonucleotides which retains significant RNAi activity compared to the unmodified control siGL2 constract in vitro, therefore, this construct is expected to have both similar RNAi activity and improved stability in vivo compared to siNA constracts having ribonucleotides.
- Figure 12 shows the results of an RNAi activity screen of chemically modifed siNA constracts.
- the screen compared various combinations of sense strand chemical modifications and antisense strand chemical modifications.
- These chemically modified siNAs were compared in the luciferase assay described herein at 1 nM and lOnM concentration using an all RNA siNA control (siGL2) having 3 '-terminal dithymidine (TT) and its co ⁇ esponding inverted control (Inv siGL2).
- the background level of luciferase expression in the HeLa cells is designated by the "cells" column.
- Sense and antisense strands of chemically modified siNA constracts are shown by RPI number (sense strand/antisense strand).
- Figure 13 shows the results of an RNAi activity screen of chemically modifed siNA constracts.
- the screen compared various combinations of sense strand chemical modifications and antisense strand chemical modifications.
- These chemically modified siNAs were compared in the luciferase assay described herein at 1 nM and lOnM concentration using an all RNA siNA control (siGL2) having 3 '-terminal dithymidine
- TT co ⁇ esponding inverted control
- Inv siGL2 co ⁇ esponding inverted control
- Figure 14 shows the results of an RNAi activity screen of chemically modifed siNA constracts including various 3 '-terminal modified siNA constracts compared to an all RNA control siNA constract using a luciferase reporter system.
- These chemically modified siNAs were compared in the luciferase assay described herein at 1 nM and lOnM concentration using an all RNA siNA control (siGL2) having 3 '-terminal dithymidine (TT) and its co ⁇ esponding inverted control (Inv siGL2).
- the background level of luciferase expression in the HeLa cells is designated by the "cells" column.
- Sense and antisense strands of chemically modified siNA constracts are shown by RPI number (sense strand/antisense strand). Sequences co ⁇ espoding to these RPI numbers are shown in Table I. As shown in the figure, the chemically modified RPI 30222/30546, 30222/30224, 30222/30551, 30222/30557 and 30222/30558 constracts retain significant RNAi activity compared to the control siNA constract.
- Figure 15 shows the results of an RNAi activity screen of chemically modifed siNA constracts.
- the screen compared various combinations of sense strand chemistries compared to a fixed antisense strand chemistry.
- These chemically modified siNAs were compared in the luciferase assay described herein at 1 nM and lOnM concentration using an all RNA siNA control (siGL2) having 3 '-terminal dithymidine (TT) and its conesponding inverted control (Inv siGL2).
- the background level of luciferase expression in the HeLa cells is designated by the "cells" column.
- Sense and antisense strands of chemically modified siNA constracts are shown by RPI number (sense strand/antisense strand). Sequences co ⁇ espoding to these RPI numbers are shown in
- a titration assay was performed to determine the lower range of siNA concentration required for RNAi activity both in a control siNA constract consisting of all RNA nucleotides containing two thymidine nucleotide overhangs and a chemically modified siNA construct comprising five phosphorothioate internucleotide linkages in both the sense and antisense strands.
- the assay was perfonned as described above, however, the siNA constracts were diluted to final concentrations between 2.5 nM and 0.025 nM. Results are shown in Figure 16.
- the chemically modified siNA constract shows a very similar concentration dependent RNAi activity profile to the control siNA constmct when compared to an inverted siNA sequence control.
- Example 7 siNA design
- siNA target sites were chosen by analyzing sequences of the target RNA and optionally prioritizing the target sites on the basis of folding (structure of any given sequence analyzed to determine siNA accessibility to the target), by using a library of siNA molecules as described in Example 4, or alternately by using an in vitro siNA system as described in Example 9 herein.
- siNA molecules were designed that could bind each target and are optionally individually analyzed by computer folding to assess whether the siNA molecule can interact with the target sequence. Varying the length of the siNA molecules can be chosen to optimize activity. Generally, a sufficient number of complementary nucleotide bases are chosen to bind to, or otherwise interact with, the target RNA, but the degree of complementarity can be modulated to accommodate siNA duplexes or varying length or base composition.
- siNA molecules can be designed to target sites within any known RNA sequence, for example those RNA sequences co ⁇ esponding to the any gene transcript.
- Chemically modified siNA constracts are designed to provide nuclease stability for systemic administration in vivo and/or improved pharmacokinetic, localization, and delivery properties while preserving the ability to mediate RNAi activity. Chemical modifications as described herein are introduced synthetically using synthetic methods described herein and those generally known in the art. The synthetic siNA constracts are then assayed for nuclease stability in serum and/or cellular/tissue extracts (e.g. liver extracts). The synthetic siNA constracts are also tested in parallel for RNAi activity using an appropriate assay, such as a luciferase reporter assay as described herein or another suitable assay that can quantity RNAi activity.
- an appropriate assay such as a luciferase reporter assay as described herein or another suitable assay that can quantity RNAi activity.
- Synthetic siNA constracts that possess both nuclease stability and RNAi activity can be further modified and re- evaluated in stability and activity assays.
- the chemical modifications of the stabilized active siNA constracts can then be applied to any siNA sequence targeting any chosen RNA and used, for example, in target screening assays to pick lead siNA compounds for therapeutic development (see for example Figure 27).
- siNA molecules can be designed to interact with various sites in the RNA message, for example, target sequences within the RNA sequences described herein.
- the sequence of one strand of the siNA molecule(s) is complementary to the target site sequences described above.
- the siNA molecules can be chemically synthesized using methods described herein.
- Inactive siNA molecules that are used as control sequences can be synthesized by scrambling the sequence of the siNA molecules such that it is not complementary to the target sequence.
- siNA constracts can by synthesized using solid phase oligonucleotide synthesis methods as described herein (see for example Usman et al, US Patent Nos.
- RNA oligonucleotides are synthesized in a stepwise fashion using the phosphoramidite chemistry as is known in the art.
- Standard phosphoramidite chemistry involves the use of nucleosides comprising any of 5'-O- dimethoxytrityl, 2'-O-tert-butyldimethylsilyl, 3'-O-2-Cyanoethyl N, N-diisopropylphos- phoroamidite groups, and exocyclic amine protecting groups (e.g. N6-benzoyl adenosine, N4 acetyl cytidine, and N2-isobutyryl guanosine).
- exocyclic amine protecting groups e.g. N6-benzoyl adenosine, N4 acetyl cytidine, and N2-isobutyryl guanosine.
- 2'-O-Silyl Ethers can be used in conjunction with acid-labile 2'-O-orthoester protecting groups in the synthesis of RNA as described by Scaringe supra.
- Differing 2' chemistries can require different protecting groups, for example 2'-deoxy-2'-amino nucleosides can utilize N-phthaloyl protection as described by Usman et al, US Patent 5,631,360, incorporated by reference herein in its entirety).
- each nucleotide is added sequentially (3'- to 5'- direction) to the solid support-bound oligonucleotide.
- the first nucleoside at the 3 '-end of the chain is covalently attached to a solid support (e.g., controlled pore glass or polystyrene) using various linkers.
- the nucleotide precursor, a ribonucleoside phosphoramidite, and activator are combined resulting in the coupling of the second nucleoside phosphoramidite onto the 5 '-end of the first nucleoside.
- the support is then washed and any unreacted 5 '-hydroxyl groups are capped with a capping reagent such as acetic anhydride to yield inactive 5 '-acetyl moieties.
- a capping reagent such as acetic anhydride to yield inactive 5 '-acetyl moieties.
- the trivalent phosphorus linkage is then oxidized to a more stable phosphate linkage.
- the 5 '-O-protecting group is cleaved under suitable conditions (e.g., acidic conditions for trityl-based groups and Fluoride for silyl-based groups). The cycle is repeated for each subsequent nucleotide.
- Modification of synthesis conditions can be used to optimize coupling efficiency, for example by using differing coupling times, differing reagent/phosphoramidite concentrations, differing contact times, differing solid supports and solid support linker chemistries depending on the particular chemical composition of the siNA to be synthesized.
- Deprotection and purification of the siNA can be performed as is generally described in Usman et al., US 5,831,071, US 6,353,098, US 6,437,117, and Bellon et al, US 6,054,576, US 6,162,909, US 6,303,773, incorporated by reference herein in their entirety or Scaringe supra,. Additionally, deprotection conditions can be modified to provide the best possible yield and purity of siNA constmcts.
- oligonucleotides comprising 2'-deoxy-2'-fluoro nucleotides can degrade under inappropriate deprotection conditions. Such oligonucleotides are deprotected using aqueous methylamine at about 35°C for 30 minutes. If the 2 '-deoxy-2' -fluoro containing oligonucleotide also comprises ribonucleotides, after deprotection with aqueous methylamine at about 35°C for 30 minutes, TEA-HF is added and the reaction maintained at about 65°C for an additional 15 minutes.
- Example 9 RNAi in vitro assay to assess siNA activity
- RNAi in vitro assay that recapitulates RNAi in a cell free system is used to evaluate siNA constracts specific to target RNA.
- the assay comprises the system described by Tuschl et al, 1999, Genes and Development, 13, 3191-3197 and Zamore et al, 2000, Cell, 101, 25-33 adapted for use with target RNA.
- a Drosophila extract derived from syncytial blastoderm is used to reconstitute RNAi activity in vitro.
- Target RNA is generated via in vitro transcription from an appropriate plasmid using T7 RNA polymerase or via chemical synthesis as described herein.
- Sense and antisense siNA strands are annealed by incubation in buffer (such as 100 mM potassium acetate, 30 mM HEPES-KOH, pH 7.4, 2 mM magnesium acetate) for 1 min. at 90°C followed by 1 hour at 37°C , then diluted in lysis buffer (for example 100 mM potassium acetate, 30 mM HEPES-KOH at pH 7.4, 2mM magnesium acetate). Annealing can be monitored by gel electrophoresis on an agarose gel in TBE buffer and stained with ethidium bromide.
- buffer such as 100 mM potassium acetate, 30 mM HEPES-KOH, pH 7.4, 2 mM magnesium acetate
- the Drosophila lysate is prepared using zero to two-hour-old embryos from Oregon R flies collected on yeasted molasses agar that are dechorionated and lysed. The lysate is centrifuged and the supematant isolated.
- the assay comprises a reaction mixture containing 50% lysate [vol/vol], RNA (10-50 pM final concentration), and 10%) [vol/vol] lysis buffer containing siNA (10 nM final concentration).
- the reaction mixture also contains 10 mM creatine phosphate, 10 ug.ml creatine phosphokinase, 100 um GTP, 100 uM UTP, 100 uM CTP, 500 uM ATP, 5 mM DTT, 0.1 U/uL RNasin (Promega), and 100 uM of each amino acid.
- the final concentration of potassium acetate is adjusted to 100 mM.
- the reactions are pre-assembled on ice and preincubated at 25° C for 10 minutes before adding RNA, then incubated at 25° C for an additional 60 minutes. Reactions are quenched with 4 volumes of 1.25 x Passive Lysis Buffer (Promega).
- Target RNA cleavage is assayed by RT-PCR analysis or other methods known in the art and are compared to control reactions in which siNA is omitted from the reaction.
- target RNA for the assay is prepared by in vitro transcription in the presence of [alpha- 3 p] CTP, passed over a G 50 Sephadex column by spin chromatography and used as target RNA without further purification.
- target RNA is 5'-32p_end labeled using T4 polynucleotide kinase enzyme.
- Assays are performed as described above and target RNA and the specific RNA cleavage products generated by RNAi are visualized on an autoradiograph of a gel. The percentage of cleavage is determined by Phosphor Imager® quantitation of bands representing intact control RNA or RNA from control reactions without siNA and the cleavage products generated by the assay.
- this assay is used to determine target sites the RNA target for siNA mediated RNAi cleavage, wherein a plurality of siNA constracts are screened for
- RNAi mediated cleavage of the RNA target for example, by analyzing the assay reaction by electrophoresis of labeled target RNA, or by northern blotting, as well as by other methodology well known in the art.
- Example 10 Nucleic acid inhibition of target RNA in vivo
- siNA molecules targeted to the target RNA are designed and synthesized as described above. These nucleic acid molecules can be tested for cleavage activity in vivo, for example, using the following procedure.
- Two formats are used to test the efficacy of siNAs targeting a particular gene transcipt.
- the reagents are tested on target expressing cells (e.g., HeLa), to determine the extent of RNA and protein inhibition.
- siNA reagents are selected against the RNA target.
- RNA inhibition is measured after delivery of these reagents by a suitable transfection agent to cells.
- Relative amounts of target RNA are measured versus actin using real-time PCR monitoring of amplification (eg., ABI 7700 Taqman®).
- a comparison is made to a mixture of oligonucleotide sequences made to unrelated targets or to a randomized siNA control with the same overall length and chemistry, but with randomly substituted nucleotides at each position.
- RNA inhibition is determined by the cell-plating format.
- Cells e.g., HeLa are seeded, for example, at 1x10 ⁇ cells per well of a six-well dish in EGM-2 (BioWhittaker) the day before transfection.
- siNA final concentration, for example 20nM
- cationic lipid e.g., final concentration 2 ⁇ g/ml
- EGM basal media Biowhittaker
- the complexed siNA is added to each well and incubated for the times indicated.
- cells are seeded, for example, at lxl 0- ⁇ in 96 well plates and siNA complex added as described.
- Total RNA is prepared from cells following siNA delivery, for example, using Qiagen RNA purification kits for 6-well or Rneasy extraction kits for 96-well assays.
- dual-labeled probes are synthesized with the reporter dye, FAM or JOE, covalently linked at the 5'-end and the quencher dye TAMRA conjugated to the 3'-end.
- RT-PCR amplifications are performed on, for example, an ABI PRISM 7700 Sequence Detector using 50 ⁇ l reactions consisting of 10 ⁇ l total RNA, 100 nM forward primer, 900 nM reverse primer, 100 nM probe, IX TaqMan PCR reaction buffer (PE- Applied Biosystems), 5.5 mM MgCl 2 , 300 ⁇ M each dATP, dCTP, dGTP, and dTTP, 10U RNase Inhibitor (Promega), 1.25U AmpliTaq Gold (PE- Applied Biosystems) and 10U M- MLN Reverse Transcriptase (Promega).
- the thermal cycling conditions can consist of 30 min at 48°C, 10 min at 95°C, followed by 40 cycles of 15 sec at 95°C and 1 min at 60°C.
- Quantitation of mR ⁇ A levels is determined relative to standards generated from serially diluted total cellular R ⁇ A (300, 100, 33, 11 ng/rxn) and normalizing to ⁇ -actin or GAPDH mR ⁇ A in parallel TaqMan reactions.
- an upper and lower primer and a fluorescently labeled probe are designed.
- Real time incorporation of SYBR Green I dye into a specific PCR product can be measured in glass capillary tubes using a lightcyler.
- a standard curve is generated for each primer pair using control cR ⁇ A. Values are represented as relative expression to GAPDH in each sample.
- Nuclear extracts can be prepared using a standard micro preparation technique (see for example Andrews and Faller, 1991, Nucleic Acids Research, 19, 2499). Protein extracts from supematants are prepared, for example using TCA precipitation. An equal volume of 20% TCA is added to the cell supernatant, incubated on ice for 1 hour and pelleted by centrifugation for 5 minutes. Pellets are washed in acetone, dried and resuspended in water. Cellular protein extracts are run on a 10%) Bis-Tris NuPage (nuclear extracts) or 4-12% Tris-Glycine (supernatant extracts) polyacrylamide gel and transferced onto nitro-cellulose membranes.
- Non-specific binding can be blocked by incubation, for example, with 5% non-fat milk for 1 hour followed by primary antibody for 16 hour at 4°C. Following washes, the secondary antibody is applied, for example (1:10,000 dilution) for 1 hour at room temperature and the signal detected with SuperS ignal reagent (Pierce).
- siNA molecules that are designed as anti-angio genie agents can be screened using animal models.
- VEGFR e.g., VEGFR1, VEGFR2, and VEGFR3 genes.
- a comeal model has been used to study angiogenesis in rat and rabbit, since recruitment of vessels can easily be followed in this normally avascular tissue (Pandey et al, 1995 Science 268: 567-569).
- angiogenesis factor e.g. bFGF or VEGF
- siNA molecules directed against VEGFR niRNAs would be delivered in the disk as well, or dropwise to the eye over the time course of the experiment.
- hypoxia has been shown to cause both increased expression of VEGF and neovascularization in the retina (Pierce et al, 1995 Proc. Natl. Acad. Sci. USA. 92: 905-909; Shweiki et al, 1992 J. Clin. Invest 91: 2235-2243).
- the cornea model is the most common and well characterized anti-angiogenic agent efficacy screening model.
- This model involves an avascular tissue into which vessels are recruited by a stimulating agent (growth factor, thermal or alkalai bum, endotoxin).
- the comeal model utilizes the intrastromal corneal implantation of a Teflon pellet soaked in a VEGF-Hydron solution to recrait blood vessels toward the pellet, which can be quantitated using standard microscopic and image analysis techniques.
- siNA molecules are applied topically to the eye or bound within Hydron on the Teflon pellet itself.
- This avascular cornea as well as the Matrigel model provide for low background assays. While the comeal model has been performed extensively in the rabbit, studies in the rat have also been conducted.
- the mouse model (Passaniti et al., supra) is a non-tissue model which utilizes
- Matrigel an extract of basement membrane (Kleinman et al., 1986) or Millipore® filter disk, which can be impregnated with growth factors and anti-angiogenic agents in a liquid form prior to injection.
- Millipore® filter disk Upon subcutaneous administration at body temperature, the
- Matrigel or Millipore® filter disk forms a solid implant.
- VEGF embedded in the Matrigel or Millipore® filter disk is used to recrait vessels within the matrix of the Matrigel or
- Millipore® filter disk which can be processed histologically for endothelial cell specific vWF (factor VIII antigen) immunohistochemistry, Trichrome-Masson stain, or hemoglobin content.
- vWF factor VIII antigen
- the Matrigel or Millipore® filter disk are avascular; however, it is not tissue.
- siNA molecules are administered within the matrix of the Matrigel or Millipore® filter disk to test their anti-angiogenic efficacy.
- delivery issues in this model as with delivery of siNA molecules by Hydron- coated Teflon pellets in the rat cornea model, may be less problematic due to the homogeneous presence of the siNA within the respective matrix.
- the Lewis lung carcinoma and B-16 murine melanoma models are well accepted models of primary and metastatic cancer and are used for initial screening of anti-cancer agents. These murine models are not dependent upon the use of immunodeficient mice, are relatively inexpensive, and minimize housing concerns. Both the Lewis lung and B-
- 16 melanoma models involve subcutaneous implantation of approximately 10 6 tumor cells from metastatically aggressive tumor cell lines (Lewis lung lines 3LL or D122, LLc- LN7; B-16-BL6 melanoma) in C57BL/6J mice.
- the Lewis lung model can be produced by the surgical implantation of tumor spheres (approximately 0.8 mm in diameter). Metastasis also may be modeled by injecting the tumor cells directly intraveneously.
- microscopic metastases can be observed approximately 14 days following implantation with quantifiable macroscopic metastatic tumors developing within 21-25 days.
- the B-16 melanoma exhibits a similar time course with tumor neovascularization beginning 4 days following implantation.
- systemic pharmacotherapy with a wide variety of agents usually begins 1-7 days following tumor implantation/inoculation with either continuous or multiple administration regimens.
- Concurrent pharmacokinetic studies can be performed to determine whether sufficient tissue levels of siNA can be achieved for pharmacodynamic effect to be expected.
- primary tumors and secondary lung metastases can be removed and subjected to a variety of in vitro studies (i.e. target RNA reduction).
- VEGFRl In utilizing these models to assess siNA activity, VEGFRl, VEGFR2, and/or
- VEGFR3 protein levels can be measured clinically or experimentally by FACS analysis.
- VEGFRl, VEGFR2, and/or VEGFR3 encoded mRNA levels can be assessed by Northern analysis, RNase-protection, primer extension analysis and/or quantitative RT-PCR.
- siNA molecules that block VEGFRl, VEGFR2, and/or VEGFR3 protein encoding mRNAs and therefore result in decreased levels of VEGFRl, VEGFR2, and/or VEGFR3 activity by more than 20% in vitro can be identified using the techniques described herein.
- Example 12 siNA-mediated inhibition of angiogenesis in vivo
- siNA molecules shown in Figure 23 have matched inverted controls which are inactive since they are not able to interact with the RNA target.
- the siNA molecules and VEGF were co-delivered using the filter disk method.
- Nitrocellulose filter disks (Millipore®) of 0.057 diameter were immersed in appropriate solutions and were surgically implanted in rat comea as described by Pandey et al, supra.
- the stimulus for angiogenesis in this study was the treatment of the filter disk with 30 ⁇ M VEGF which is implanted within the cornea's stroma. This dose yields reproducible neovascularization stemming from the pericorneal vascular plexus growing toward the disk in a dose-response study 5 days following implant. Filter disks treated only with the vehicle for VEGF show no angiogenic response.
- the siNA were co- adminstered with VEGF on a disk in three different siNA concentrations.
- VEGF receptors can be stimulated.
- Applicant has observed that in low VEGF doses, the neovascular response reverts to normal suggesting that the VEGF stimulus is essential for maintaining the angiogenic response. Blocking the production of VEGF receptors using simultaneous administration of anti-VEGF-R mRNA siNA could attenuate the normal neovascularization induced by the filter disk treated with VEGF.
- Animals are housed in groups of two. Feed, water, temperature and humidity are determined according to Pharmacology Testing Facility performance standards (SOP's) which are in accordance with the 1996 Guide for the Care and Use of Laboratory Animals (NRC). Animals are acclimated to the facility for at least 7 days prior to experimentation. During this time, animals are observed for overall health and sentinels are bled for baseline serology.
- SOP's Pharmacology Testing Facility performance standards
- NRC Laboratory Animals
- VEGF and siNAs were prepared as a IX solution for final concentrations shown in the experimental groups described in Table III.
- siNA sense and antisense strands are annealed for 1 minute in H 2 O at
- duplexed siNA 1.67mg/mL/strand followed by a 1 hour incubation at 37°C producing 3.34 mg/mL of duplexed siNA.
- 6 ⁇ Ls of the 3.34 mg/mL duplex is injected into the eye (see below).
- the 3.34 mg/mL duplex siNA can then be serially diluted for dose response assays.
- nitrocellulose filter membranes 0.45 ⁇ m pore diameter nitrocellulose filter membranes (Millipore Corporation), were soaked for 30 min in 1 ⁇ L of 75 ⁇ M VEGF in 82 mM TrisHCl (pH 6.9) in covered petri dishes on ice. Filter disks soaked only with the vehicle for NEGF (83 mM Tris-Cl pH
- the rat comeal model used in this study was a modified from Koch et al. Supra and Pandey et al, supra. Briefly, corneas were irrigated with 0.5% povidone iodine solution followed by normal saline and two drops of 2% lidocaine. Under a dissecting microscope (Leica MZ-6), a stromal pocket was created and a presoaked filter disk (see above) was inserted into the pocket such that its edge was 1 mm from the comeal limbus.
- test solution saliva, inverted control or sterile water vehicle
- test solution inverted control or sterile water vehicle
- test solution inverted control or sterile water vehicle
- the injector was then removed, serially rinsed in 70%> ethanol and sterile water and immersed in sterile water between each injection.
- closure of the eyelid was maintained using microaneurism clips until the animal began to recover gross motor activity. Following treatment, animals were warmed on a heating pad at 37 C.
- ⁇ SA neovascular surface area
- group mean percent inhibition of VEGF-induced angiogenesis was subjected to a one-way analysis of variance. This was followed by two post-hoc tests for significance including Dunnett's
- a chemically modified version of the VEGFrl site 4229 active siNA comprising a sense strand having 2 '-deoxy-2' -fluoro pyrimidines and ribo purines with 5' and 3' terminal inverted deoxyabasic residues and an antisense strand having having 2'- deoxy-2' -fluoro pyrimidines and ribo purines with a terminal 3 '-phosphorothioate internucleotide linkage (RPI 30196/30416), showed similar inhibition.
- This result shows siNA molecules having different chemically modified composition, such as the modifications described herein, are capable of significantly inhibiting angiogenesis in vivo.
- the human hepatocellular carcinoma cell line Hep G2 was grown in Dulbecco's modified Eagle media supplemented with 10% fetal calf serum, 2 mM glutamine, 0.1 mM nonessential amino acids, 1 mM sodium pyruvate, 25 mM Hepes, 100 units penicillin, and 100 ⁇ g/ml streptomycin.
- To generate a replication competent cDNA prior to transfection the HBV genomic sequences are excised from the bacterial plasmid sequence contained in the psHBV-1 vector. Other methodsknown in the art can be used to generate a replication competent cDNA. This was done with an EcoRI and Hind III restriction digest. Following completion of the digest, a ligation was performed under dilute conditions (20 ⁇ g/ml) to favor intermolecular ligation. The total ligation mixture was then concentrated using Qiagen spin columns.
- HBV proteins results in the expression of HBV proteins and the production of virions.
- siNA duplexes targeting different sites within HBV pregenomic RNA were co-transfected with HBV genomic DNA once at 25 nM with lipid at 12.5 ug/ml into Hep G2 cells, and the subsequent levels of secreted HBV surface antigen (HBsAg) were analyzed by ELISA (see Figure 24). Inverted sequence duplexes were used as negative controls.
- HbsAg levels were measured following transfection with siNA.
- hnmulon 4 (Dynax) microtiter wells were coated overnight at 4° C with anti- HBsAg Mab (Biostride B88-95-31ad,ay) at 1 ⁇ g/ml in Carbonate Buffer (Na2CO3 15 mM, NaHCO3 35 mM, pH 9.5).
- the wells were then washed 4x with PBST (PBS, 0.05% Tween® 20) and blocked for 1 hr at 37° C with PBST, 1% BSA. Following washing as above, the wells were dried at 37° C for 30 min.
- PBST PBS, 0.05% Tween® 20
- Biotinylated goat ant-HBsAg (Accurate YVS1807) was diluted 1:1000 in PBST and incubated in the wells for 1 hr. at 37° C. The wells were washed 4x with PBST. Streptavidin/Alkaline Phosphatase Conjugate (Pierce 21324) was diluted to 250 ng/ml in PBST, and incubated in the wells for 1 hr. at 37° C. After washing as above, p-nitrophenyl phosphate substrate (Pierce 37620) was added to the wells, which were then incubated for 1 hour at 37° C. The optical density at 405 nm was then dete ⁇ nined.
- results of the HBV screen study are summarized in Figure 24, whereas the results of a dose response assay using lead siNA constracts targeting sites 262 and 1580 of the HBV pregenomic RNA are shown in Figure 25.
- the siNA constracts targeting sites 262 and 1580 of HBV RNA provides significant dose response inhibition of viral replication/activity when compared to inverted siNA controls.
- the "Stab7/8" (Table IV) constructs comprise a sense strand having 2 '-deoxy-2' -fluoro pyrimidine nucleotides and 2 '-deoxy purine nucleotides with 5' and 3' terminal inverted deoxyabasic residues and an antisense strand having 2 '-deoxy-2' -fluoro pyrimidine nucleotides and 2'-O-methyl purine nucleotides with a terminal 3' phosphorothioate linkage.
- the "Stab 7/11 (Table IV) constracts comprise a sense strand having 2'-deoxy-2'-fluoro pyrimidine nucleotides and 2 '-deoxy purine nucleotides with 5' and 3' terminal inverted deoxyabasic residues and an antisense strand having 2 '-deoxy-2' -fluoro pyrimidine nucleotides and 2 '-deoxy purine nucleotides with a terminal 3' phosphorothioate linkage (see for example Table I).
- the chemically stabilized siNA constmcts both show significant inhibition of HBV antigen in a dose dependent manner compared to matched inverted contols.
- siNA constracts having different stabilization chemistries were compared to an unstabilized siRNA constract in a dose response time course HBsAg assay, the results of which are shown in Figures 28-31.
- the different constracts were compared to an unstabilized ribonucleotide control siRNA constract (RPI#30287/30298) at different concentrations (5nM, 10 nM, 25 nM, 50 nM, and 100 nM) over the course of nine days.
- Activity based on HBsAg levels was determined at day 3, day 6, and day 9.
- the "Stab 4/5" (Table IV) constracts comprise a sense strand (RPI#30355) having 2'- deoxy-2' -fluoro pyrimidine nucleotides and purine ribonucleotides with 5' and 3' terminal inverted deoxyabasic residues and an antisense strand (RPI#30366) having 2'- deoxy-2' -fluoro pyrimidine nucleotides and purine ribonucleotides with a terminal 3' phosphorothioate linkage (data shown in Figure 28).
- the "Stab7/8" (Table IV) constmcts comprise a sense strand (RPI#30612) having 2 '-deoxy-2' -fluoro pyrimidine nucleotides and 2 '-deoxy purine nucleotides with 5' and 3' terminal inverted deoxyabasic residues and an antisense strand (RPI#30620) having 2 '-deoxy-2' -fluoro pyrimidine nucleotides and 2'-O-methyl purine nucleotides with a terminal 3' phosphorothioate linkage (data shown in Figure 29).
- the "Stab7/ll (Table IV) constracts comprise a sense (RPI#30612) strand having 2 '-deoxy-2' -fluoro pyrimidine nucleotides and 2 '-deoxy purine nucleotides with 5' and 3' terminal inverted deoxyabasic residues and an antisense strand (RPI#31175) having 2 '-deoxy-2' -fluoro pyrimidine nucleotides and 2'-deoxy purine nucleotides with a terminal 3' phosphorothioate linkage (data shown in Figure 30).
- the "Stab9/10 (Table IV) constracts comprise a sense (RPI#31335) strand having ribonucleotides with 5' and 3' terminal inverted deoxyabasic residues and an antisense strand (RPI#31337) having ribonucleotides with a terminal 3' phosphorothioate linkage (data shown in Figure 31).
- the chemically stabilized siNA constmcts all show significantly greater inhibition of HBV antigen in a dose dependent manner over the time course experiment compared to the unstabilized siRNA constract.
- siNA duplexes in HeLa cells. Seven siNA constructs were designed that target three regions in the highly conserved 5' untranslated region (UTR) of HCV RNA. The siNAs were screened in two cell culture systems dependent upon the 5'-UTR of HCV; one requires translation of an HCV/luciferase gene, while the other involves replication of a chimeric HCV/poliovirus (PV) (see Blatt et al, USSN 09/740,332, filed December 18, 2000, incorporated by reference herein).
- PV chimeric HCV/poliovirus
- siNAs (29579/29586; 29578/29585) targeting the same region (shifted by one nucleotide) are active in both systems (see Figure 32) as compared with inverse control siNA (29593/29600).
- inverse control siNA 29593/29600.
- IC50 ⁇ 2.5 nM
- F 2'-fluoro
- siNA activity is compared to relevant controls (untreated cells, scrambled/inactive control sequences, or transfection controls).
- relevant controls untreated cells, scrambled/inactive control sequences, or transfection controls.
- siNA constracts of the invention provide potent inhibition of HCV RNA in the HCV/poliovims chimera system.
- siNA constructs, inlcuding chemically modified, nuclease resistant siNA molecules represent an important class of therapeutic agents for treating chronic HCV infection.
- a HCV replicaon system was used to test the efficacy of siNAs targeting HCV RNA.
- the reagents are tested in cell culture using Huh7 cells (see for example Randall et al, 2003, PNAS USA, 100, 235-240) to determine the extent of RNA and protein inhibition.
- siNA were selected against the HCV target as described herein.
- RNA inhibition was measured after delivery of these reagents by a suitable transfection agent to Huh7 cells.
- Relative amounts of target RNA are measured versus actin using real-time PCR monitoring of amplification (eg., ABI 7700 Taqman®).
- a comparison is made to a mixture of oligonucleotide sequences designed to target unrelated targets or to a randomized siNA control with the same overall length and chemistry, but with randomly substituted nucleotides at each position.
- Primary and secondary lead reagents were chosen for the target and optimization performed. After an optimal transfection agent concentration is chosen, a RNA time-course of inhibition is performed with the lead siNA molecule.
- a cell-plating format can be used to determine RNA inhibition.
- a non-limiting example of a multiple target screen to assay siNA mediated inhibition of HCV RNA is shown in Figure 38.
- siNA reagents (Table I) were transfected at 25 nM into Huh7 cells and HCV RNA quantitated compared to untreated cells ("cells" column in the figure) and cells transfected with lipofectamine ("LFA2K” column in the figure). As shown in the Figure, several siNA constmcts show significant inhibition of HCV RNA expression in the Huh7 replicon system.
- compositions and methods of the invention are used to discover genes involved in a process of interest within mammalian cells, such as cell growth, proliferation, apoptosis, morphology, angiogenesis, differentiation, migration, viral multiplication, drug resistance, signal transduction, cell cycle regulation, or temperature sensitivity or other process.
- a randomized siNA library is generated. These constracts are inserted into a vector capable of expressing a siNA from the library inside mammalian cells. Altemately, a pool of synthetic siNA molecules is generated.
- a readily assayable reporter system is constructed in which a reporter molecule is co-expressed when a particular protein of interest is expressed.
- the reporter system consists of a plasmid constract bearing a gene coding for a reporter gene, such as Green Fluorescent Protein (GFP) or other reporter proteins known and readily available in the art.
- GFP Green Fluorescent Protein
- the promoter region of the GFP gene is replaced by a portion of a promoter for the protein of interest sufficient to direct efficient transcription of the GFP gene.
- the plasmid can also contain a drag resistance gene, such as neomycin resistance, in order to select cells containing the plasmid.
- a cell line is selected as host for target discovery.
- the cell line is preferably known to express the protein of interest, such that upstream genes controlling the expression of the protein can be identified when modulated by a siNA constract expressed therein.
- the cells preferably retain protein expression characteristics in culture.
- the reporter plasmid is transfected into cells, for example, using a cationic lipid formulation. Following transfection, the cells are subjected to limiting dilution cloning, for example, under selection by 600 ⁇ g/mL Geneticin. Cells retaining the plasmid survive the Geneticin treatment and form colonies derived from single surviving cells. The resulting clonal cell lines are screened by flow cytometry for the capacity to upregulate GFP production.
- PCM Pseudomonas conditioned medium
- PMA phorbol myristate acetate
- a siNA library was constmcted with oligonucletides containing hairpin siNA constracts having randomized antisense regions and self complementary sense regions.
- the library is generated synthesizing siNA constracts having randomized sequence.
- the siNA libraries are constructed as described in Usman et al, USSN
- Oligo sequence 5' and 3' of the siNA contains restriction endonuclease cleavage sites for cloning.
- the 3' trailing sequence fonns a stem-loop for priming DNA polymerase extension to form a hairpin structure.
- the hairpin DNA constmct is melted at 90°C allowing DNA polymerase to generate a dsDNA constract.
- the double-stranded siNA library is cloned into, for example, a U6+27 transcription unit located in the 5 ' LTR region of a retro viral vector containing the human nerve growth factor receptor (hNGFr) reporter gene.
- hNGFr human nerve growth factor receptor
- the siNA is transcribed by RNA polymerase III from U6+27 and by RNA polymerase II activity directed by the 5' LTR.
- the siNA library is packaged into retro viral particles that are used to infect and transduce clonal cells selected above.
- Assays of the hNGFr reporter are used to indicate the percentage of cells that incorporated the siNA constract.
- randomized region is meant a region of completely random sequence and/or partially random sequence.
- completely random sequence is meant a sequence wherein theoretically there is equal representation of A, T, G and C nucleotides or modified derivatives thereof, at each position in the sequence.
- partially random sequence is meant a sequence wherein there is an unequal representation of A, T, G and C nucleotides or modified derivatives thereof, at each position in the sequence.
- a partially random sequence can therefore have one or more positions of complete randomness and one or more positions with defined nucleotides.
- Sorting of siNA library-containing cells is performed to enrich for cells that produce less reporter GFP after treatment with the promoter inducers PCM and PMA.
- siNA can directly target the mucin/GFP transcript resulting in reduced GFP expression.
- Cells are seeded at a certain density, such as 1 x 10 6 per 150 cm 2 style cell culture flasks and grown in the appropriate cell culture medium with fetal bovine serum.
- the cell culture medium is replaced with serum-free medium.
- the cells are treated with serum-containing medium supplemented with PCM (to 40%) and PMA (to 50 nM) to induced GFP production.
- PCM serum-containing medium
- PMA to 50 nM
- cells are monitored for GFP level on, for example, a FACStar Plus cell sorter. Sorting is performed if ⁇ 90% of siNA library cells from an unsorted control sample were induced to produce GFP above background levels. Two cell fractions are collected in each round of sorting. Following the appropriate round of sorting, the Ml fraction is selected to generate a database of siNA molecules present in the sorted cells.
- Genomic DNA is obtained from sorted siNA library cells by standard methods. Nested polymerase chain reaction (PCR) primers that hybridized to the retroviral vector 5' and 3' of the siNA are used to recover and amplify the siNA sequences from the particular clone of library cell DNA. The PCR product is ligated into a bacterial cloning vector. The recovered siNA library in plasmid form can be used to generate a database of siNA sequences. For example, the library is cloned into E. coli. DNA is prepared by plasmid isolation from bacterial colonies or by direct colony PCR and siNA sequence is determined. A second method can use the siNA library to transfect cloned cells.
- PCR polymerase chain reaction
- TST Target Sequence Tag
- Bioinformatics The antisense region sequences of the isolated siNA constracts are compared to public and private gene data banks. Gene matches are compiled according to perfect and imperfect matches. Potential gene targets are categorized by the number of different siNA sequences matching each gene. Genes with more than one perfect siNA match are selected for Target Validation studies.
- siNA reagents are designed to the target gene cDNA sequence from Genbank.
- the siNA reagents are complexed with a cationic lipid formulation prior to administration to cloned cells at appropriate concentrations (e.g. 5-50 nM or less).
- Cells are treated with siNA reagents, for example from 72 to 96 hours.
- PCM to 40 %
- PMA to 50 nM
- Reduced GFP expression in siNA treated cells is taken as evidence for validation of the target gene. Knockdown of target RNA in siNA treated cells can co ⁇ elate with reduced endogenous RNA and reduced GFP RNA to complete validation of the target.
- Example 16 Screening siNA constracts for improved pharmacokineti.es
- siNA constracts are screened in vivo for improved pharmacokinetic properties compared to all RNA or unmodified siNA constracts.
- Chemical modifications are introduced into the siNA constract based on educated design parameters (e.g. introducing 2'-mofications, base modifications, backbone modifications, terminal cap modifications, or covalently attached conjugates etc).
- the modified constmct in tested in an appropriate system (e.g human serum for nuclease resistance, shown, or an animal model for PK/delivery parameters).
- the siNA constract is tested for RNAi activity, for example in a cell culture system such as a luciferase reporter assay).
- siNA constracts are then identified which possess a particular characteristic while maintaining RNAi activity, and can be further modified and assayed once again. This same approach can be used to identify siNA-conjugate molecules with improved phannacokinetic profiles, delivery, localized delivery, cellular uptake, and RNAi activity.
- siNA molecules of the invention can be used to treat a variety of diseases and conditions through modulation of gene expression.
- chemically modified siNA molecules can be designed to modulate the expression any number of target genes, including but not limited to genes associated with cancer, metabolic diseases, infectious diseases such as viral, bacterial or fungal infections, neurologic diseases, musculoskeletal diseases, diseases of the immune system, diseases associated with signaling pathways and cellular messengers, and diseases associated with transport systems including molecular pumps and channels.
- Non-limiting examples of various viral genes that can be targeted using siNA molecules of the invention include Hepatitis C Vims (HCV, for example Genbank Accession Nos: D11168, D50483.1, L38318 and S82227), Hepatitis B Virus (HBV, for example GenBank Accession No. AF100308.1), Human Immunodeficiency Virus type 1 (HIN-1, for example GenBank Accession No. U51188), Human Immunodeficiency Virus type 2 (HIV-2, for example GenBank Accession No. X60667), West Nile Virus (WNV for example GenBank accession No. NC_001563), cytomegaloviras (CMV for example GenBank Accession No.
- NC_001347 respiratory syncytial virus
- RSV respiratory syncytial virus
- influenza virus for example GenBank Accession No. AF037412
- rhinoviras for example, GenBank accession numbers: D00239, X02316, X01087, L24917, M16248, K02121, X01087)
- papillomaviras for example GenBank Accession No. NC_001353
- Herpes Simplex Virus HSV for example GenBank Accession No. NC_001345
- other viruses such as HTLV (for example GenBank Accession No. AJ430458).
- siNA molecules Due to the high sequence variability of many viral genomes, selection of siNA molecules for broad therapeutic applications would likely involve the conserved regions of the viral genome.
- conserved regions of the viral genomes include but are not limited to 5 '-Non Coding Regions (NCR), 3'- Non Coding Regions (NCR) LTR regions and/or internal ribosome entry sites (TRES).
- NCR Nonlimiting examples of conserved regions of the viral genomes include but are not limited to 5 '-Non Coding Regions (NCR), 3'- Non Coding Regions (NCR) LTR regions and/or internal ribosome entry sites (TRES).
- siNA molecules designed against conserved regions of various viral genomes will enable efficient inhibition of viral replication in diverse patient populations and may ensure the effectiveness of the siNA molecules against viral quasi species which evolve due to mutations in the non-conserved regions of the viral genome.
- Non-limiting examples of human genes that can be targeted using siNA molecules of the invention using methods described herein include any human RNA sequence, for example those commonly refe ⁇ ed to by Genbank Accession Number. These RNA sequences can be used to design siNA molecules that inhibit gene expression and therefore abrogate diseases, conditions, or infections associated with expression of those genes.
- Such non-limiting examples of human genes that can be targeted using siNA molecules of the invention include VEGFr (VEGFRl for example GenBank Accession No. XM_067723, VEGFR2 for example GenBank Accession No.
- telomere for example GenBank Accession No. NM_003219
- telomerase RNA for example GenBank Accession No. U86046
- NFkappaB for example GenBank Accession No. NM_005228
- NOGO for example GenBank Accession No. AB020693
- NOGOr for example GenBank Accession No. XM_015620
- RAS for example GenBank Accession No.
- RAF for example GenBank Accession No. XMJ333884
- CD20 for example GenBank Accession No. X07203
- METAP2 for example GenBank Accession No. NM_003219
- CLCA1 for example GenBank Accession No. NM_001285
- phospholamban for example GenBank Accession No. NM_002667
- PTP1B for example GenBank Accession No. M31724
- PCNA for example GenBank Accession No. NM_002592.1
- PKC-alpha for example GenBank Accession No. NM 002737
- the genes described herein are provided as non-limiting examples of genes that can be targeted using siNA molecules of the invention. Additional examples of such genes are described by accession number in Beigelman et al, USSN 60/363,124, filed March 11, 2002 and incorporated by reference herein in its entirety.
- siNA molecule of the mvention can also be used in a variety of agricultural applications involving modulation of endogenous or exogenous gene expression in plants using siNA, including use as insecticidal, antiviral and anti-fungal agents or modulate plant traits such as oil and starch profiles and stress resistance.
- siNA molecules of the invention can be used in a variety of diagnostic applications, such as in the identification of molecular targets (e.g., RNA) in a variety of applications, for example, in clinical, industrial, environmental, agricultural and/or research settings.
- diagnostic use of siNA molecules involves utilizing reconstituted
- siNA molecules of this invention can be used as diagnostic tools to examine genetic drift and mutations within diseased cells or to detect the presence of endogenous or exogenous, for example viral, RNA in a cell.
- the close relationship between siNA activity and the stracture of the target RNA allows the detection of mutations in any region of the molecule, which alters the base-pairing and three-dimensional stracture of the target
- RNA By using multiple siNA molecules described in this invention, one can map nucleotide changes, which are important to RNA stracture and function in vitro, as well as in cells and tissues. Cleavage of target RNAs with siNA molecules can be used to inhibit gene expression and define the role of specified gene products in the progression of disease or infection. In this manner, other genetic targets can be defined as important mediators of the disease. These experiments will lead to better treatment of the disease progression by affording the possibility of combination therapies (e.g., multiple siNA molecules targeted to different genes, siNA molecules coupled with known small molecule inhibitors, or intermittent treatment with combinations siNA molecules and/or other chemical or biological molecules).
- combination therapies e.g., multiple siNA molecules targeted to different genes, siNA molecules coupled with known small molecule inhibitors, or intermittent treatment with combinations siNA molecules and/or other chemical or biological molecules.
- siNA molecules of this invention include detection of the presence of mRNAs associated with a disease, infection, or related condition. Such RNA is detected by determining the presence of a cleavage product after treatment with a siNA using standard methodologies, for example, fluorescence resonance emission transfer (FRET).
- FRET fluorescence resonance emission transfer
- siNA molecules that cleave only wild-type or mutant forms of the target RNA are used for the assay.
- the first siNA molecules i.e., those that cleave only wild-type forms of target RNA
- the second siNA molecules i.e., those that cleave only mutant forms of target RNA
- synthetic substrates of both wild-type and mutant RNA are cleaved by both siNA molecules to demonstrate the relative siNA efficiencies in the reactions and the absence of cleavage of the "non-targeted" RNA species.
- the cleavage products from the synthetic substrates also serve to generate size markers for the analysis of wild-type and mutant RNAs in the sample population.
- each analysis requires two siNA molecules, two substrates and one unknown sample, which is combined into six reactions.
- the presence of cleavage products is determined using an RNase protection assay so that full-length and cleavage fragments of each RNA can be analyzed in one lane of a polyacrylamide gel. It is not absolutely required to quantify the results to gain insight into the expression of mutant RNAs and putative risk of the desired phenotypic changes in target cells.
- the expression of mRNA whose protein product is implicated in the development of the phenotype is adequate to establish risk.
- Tandem synthesis utilizes double coupling of linker molecule Table III Table IV
- Non-limiting examples of Stabilization Chemistries for chemically modified siNA constructs CAP any terminal cap, see for example Figure 10.
- All Stab 1-11 chemistries can comprise 3 '-terminal thymidine (TT) residues
- All Stab 1-11 chemistries typically comprise 21 nucleotides, but can vary as described herein.
Abstract
Description
Claims
Priority Applications (231)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| JP2003569811A JP2005517438A (en) | 2002-02-20 | 2003-02-20 | RNA interference-mediated inhibition of gene expression using chemically modified short interfering nucleic acids (siNA) |
| GB0406022A GB2397818B (en) | 2002-02-20 | 2003-02-20 | Rna interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| EP03716126A EP1458741B1 (en) | 2002-02-20 | 2003-02-20 | Rna interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid |
| CA002459532A CA2459532A1 (en) | 2002-02-20 | 2003-02-20 | Rna interference by modified short interfering nucleic acid |
| AU2003221258A AU2003221258B2 (en) | 2002-02-20 | 2003-02-20 | RNA interference by modified short interfering nucleic acid |
| US10/424,339 US20060127891A1 (en) | 2002-02-20 | 2003-04-25 | RNA interference mediated inhibition of MAP kinase gene expression or expression of genes involved in MAP kinase pathway using short interfering nucleic acid (siNA) |
| US10/427,160 US7833992B2 (en) | 2001-05-18 | 2003-04-30 | Conjugates and compositions for cellular delivery |
| US10/444,853 US8202979B2 (en) | 2002-02-20 | 2003-05-23 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid |
| PCT/US2003/018911 WO2003106476A1 (en) | 2002-02-20 | 2003-06-16 | Nucleic acid mediated inhibition of enterococcus infection and cytolysin toxin activity |
| US10/607,933 US20080032942A1 (en) | 2000-08-30 | 2003-06-27 | RNA interference mediated treatment of Alzheimer's disease using short interfering nucleic acid (siNA) |
| US10/652,791 US20050106726A1 (en) | 2002-02-20 | 2003-08-29 | RNA interference mediated inhibition of platelet-derived endothelial cell growth factor (ECGF1) gene expression using short interfering nucleic acid (siNA) |
| US10/693,059 US20080039414A1 (en) | 2002-02-20 | 2003-10-23 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US10/720,448 US8273866B2 (en) | 2002-02-20 | 2003-11-24 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (SINA) |
| US10/724,270 US20050080031A1 (en) | 2001-05-18 | 2003-11-26 | Nucleic acid treatment of diseases or conditions related to levels of Ras, HER2 and HIV |
| US10/727,780 US20050233329A1 (en) | 2002-02-20 | 2003-12-03 | Inhibition of gene expression using duplex forming oligonucleotides |
| US10/757,803 US20050020525A1 (en) | 2002-02-20 | 2004-01-14 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US10/780,447 US7491805B2 (en) | 2001-05-18 | 2004-02-13 | Conjugates and compositions for cellular delivery |
| US10/783,128 US20050096284A1 (en) | 2002-02-20 | 2004-02-20 | RNA interference mediated treatment of polyglutamine (polyQ) repeat expansion diseases using short interfering nucleic acid (siNA) |
| US10/798,090 US20050014172A1 (en) | 2002-02-20 | 2004-03-11 | RNA interference mediated inhibition of muscarinic cholinergic receptor gene expression using short interfering nucleic acid (siNA) |
| US10/800,487 US20050048529A1 (en) | 2002-02-20 | 2004-03-15 | RNA interference mediated inhibition of intercellular adhesion molecule (ICAM) gene expression using short interfering nucleic acid (siNA) |
| US10/824,036 US20050191638A1 (en) | 2002-02-20 | 2004-04-14 | RNA interference mediated treatment of polyglutamine (polyQ) repeat expansion diseases using short interfering nucleic acid (siNA) |
| US10/825,485 US20060160757A1 (en) | 2002-02-20 | 2004-04-15 | RNA interference mediated inhibition hairless of (HR) gene expression using short interfering nucleic acid (siNA) |
| US10/826,966 US20050032733A1 (en) | 2001-05-18 | 2004-04-16 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (SiNA) |
| US10/830,569 US20050054598A1 (en) | 2002-02-20 | 2004-04-23 | RNA interference mediated inhibition hairless (HR) gene expression using short interfering nucleic acid (siNA) |
| US10/831,620 US20050148530A1 (en) | 2002-02-20 | 2004-04-23 | RNA interference mediated inhibition of vascular endothelial growth factor and vascular endothelial growth factor receptor gene expression using short interfering nucleic acid (siNA) |
| US10/832,522 US20050233996A1 (en) | 2002-02-20 | 2004-04-26 | RNA interference mediated inhibition of hairless (HR) gene expression using short interfering nucleic acid (siNA) |
| US10/840,731 US20050137153A1 (en) | 2002-02-20 | 2004-05-06 | RNA interference mediated inhibition of alpha-1 antitrypsin (AAT) gene expression using short interfering nucleic acid (siNA) |
| US10/844,076 US7176304B2 (en) | 2002-02-20 | 2004-05-11 | RNA interference mediated inhibition of vascular endothelial growth factor and vascular endothelial growth factor receptor gene expression using short interfering nucleic acid (siNA) |
| US10/844,072 US20050159376A1 (en) | 2002-02-20 | 2004-05-12 | RNA interference mediated inhibition 5-alpha reductase and androgen receptor gene expression using short interfering nucleic acid (siNA) |
| US10/557,542 US20070032441A1 (en) | 2001-05-18 | 2004-05-24 | Rna interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (sina) |
| US10/861,060 US20050137155A1 (en) | 2001-05-18 | 2004-06-03 | RNA interference mediated treatment of Parkinson disease using short interfering nucleic acid (siNA) |
| US10/864,044 US20050171040A1 (en) | 2001-05-18 | 2004-06-09 | RNA interference mediated inhibition of cholesteryl ester transfer protein (CEPT) gene expression using short interfering nucleic acid (siNA) |
| US10/863,973 US20050143333A1 (en) | 2001-05-18 | 2004-06-09 | RNA interference mediated inhibition of interleukin and interleukin receptor gene expression using short interfering nucleic acid (SINA) |
| US10/869,638 US20050119211A1 (en) | 2001-05-18 | 2004-06-16 | RNA mediated inhibition connexin gene expression using short interfering nucleic acid (siNA) |
| US10/871,222 US20050119212A1 (en) | 2001-05-18 | 2004-06-18 | RNA interference mediated inhibition of FAS and FASL gene expression using short interfering nucleic acid (siNA) |
| US10/562,561 US20060247194A1 (en) | 2000-08-30 | 2004-06-25 | Rna interference mediated treatment of alzheimer's disease using short interfering nucleic acid (sina) |
| US10/877,889 US20050209179A1 (en) | 2000-08-30 | 2004-06-25 | RNA interference mediated treatment of Alzheimer's disease using short interfering nucleic acid (siNA) |
| US10/879,867 US20050124566A1 (en) | 2001-05-18 | 2004-06-28 | RNA interference mediated inhibition of myostatin gene expression using short interfering nucleic acid (siNA) |
| US10/881,118 US20050130181A1 (en) | 2001-05-18 | 2004-06-30 | RNA interference mediated inhibition of wingless gene expression using short interfering nucleic acid (siNA) |
| US10/881,580 US20060142225A1 (en) | 2001-05-18 | 2004-06-30 | RNA interference mediated inhibition of cyclin dependent kinase-2 (CDK2) gene expression using short interfering nucleic acid (siNA) |
| US10/883,218 US20050124567A1 (en) | 2001-05-18 | 2004-07-01 | RNA interference mediated inhibition of TRPM7 gene expression using short interfering nucleic acid (siNA) |
| US10/888,226 US20050124568A1 (en) | 2001-05-18 | 2004-07-09 | RNA interference mediated inhibition of acetyl-CoA-carboxylase gene expression using short interfering nucleic acid (siNA) |
| US10/888,269 US20050222064A1 (en) | 2002-02-20 | 2004-07-09 | Polycationic compositions for cellular delivery of polynucleotides |
| US10/893,010 US20050164224A1 (en) | 2001-05-18 | 2004-07-16 | RNA interference mediated inhibition of cyclin D1 gene expression using short interfering nucleic acid (siNA) |
| US10/892,922 US20050124569A1 (en) | 2001-05-18 | 2004-07-16 | RNA interference mediated inhibition of CXCR4 gene expression using short interfering nucleic acid (siNA) |
| US10/894,475 US20050070497A1 (en) | 2001-05-18 | 2004-07-19 | RNA interference mediated inhibtion of tyrosine phosphatase-1B (PTP-1B) gene expression using short interfering nucleic acid (siNA) |
| US10/897,902 US20050176663A1 (en) | 2001-05-18 | 2004-07-23 | RNA interference mediated inhibition of protein tyrosine phosphatase type IVA (PRL3) gene expression using short interfering nucleic acid (siNA) |
| US10/898,311 US20050277608A1 (en) | 2001-05-18 | 2004-07-23 | RNA interference mediated inhibtion of vitamin D receptor gene expression using short interfering nucleic acid (siNA) |
| US10/898,660 US20050196765A1 (en) | 2001-05-18 | 2004-07-23 | RNA interference mediated inhibition of checkpoint Kinase-1 (CHK-1) gene expression using short interfering nucleic acid (siNA) |
| US10/903,128 US20050182006A1 (en) | 2001-05-18 | 2004-07-30 | RNA interference mediated inhibition of protein kinase C alpha (PKC-alpha) gene expression using short interfering nucleic acid (siNA) |
| US10/567,888 US20070093437A1 (en) | 2001-05-18 | 2004-08-06 | Rna interference mediated inhibition of xiap gene expression using short interfering nucleic acid (sina) |
| US10/915,896 US20050159378A1 (en) | 2001-05-18 | 2004-08-11 | RNA interference mediated inhibition of Myc and/or Myb gene expression using short interfering nucleic acid (siNA) |
| US10/916,030 US20050159379A1 (en) | 2001-05-18 | 2004-08-11 | RNA interference mediated inhibition of gastric inhibitory polypeptide (GIP) and gastric inhibitory polypeptide receptor (GIPR) gene expression using short interfering nucleic acid (siNA) |
| US10/916,095 US20050158735A1 (en) | 2001-05-18 | 2004-08-11 | RNA interference mediated inhibition of proliferating cell nuclear antigen (PCNA) gene expression using short interfering nucleic acid (siNA) |
| US10/918,896 US20050164966A1 (en) | 2001-05-18 | 2004-08-16 | RNA interference mediated inhibition of type 1 insulin-like growth factor receptor gene expression using short interfering nucleic acid (siNA) |
| US10/918,987 US20050203040A1 (en) | 2001-05-18 | 2004-08-16 | RNA interference mediated inhibition of vascular cell adhesion molecule (VCAM) gene expression using short interfering nucleic acid (siNA) |
| US10/918,969 US20050153914A1 (en) | 2001-05-18 | 2004-08-16 | RNA interference mediated inhibition of MDR P-glycoprotein gene expression using short interfering nucleic acid (siNA) |
| US10/919,866 US20050176664A1 (en) | 2001-05-18 | 2004-08-17 | RNA interference mediated inhibition of cholinergic muscarinic receptor (CHRM3) gene expression using short interfering nucleic acid (siNA) |
| US10/919,584 US20050233997A1 (en) | 2001-05-18 | 2004-08-17 | RNA interference mediated inhibition of matrix metalloproteinase 13 (MMP13) gene expression using short interfering nucleic acid (siNA) |
| US10/919,964 US20050176665A1 (en) | 2001-05-18 | 2004-08-17 | RNA interference mediated inhibition of hairless (HR) gene expression using short interfering nucleic acid (siNA) |
| US10/922,544 US20050153915A1 (en) | 2001-05-18 | 2004-08-19 | RNA interference mediated inhibition of early growth response gene expression using short interfering nucleic acid (siNA) |
| US10/923,640 US20050136436A1 (en) | 2001-05-18 | 2004-08-19 | RNA interference mediated inhibition of G72 and D-amino acid oxidase (DAAO) gene expression using short interfering nucleic acid (siNA) |
| US10/922,034 US20050164967A1 (en) | 2001-05-18 | 2004-08-19 | RNA interference mediated inhibition of platelet-derived endothelial cell growth factor (ECGF1) gene expression using short interfering nucleic acid (siNA) |
| US10/922,626 US20050159380A1 (en) | 2001-05-18 | 2004-08-19 | RNA interference mediated inhibition of angiopoietin gene expression using short interfering nucleic acid (siNA) |
| US10/923,580 US20050159382A1 (en) | 2001-05-18 | 2004-08-19 | RNA interference mediated inhibition of polycomb group protein EZH2 gene expression using short interfering nucleic acid (siNA) |
| US10/921,554 US20060142226A1 (en) | 2001-05-18 | 2004-08-19 | RNA interference mediated inhibition of cholesteryl ester transfer protein (CETP) gene expression using short interfering nucleic acid (siNA) |
| US10/923,476 US20050288242A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of RAS gene expression using short interfering nucleic acid (siNA) |
| US10/922,761 US20050267058A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of placental growth factor gene expression using short interfering nucleic acid (sINA) |
| US10/923,330 US20050153916A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of telomerase gene expression using short interfering nucleic acid (siNA) |
| US10/923,181 US20050187174A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of intercellular adhesion molecule (ICAM) gene expression using short interfering nucleic acid (siNA) |
| US10/923,473 US20050191618A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of human immunodeficiency virus (HIV) gene expression using short interfering nucleic acid (siNA) |
| US10/923,516 US20050176025A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of B-cell CLL/Lymphoma-2 (BCL-2) gene expression using short interfering nucleic acid (siNA) |
| US10/923,329 US20050164968A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of ADAM33 gene expression using short interfering nucleic acid (siNA) |
| US10/922,340 US20050170371A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of 5-alpha reductase and androgen receptor gene expression using short interfering nucleic acid (siNA) |
| US10/923,270 US20050233344A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of platelet derived growth factor (PDGF) and platelet derived growth factor receptor (PDGFR) gene expression using short interfering nucleic acid (siNA) |
| US10/923,470 US20050227935A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of TNF and TNF receptor gene expression using short interfering nucleic acid (siNA) |
| US10/923,142 US20050182008A1 (en) | 2000-02-11 | 2004-08-20 | RNA interference mediated inhibition of NOGO and NOGO receptor gene expression using short interfering nucleic acid (siNA) |
| US10/923,380 US20050196767A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of GRB2 associated binding protein (GAB2) gene expression using short interfering nucleic acis (siNA) |
| US10/922,675 US20050182007A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of interleukin and interleukin receptor gene expression using short interfering nucleic acid (SINA) |
| US10/923,182 US20050176666A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of GPRA and AAA1 gene expression using short interfering nucleic acid (siNA) |
| US10/922,554 US20080188430A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of hypoxia inducible factor 1 (HIF1) gene expression using short interfering nucleic acid (siNA) |
| US10/923,115 US20050079610A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of Fos gene expression using short interfering nucleic acid (siNA) |
| US10/923,354 US20050176024A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of epidermal growth factor receptor (EGFR) gene expression using short interfering nucleic acid (siNA) |
| US10/923,379 US20050239731A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of MAP kinase gene expression using short interfering nucleic acid (siNA) |
| US10/923,522 US20050159381A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of chromosome translocation gene expression using short interfering nucleic acid (siNA) |
| US10/923,475 US20050227936A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of TGF-beta and TGF-beta receptor gene expression using short interfering nucleic acid (siNA) |
| US10/923,451 US20050256068A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of stearoyl-CoA desaturase (SCD) gene expression using short interfering nucleic acid (siNA) |
| US10/923,536 US20070042983A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of gene expression using short interfering nucleic acid (siNA) |
| US10/923,201 US20050182009A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of NF-Kappa B / REL-A gene expression using short interfering nucleic acid (siNA) |
| US10/942,560 US20050209180A1 (en) | 2001-05-18 | 2004-09-15 | RNA interference mediated inhibition of hepatitis C virus (HCV) expression using short interfering nucleic acid (siNA) |
| US10/944,611 US20050233998A1 (en) | 2001-05-18 | 2004-09-16 | RNA interference mediated inhibition of vascular endothelial growth factor and vascular endothelial growth factor receptor gene expression using short interfering nucleic acid (siNA) |
| US10/962,898 US20050222066A1 (en) | 2001-05-18 | 2004-10-12 | RNA interference mediated inhibition of vascular endothelial growth factor and vascular endothelial growth factor receptor gene expression using short interfering nucleic acid (siNA) |
| HK04107996.5A HK1065049A1 (en) | 2002-02-20 | 2004-10-15 | Rna interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid |
| US10/981,966 US20050266422A1 (en) | 2002-02-20 | 2004-11-05 | Fluoroalkoxy, nucleosides, nucleotides, and polynucleotides |
| US11/001,347 US20050261219A1 (en) | 2001-05-18 | 2004-12-01 | RNA interference mediated inhibition of interleukin and interleukin receptor gene expression using short interfering nucleic acid (siNA) |
| US11/011,913 US20050209182A1 (en) | 2002-02-20 | 2004-12-14 | Nucleic acid mediated inhibition of enterococcus infection and cytolysin toxin activity |
| US11/014,373 US20050196781A1 (en) | 2001-05-18 | 2004-12-15 | RNA interference mediated inhibition of STAT3 gene expression using short interfering nucleic acid (siNA) |
| US11/031,668 US20060019913A1 (en) | 2001-05-18 | 2005-01-06 | RNA interference mediated inhibtion of protein tyrosine phosphatase-1B (PTP-1B) gene expression using short interfering nucleic acid (siNA) |
| US11/035,813 US20060025361A1 (en) | 2001-05-18 | 2005-01-14 | RNA interference mediated inhibition of protein tyrosine phosphatase-1B (PTP-1B) gene expression using short interfering nucleic acid (siNA) |
| US11/054,047 US20050287128A1 (en) | 2001-05-18 | 2005-02-09 | RNA interference mediated inhibition of TGF-beta and TGF-beta receptor gene expression using short interfering nucleic acid (siNA) |
| US11/058,582 US20050260620A1 (en) | 2001-05-18 | 2005-02-15 | RNA interference mediated inhibition of retinolblastoma (RBI) gene expression using short interfering nucleic acid (siNA) |
| US11/063,415 US20050277133A1 (en) | 2001-05-18 | 2005-02-22 | RNA interference mediated treatment of polyglutamine (polyQ) repeat expansion diseases using short interfering nucleic acid (siNA) |
| US11/098,303 US20050282188A1 (en) | 2001-05-18 | 2005-04-04 | RNA interference mediated inhibition of gene expression using short interfering nucleic acid (siNA) |
| US11/140,328 US20060019917A1 (en) | 2001-05-18 | 2005-05-27 | RNA interference mediated inhibition of stromal cell-derived factor-1 (SDF-1) gene expression using short interfering nucleic acid (siNA) |
| US11/205,646 US20080161256A1 (en) | 2001-05-18 | 2005-08-17 | RNA interference mediated inhibition of gene expression using short interfering nucleic acid (siNA) |
| US11/217,936 US20060148743A1 (en) | 2001-05-18 | 2005-09-01 | RNA interference mediated inhibition of histone deacetylase (HDAC) gene expression using short interfering nucleic acid (siNA) |
| US11/234,730 US20070270579A1 (en) | 2001-05-18 | 2005-09-23 | RNA interference mediated inhibition of gene expression using short interfering nucleic acid (siNA) |
| US11/259,603 US20060241075A1 (en) | 2001-05-18 | 2005-10-26 | RNA interference mediated inhibition of desmoglein gene expression using short interfering nucleic acid (siNA) |
| US11/265,730 US20070179104A1 (en) | 2001-05-18 | 2005-11-02 | RNA interference mediated inhibition of winged helix nude (WHN) gene expression using short interfering nucleic acid (siNA) |
| US11/299,254 US20060217331A1 (en) | 2001-05-18 | 2005-12-08 | Chemically modified double stranded nucleic acid molecules that mediate RNA interference |
| US11/299,391 US7517864B2 (en) | 2001-05-18 | 2005-12-09 | RNA interference mediated inhibition of vascular endothelial growth factor and vascular endothelial growth factor receptor gene expression using short interfering nucleic acid (siNA) |
| US11/311,826 US20060211642A1 (en) | 2001-05-18 | 2005-12-19 | RNA inteference mediated inhibition of hepatitis C virus (HVC) gene expression using short interfering nucleic acid (siNA) |
| US11/332,655 US20060276422A1 (en) | 2001-05-18 | 2006-01-13 | RNA interference mediated inhibition of B7-H1 gene expression using short interfering nucleic acid (siNA) |
| US11/358,443 US20060217334A1 (en) | 2002-02-20 | 2006-02-21 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US11/358,540 US20060247428A1 (en) | 2002-02-20 | 2006-02-21 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US11/358,807 US20060217335A1 (en) | 2002-02-20 | 2006-02-21 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US11/358,862 US20060287266A1 (en) | 2002-02-20 | 2006-02-21 | RNA interference mediated ihibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US11/358,932 US20060217337A1 (en) | 2002-02-20 | 2006-02-21 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US11/358,924 US20060247429A1 (en) | 2002-02-20 | 2006-02-21 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US11/358,863 US20060217336A1 (en) | 2002-02-20 | 2006-02-21 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US11/369,108 US20070160980A1 (en) | 2001-05-18 | 2006-03-06 | RNA interference mediated inhibition of gene expression using short interfering nucleic acid (siNA) |
| US11/395,833 US20060287267A1 (en) | 2001-05-18 | 2006-03-31 | RNA interference mediated inhibition of respiratory syncytial virus (RSV) expression using short interfering nucleic acid (siNA) |
| US11/448,115 US20060216747A1 (en) | 2001-05-18 | 2006-06-06 | RNA interference mediated inhibition of checkpoint kinase-1 (CHK-1) gene expression using short interfering nucleic acid (siNA) |
| US11/450,856 US20060270623A1 (en) | 2001-05-18 | 2006-06-09 | RNA interference mediated treatment of polyglutamine (polyQ) repeat expansion diseases using short interfering nucleic acid (siNA) |
| US11/455,205 US20070049543A1 (en) | 2001-05-18 | 2006-06-16 | RNA interference mediated inhibition of 11 beta-hydroxysteroid dehydrogenase-1 (11 beta-HSD-1) gene expression using short interfering nucleic acid siNA |
| US11/487,788 US20070173473A1 (en) | 2001-05-18 | 2006-07-17 | RNA interference mediated inhibition of proprotein convertase subtilisin Kexin 9 (PCSK9) gene expression using short interfering nucleic acid (siNA) |
| US11/488,374 US20080249040A1 (en) | 2001-05-18 | 2006-07-18 | RNA interference mediated inhibition of sterol regulatory element binding protein 1 (SREBP1) gene expression using short interfering nucleic acid (siNA) |
| US11/499,520 US20070004663A1 (en) | 2002-02-20 | 2006-08-04 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US11/499,633 US20070004665A1 (en) | 2002-02-20 | 2006-08-04 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US11/499,521 US7989612B2 (en) | 2002-02-20 | 2006-08-04 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US11/499,529 US7923547B2 (en) | 2002-09-05 | 2006-08-04 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US11/499,828 US7956176B2 (en) | 2002-09-05 | 2006-08-04 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US11/499,533 US20060293272A1 (en) | 2002-02-20 | 2006-08-04 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US11/502,885 US20060292691A1 (en) | 2002-02-20 | 2006-08-11 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US11/502,893 US20060275903A1 (en) | 2002-02-20 | 2006-08-11 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US11/502,875 US20070004667A1 (en) | 2002-02-20 | 2006-08-11 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US11/502,876 US20060281175A1 (en) | 2002-02-20 | 2006-08-11 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US11/592,039 US20070185049A1 (en) | 2001-05-18 | 2006-11-02 | RNA interference mediated inhibition of histone deacetylase (HDAC) gene expression using short interfering nucleic acid (siNA) |
| US11/676,124 US8846894B2 (en) | 2002-02-20 | 2007-02-16 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US11/684,465 US20070161596A1 (en) | 2000-08-30 | 2007-03-09 | RNA INTERFERENCE MEDIATED TREATMENT OF ALZHEIMER'S DISEASE USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US11/748,029 US20090239931A1 (en) | 2001-05-18 | 2007-05-14 | RNA INTERFERENCE MEDIATED INHIBITION OF PROTEIN TYROSINE PHOSPHATASE-1B (PTP-1B) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US11/756,240 US20090299045A1 (en) | 2001-05-18 | 2007-05-31 | RNA Interference Mediated Inhibition Of Interleukin and Interleukin Gene Expression Using Short Interfering Nucleic Acid (siNA) |
| US11/879,470 US20080033156A1 (en) | 2002-02-20 | 2007-07-17 | Polycationic compositions for cellular delivery of polynucleotides |
| US11/851,090 US20080249294A1 (en) | 2001-05-18 | 2007-09-06 | RNA Interference Mediated Inhibition of Gene Expression Using Short Interfering Nucleic Acid (siNA) |
| US12/105,010 US8232383B2 (en) | 2002-02-20 | 2008-04-17 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US12/167,045 US20090137509A1 (en) | 2002-02-20 | 2008-07-02 | RNA INTERFERENCE MEDIATED INHIBITION OF PROLIFERATION CELL NUCLEAR ANTIGEN (PCNA) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US12/167,012 US7928218B2 (en) | 2002-02-20 | 2008-07-02 | RNA interference mediated inhibition of polycomb group protein EZH2 gene expression using short interfering nucleic acid (siNA) |
| US12/166,991 US20090137507A1 (en) | 2002-02-20 | 2008-07-02 | RNA INTERFERENCE MEDIATED INHIBITION OF ANGIOPOIETIN GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US12/169,519 US20090156533A1 (en) | 2001-05-18 | 2008-07-08 | RNA INTERFERENCE MEDIATED INHIBITION OF STROMAL CELL-DERIVED FACTOR-1 (SDF-1) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US12/170,290 US7662951B2 (en) | 2000-08-30 | 2008-07-09 | RNA interference mediated treatment of Alzheimer's disease using short interfering nucleic acid (siNA) |
| US12/170,310 US7897752B2 (en) | 2002-02-20 | 2008-07-09 | RNA interference mediated inhibition of telomerase gene expression using short interfering nucleic acid (siNA) |
| US12/171,111 US7928219B2 (en) | 2002-02-20 | 2008-07-10 | RNA interference mediated inhibition of placental growth factor gene expression using short interfering nucleic acid (SINA) |
| US12/171,034 US20090137510A1 (en) | 2002-02-20 | 2008-07-10 | RNA INTERFERENCE MEDIATED INHIBITION OF NF-KAPPA B/ REL-A GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US12/174,526 US7691999B2 (en) | 2002-02-20 | 2008-07-16 | RNA interference mediated inhibition of NOGO and NOGO receptor gene expression using short interfering nucleic acid (siNA) |
| US12/175,392 US20090192105A1 (en) | 2002-02-20 | 2008-07-17 | RNA INTERFERENCE MEDIATED INHIBITION OF INTERCELLULAR ADHESION MOLECULE (ICAM) GENE EXPRESSION USING SHORT INTERFERING NUCELIC ACID (siNA) |
| US12/175,385 US7659389B2 (en) | 2001-05-18 | 2008-07-17 | RNA interference mediated inhibition of MYC and/or MYB gene expression using short interfering nucleic acid (siNA) |
| US12/175,367 US7795422B2 (en) | 2002-02-20 | 2008-07-17 | RNA interference mediated inhibition of hypoxia inducible factor 1 (HIF1) gene expression using short interfering nucleic acid (siNA) |
| US12/185,652 US7910724B2 (en) | 2002-02-20 | 2008-08-04 | RNA interference mediated inhibition of Fos gene expression using short interfering nucleic acid (siNA) |
| US12/185,678 US20090099117A1 (en) | 2002-02-20 | 2008-08-04 | RNA INTERFERENCE MEDIATED INHIBITION OF MYOSTATIN GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US12/192,869 US20090099119A1 (en) | 2001-05-18 | 2008-08-15 | RNA INTERFERENCE MEDIATED INHIBITION OF RAS GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US12/192,853 US8017761B2 (en) | 2001-05-18 | 2008-08-15 | RNA interference mediated inhibition of Stearoyl-CoA desaturase (SCD) gene expression using short interfering nucelic acid (siNA) |
| US12/192,878 US20090247613A1 (en) | 2002-02-20 | 2008-08-15 | RNA INTERFERENCE MEDIATED INHIBITION OF B-CELL CLL/LYMPHOMA-2 (BCL2) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US12/200,736 US8067575B2 (en) | 2002-02-20 | 2008-08-28 | RNA interference mediated inhibition of cyclin D1 gene expression using short interfering nucleic acid (siNA) |
| US12/200,750 US20090137513A1 (en) | 2002-02-20 | 2008-08-28 | RNA Interference Mediated Inhibition of Acetyl-CoA-Carboxylase Gene Expression Using Short Interfering Nucleic Acid (siNA) |
| US12/201,759 US20090023676A1 (en) | 2002-02-20 | 2008-08-29 | RNA Interference Mediated Inhibition of MAP Kinase Gene Expression or Expression of Genes Involved in MAP Kinase Pathway Using Short Interfering Nucleic Acid (SiNA) |
| US12/203,029 US7662952B2 (en) | 2002-02-20 | 2008-09-02 | RNA interference mediated inhibition of GRB2 associated binding protein (GAB2) gene expression using short interfering nucleic acid (siNA) |
| US12/203,055 US7700760B2 (en) | 2002-02-20 | 2008-09-02 | RNA interference mediated inhibition of vascular cell adhesion molecule (VCAM) gene expression using short interfering nucleic acid (siNA) |
| US12/203,669 US7667029B2 (en) | 2002-02-20 | 2008-09-03 | RNA interference mediated inhibition of checkpoint kinase-1 (CHK-1) gene expression using short interfering nucleic acid (siNA) |
| US12/203,707 US7683165B2 (en) | 2002-02-20 | 2008-09-03 | RNA interference mediated inhibition of interleukin and interleukin receptor gene expression using short interfering nucleic acid (siNA) |
| US12/203,731 US7659390B2 (en) | 2002-02-20 | 2008-09-03 | RNA interference mediated inhibition of muscarinic colinergic receptor gene expression using short interfering nucleic acid (siNA) |
| US12/204,612 US7667030B2 (en) | 2002-02-20 | 2008-09-04 | RNA interference mediated inhibition of matrix metalloproteinase 13 (MMP13) gene expression using short interfering nucleic acid (siNA) |
| US12/204,637 US7683166B2 (en) | 2002-02-20 | 2008-09-04 | RNA interference mediated inhibition of interleukin and interleukin receptor gene expression using short interfering nucleic acid (siNA) |
| US12/204,572 US7678897B2 (en) | 2002-02-20 | 2008-09-04 | RNA interference mediated inhibition of platelet-derived endothelial cell growth factor (ECGF1) gene expression using short interfering nucleic acid (siNA) |
| US12/205,113 US7897753B2 (en) | 2002-02-20 | 2008-09-05 | RNA interference mediated inhibition of XIAP gene expression using short interfering nucleic acid (siNA) |
| US12/205,558 US20090093439A1 (en) | 2002-02-20 | 2008-09-05 | RNA INTERFERENCE MEDIATED INHIBITION OF CHROMOSOME TRANSLOCATION GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US12/247,971 US8258288B2 (en) | 2002-02-20 | 2008-10-08 | RNA interference mediated inhibition of respiratory syncytial virus (RSV) expression using short interfering nucleic acid (siNA) |
| US12/253,156 US7964578B2 (en) | 2001-05-18 | 2008-10-16 | Conjugates and compositions for cellular delivery |
| US12/334,181 US20090264504A1 (en) | 2001-05-18 | 2008-12-12 | RNA INTERFERENCE MEDIATED INHIBITION OF HUMAN IMMUNODEFICIENCY VIRUS (HIV) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US12/334,163 US8013143B2 (en) | 2002-02-20 | 2008-12-12 | RNA interference mediated inhibition of CXCR4 gene expression using short interfering nucleic acid (siNA) |
| US12/334,146 US20090306182A1 (en) | 2002-02-20 | 2008-12-12 | RNA INTERFERENCE MEDIATED INHIBITION OF MAP KINASE GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US12/334,224 US20090253774A1 (en) | 2002-02-20 | 2008-12-12 | RNA INTERFERENCE MEDIATED INHIBITION OF PLATELET DERIVED GROWTH FACTOR (PDGF) AND PLATELET DERIVED GROWTH FACTOR RECEPTOR (PDGFR) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US12/334,197 US20090253773A1 (en) | 2002-02-20 | 2008-12-12 | RNA INTERFERENCE MEDIATED INHIBITION OF TNF AND TNF RECEPTOR GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US12/418,477 US7935812B2 (en) | 2002-02-20 | 2009-04-03 | RNA interference mediated inhibition of hepatitis C virus (HCV) expression using short interfering nucleic acid (siNA) |
| US12/635,619 US7893248B2 (en) | 2002-02-20 | 2009-12-10 | RNA interference mediated inhibition of Myc and/or Myb gene expression using short interfering nucleic acid (siNA) |
| US12/635,228 US7858771B2 (en) | 2002-02-20 | 2009-12-10 | RNA interference mediated inhibition of muscarinic colinergic receptor gene expression using short interfering nucleic acid (siNA) |
| US12/640,668 US7956178B2 (en) | 2002-02-20 | 2009-12-17 | RNA interference mediated inhibition of GRB2 associated binding protein (GAB2) gene expression using short interfering nucleic acid (siNA) |
| US12/640,489 US7855284B2 (en) | 2002-02-20 | 2009-12-17 | RNA interference mediated inhibition of checkpoint kinase-1 (CHK-1) gene expression using short interfering nucleic acid (siNA) |
| US12/640,411 US8017765B2 (en) | 2000-08-30 | 2009-12-17 | RNA interference mediated treatment of alzheimer's disease using short interfering nucleic acid (siNA) |
| US12/640,743 US8013146B2 (en) | 2002-02-20 | 2009-12-17 | RNA interference mediated inhibition of matrix metalloproteinase 13 (MMP13) gene expression using short interfering nucleic acid (siNA) |
| US12/683,349 US7910725B2 (en) | 2002-02-20 | 2010-01-06 | RNA interference mediated inhibition of interleukin and interleukin receptor gene expression using short interfering nucleic acid (siNA) |
| US12/683,996 US7923549B2 (en) | 2002-02-20 | 2010-01-07 | RNA interference mediated inhibition of interleukin and interleukin receptor gene expression using short interfering nucleic acid (siNA) |
| US12/689,714 US7897755B2 (en) | 2002-02-20 | 2010-01-19 | RNA interference mediated inhibition of platelet-derived endothelial cell growth factor (ECGF1) gene expression using short interfering nucleic acid (siNA) |
| US12/693,787 US8153778B2 (en) | 2002-02-20 | 2010-01-26 | RNA interference mediated inhibition of vascular cell adhesion molecule (VCAM) gene expression using short interfering nucleic acid (siNA) |
| US12/693,900 US7897756B2 (en) | 2002-02-20 | 2010-01-26 | RNA interference mediated inhibition of NOGO and NOGO receptor gene expression using short interfering nucleic acid (siNA) |
| US12/712,985 US7897757B2 (en) | 2002-02-20 | 2010-02-25 | RNA interference mediated inhibition of protein tyrosine phosphatase-1B (PTP-1B) gene expression using short interfering nucleic acid (siNA) |
| US12/717,511 US20100227911A1 (en) | 2002-02-20 | 2010-03-04 | RNA INTERFERENCE MEDIATED INHIBITION OF CHROMOSOME TRANSLOCATION GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US12/724,651 US7928220B2 (en) | 2002-02-20 | 2010-03-16 | RNA interference mediated inhibition of stromal cell-derived factor-1 (SDF-1) gene expression using short interfering nucleic acid (siNA) |
| US12/748,075 US20100240730A1 (en) | 2002-02-20 | 2010-03-26 | RNA Interference Mediated Inhibition of Gene Expression Using Chemically Modified Short Interfering Nucleic Acid (siNA) |
| US12/765,450 US7977472B2 (en) | 2002-02-20 | 2010-04-22 | RNA interference mediated inhibition of myostatin gene expression using short interfering nucleic acid (siNA) |
| US12/765,818 US20100228018A1 (en) | 2002-02-20 | 2010-04-22 | RNA INTERFERENCE MEDIATED INHIBITION OF PROLIFERATING CELL NUCLEAR ANTIGEN (PCNA) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US12/777,816 US8008473B2 (en) | 2002-02-20 | 2010-05-11 | RNA interference mediated inhibition of TNF and TNF receptor gene expression using short interfering nucleic acid (siNA) |
| US12/777,767 US8008472B2 (en) | 2001-05-29 | 2010-05-11 | RNA interference mediated inhibition of human immunodeficiency virus (HIV) gene expression using short interfering nucleic acid (siNA) |
| US12/777,913 US7985853B2 (en) | 2002-02-20 | 2010-05-11 | RNA interference mediated inhibition of platelet derived growth factor (PDGF) and platelet derived growth factor receptor (PDGFR) gene expression using short interfering nucleic acid (siNA) |
| US12/838,183 US7943757B2 (en) | 2002-02-20 | 2010-07-16 | RNA interference mediated inhibition of intercellular adhesion molecule (ICAM) gene expression using short interfering nucleic acid (siNA) |
| US12/891,634 US20110124853A1 (en) | 2001-05-18 | 2010-09-27 | Conjugates and Compositions for Cellular Delivery |
| US12/949,886 US8076472B2 (en) | 2002-02-20 | 2010-11-19 | RNA interference mediated inhibition of muscarinic colinergic receptor gene expression using short interfering nucleic acid (siNA) |
| US13/081,362 US8242257B2 (en) | 2002-09-05 | 2011-04-06 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US13/081,357 US8236944B2 (en) | 2002-09-05 | 2011-04-06 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US13/105,693 US8268986B2 (en) | 2002-09-05 | 2011-05-11 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US13/165,635 US20120004403A1 (en) | 2002-02-20 | 2011-06-21 | RNA INTERFERENCE MEDIATED INHIBITION OF TNF AND TNF RECEPTOR GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US13/471,857 US8648185B2 (en) | 2002-02-20 | 2012-05-15 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US13/480,655 US8618277B2 (en) | 2002-02-20 | 2012-05-25 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US14/083,525 US20140288148A1 (en) | 2002-02-20 | 2013-11-19 | RNA INTERFERENCE MEDIATED INHIBITION OF GENE EXPRESSION USING CHEMICALLY MODIFIED SHORT INTERFERING NUCLEIC ACID (siNA) |
| US14/458,578 US20150105445A1 (en) | 2001-05-18 | 2014-08-13 | RNA INTERFERENCE MEDIATED INHIBITION OF GENE EXPRESSION USING CHEMICALLY MODIFIED SHORT INTERFERING NUCLEIC ACID (siNA) |
| US14/514,112 US9181551B2 (en) | 2002-02-20 | 2014-10-14 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US14/712,733 US9994853B2 (en) | 2001-05-18 | 2015-05-14 | Chemically modified multifunctional short interfering nucleic acid molecules that mediate RNA interference |
| US14/731,346 US9771588B2 (en) | 2002-02-20 | 2015-06-04 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US14/861,805 US9657294B2 (en) | 2002-02-20 | 2015-09-22 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US15/084,865 US9732344B2 (en) | 2002-02-20 | 2016-03-30 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US15/090,421 US20160272975A1 (en) | 2001-05-18 | 2016-04-04 | Chemically modified short interfering nucleic acid molecules that mediate rna interference |
| US15/483,354 US9738899B2 (en) | 2002-02-20 | 2017-04-10 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US15/617,814 US9957517B2 (en) | 2002-02-20 | 2017-06-08 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US15/637,579 US10000754B2 (en) | 2002-02-20 | 2017-06-29 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US15/677,719 US10351852B2 (en) | 2002-02-20 | 2017-08-15 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US15/977,746 US20180245083A1 (en) | 2002-02-20 | 2018-05-11 | RNA INTERFERENCE MEDIATED INHIBITION OF GENE EXPRESSION USING CHEMICALLY MODIFIED SHORT INTERFERING NUCLEIC ACID (siNA) |
| US16/136,191 US20190010497A1 (en) | 2002-02-20 | 2018-09-19 | RNA INTERFERENCE MEDIATED INHIBITION OF GENE EXPRESSION USING CHEMICALLY MODIFIED SHORT INTERFERING NUCLEIC ACID (siNA) |
| US16/183,530 US10889815B2 (en) | 2002-02-20 | 2018-11-07 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US16/295,274 US20190194658A1 (en) | 2002-02-20 | 2019-03-07 | RNA INTERFERENCE MEDIATED INHIBITION OF GENE EXPRESSION USING CHEMICALLY MODIFIED SHORT INTERFERING NUCLEIC ACID (siNA) |
| US16/381,288 US10662428B2 (en) | 2002-02-20 | 2019-04-11 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US17/105,174 US20220056441A1 (en) | 2002-02-20 | 2020-11-25 | Rna interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (sina) |
| US17/497,747 US20220275366A1 (en) | 2001-05-18 | 2021-10-08 | Rna interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (sina) |
Applications Claiming Priority (14)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| US35858002P | 2002-02-20 | 2002-02-20 | |
| US60/358,580 | 2002-02-20 | ||
| US33612402P | 2002-03-11 | 2002-03-11 | |
| US60/363,124 | 2002-03-11 | ||
| US38678202P | 2002-06-06 | 2002-06-06 | |
| US60/386,782 | 2002-06-06 | ||
| US40678402P | 2002-08-29 | 2002-08-29 | |
| US60/406,784 | 2002-08-29 | ||
| US40837802P | 2002-09-05 | 2002-09-05 | |
| US60/408,378 | 2002-09-05 | ||
| US40929302P | 2002-09-09 | 2002-09-09 | |
| US60/409,293 | 2002-09-09 | ||
| US44012903P | 2003-01-15 | 2003-01-15 | |
| US60/440,129 | 2003-01-15 |
Related Parent Applications (4)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/US2003/005028 Continuation-In-Part WO2003074654A2 (en) | 2000-02-11 | 2003-02-20 | Rna interference mediated inhibition of gene expression using short interfering nucleic acid (sina) |
| US10/444,853 Continuation-In-Part US8202979B2 (en) | 2000-02-11 | 2003-05-23 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid |
| US11/299,391 Continuation-In-Part US7517864B2 (en) | 2001-05-18 | 2005-12-09 | RNA interference mediated inhibition of vascular endothelial growth factor and vascular endothelial growth factor receptor gene expression using short interfering nucleic acid (siNA) |
| US11/499,521 Continuation-In-Part US7989612B2 (en) | 2002-02-20 | 2006-08-04 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
Related Child Applications (18)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/US2003/002510 Continuation-In-Part WO2003072590A1 (en) | 2001-05-18 | 2003-01-28 | Rna interference mediated inhibition of map kinase genes |
| US41701203A Continuation-In-Part | 2000-08-30 | 2003-04-16 | |
| US10/424,339 Continuation-In-Part US20060127891A1 (en) | 2001-05-18 | 2003-04-25 | RNA interference mediated inhibition of MAP kinase gene expression or expression of genes involved in MAP kinase pathway using short interfering nucleic acid (siNA) |
| US10/427,160 Continuation-In-Part US7833992B2 (en) | 2000-02-11 | 2003-04-30 | Conjugates and compositions for cellular delivery |
| US10/444,835 Continuation-In-Part US20040249871A1 (en) | 2003-05-22 | 2003-05-22 | System and method for automatically removing documents from a knowledge repository |
| US10/444,853 Continuation-In-Part US8202979B2 (en) | 2000-02-11 | 2003-05-23 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid |
| US10/607,933 Continuation-In-Part US20080032942A1 (en) | 2000-08-30 | 2003-06-27 | RNA interference mediated treatment of Alzheimer's disease using short interfering nucleic acid (siNA) |
| US10/652,791 Continuation-In-Part US20050106726A1 (en) | 2000-08-30 | 2003-08-29 | RNA interference mediated inhibition of platelet-derived endothelial cell growth factor (ECGF1) gene expression using short interfering nucleic acid (siNA) |
| US10/693,059 Continuation-In-Part US20080039414A1 (en) | 2000-02-11 | 2003-10-23 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US10/724,270 Continuation-In-Part US20050080031A1 (en) | 2001-05-18 | 2003-11-26 | Nucleic acid treatment of diseases or conditions related to levels of Ras, HER2 and HIV |
| US10/757,803 Continuation-In-Part US20050020525A1 (en) | 2000-02-11 | 2004-01-14 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US10/780,447 Continuation-In-Part US7491805B2 (en) | 2000-02-11 | 2004-02-13 | Conjugates and compositions for cellular delivery |
| US10/783,128 Continuation-In-Part US20050096284A1 (en) | 2002-02-20 | 2004-02-20 | RNA interference mediated treatment of polyglutamine (polyQ) repeat expansion diseases using short interfering nucleic acid (siNA) |
| US10/824,036 Continuation-In-Part US20050191638A1 (en) | 2001-05-18 | 2004-04-14 | RNA interference mediated treatment of polyglutamine (polyQ) repeat expansion diseases using short interfering nucleic acid (siNA) |
| US10/826,966 Continuation-In-Part US20050032733A1 (en) | 2000-02-11 | 2004-04-16 | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (SiNA) |
| US10/840,731 Continuation-In-Part US20050137153A1 (en) | 2002-02-20 | 2004-05-06 | RNA interference mediated inhibition of alpha-1 antitrypsin (AAT) gene expression using short interfering nucleic acid (siNA) |
| US10/923,536 Continuation-In-Part US20070042983A1 (en) | 2001-05-18 | 2004-08-20 | RNA interference mediated inhibition of gene expression using short interfering nucleic acid (siNA) |
| US11/011,913 Continuation-In-Part US20050209182A1 (en) | 2002-02-20 | 2004-12-14 | Nucleic acid mediated inhibition of enterococcus infection and cytolysin toxin activity |
Publications (2)
| Publication Number | Publication Date |
|---|---|
| WO2003070918A2 true WO2003070918A2 (en) | 2003-08-28 |
| WO2003070918A3 WO2003070918A3 (en) | 2004-07-08 |
Family
ID=56290382
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/US2003/005346 WO2003070918A2 (en) | 2000-02-11 | 2003-02-20 | Rna interference by modified short interfering nucleic acid |
Country Status (1)
| Country | Link |
|---|---|
| WO (1) | WO2003070918A2 (en) |
Cited By (237)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| EP1423406A2 (en) * | 2002-02-20 | 2004-06-02 | Sirna Therapeutics, Inc. | Rna interference mediated inhibition of gene expression using short interfering nucleic acid (sina) |
| EP1430157A2 (en) * | 2002-02-20 | 2004-06-23 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF HEPATITIS C VIRUS (HCV) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| WO2005021749A1 (en) * | 2003-08-28 | 2005-03-10 | Novartis Ag | Interfering rna duplex having blunt-ends and 3’-modifications |
| WO2005040379A2 (en) * | 2003-10-23 | 2005-05-06 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF RAS GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| WO2005045032A2 (en) * | 2003-10-20 | 2005-05-19 | Sima Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF EARLY GROWTH RESPONSE GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| WO2005045036A2 (en) * | 2002-02-20 | 2005-05-19 | Sirna Therapeutics, Inc. | Rna interference mediated inhibition of hairless (hr) gene expression using short interfering nucleic acid (sina) |
| WO2005045040A2 (en) * | 2003-10-23 | 2005-05-19 | Sirna Therapeutics, Inc. | RNA INTERFERANCE MEDIATED INHIBITION OF CHOLINERGIC MUSCARINIC RECEPTOR (CHRM3) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID /siNA) |
| WO2005045037A2 (en) * | 2003-10-23 | 2005-05-19 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF 5-ALPHA REDUCTASE AND ANDROGEN RECEPTOR GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| WO2005045039A2 (en) * | 2003-10-23 | 2005-05-19 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF INTERCELLULAR ADHESION MOLECULE (ICAM) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| WO2005045041A2 (en) * | 2003-10-23 | 2005-05-19 | Sirna Therapeutics, Inc. | Rna interference mediated inhibition of cholesteryl ester transfer protein (cetp) gene expression using short interfering nucleic acid (sina) |
| WO2005045038A2 (en) * | 2003-10-23 | 2005-05-19 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF GPRA AND AAA1 GENE EXPRESSION USING SHORT NUCLEIC ACID (siNA) |
| WO2005045034A2 (en) * | 2003-10-23 | 2005-05-19 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED TREATMENT OF PARKINSON DISEASE USING SHORT INTERERING NUCLEIC ACID (siNA) |
| WO2005056021A1 (en) * | 2003-12-04 | 2005-06-23 | University Of South Florida | Polynucleotides for reducing respiratory syncytial virus gene expression |
| WO2005059134A1 (en) * | 2003-12-17 | 2005-06-30 | Index Pharmaceuticals Ab | COMPOUNDS AND METHODS FOR RNA INTERFERENCE OF THE p65 SUBUNIT OF NF-KAPPA-B |
| WO2005073378A1 (en) * | 2004-01-30 | 2005-08-11 | Santaris Pharma A/S | MODIFIED SHORT INTERFERING RNA (MODIFIED siRNA) |
| EP1563070A2 (en) * | 2002-11-05 | 2005-08-17 | Isis Pharmaceuticals, Inc. | 2'-substituted oligomeric compounds and compositions for use in gene modulations |
| WO2005080568A1 (en) * | 2004-02-20 | 2005-09-01 | Index Pharmaceuticals Ab | Methods and compositions for treatment or prevention of secondary ischemic injury |
| EP1572128A2 (en) * | 2002-02-20 | 2005-09-14 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF HIV GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| EP1580264A1 (en) * | 2002-10-29 | 2005-09-28 | Oriental Yeast Co., Ltd. | Construction of knockdown animal by transferring double-stranded rna expression vector |
| WO2005078097A3 (en) * | 2004-02-10 | 2005-10-27 | Sirna Therapeutics Inc | RNA INTERFERENCE MEDIATED INHIBITION OF GENE EXPRESSION USING MULTIFUNCTIONAL SHORT INTERFERING NUCLEIC ACID (Multifunctional siNA) |
| WO2005014806A3 (en) * | 2003-06-12 | 2005-12-01 | Nucleonics Inc | Conserved hbv and hcv sequences useful for gene silencing |
| WO2006025154A1 (en) * | 2004-08-30 | 2006-03-09 | Gifu University | Modified oligonucleotides |
| EP1636342A2 (en) * | 2003-06-20 | 2006-03-22 | Isis Pharmaceuticals, Inc. | Oligomeric compounds for use in gene modulation |
| EP1644475A2 (en) * | 2003-06-20 | 2006-04-12 | Isis Pharmaceuticals, Inc. | DOUBLE STRANDED COMPOSITIONS COMPRISING A 3’-ENDO MODIFIED STRAND FOR USE IN GENE MODULATION |
| WO2005121372A3 (en) * | 2004-06-03 | 2006-04-13 | Isis Pharmaceuticals Inc | Double strand compositions comprising differentially modified strands for use in gene modulation |
| WO2006060598A2 (en) * | 2004-12-01 | 2006-06-08 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF INTERLEUKIN AND INTERLEUKIN RECEPTOR GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| EP1681347A1 (en) * | 2005-01-18 | 2006-07-19 | Metanomics GmbH & Co. KGaA | Improved methods for double-stranded RNA mediated gene silencing |
| WO2006078798A2 (en) * | 2005-01-18 | 2006-07-27 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF RETINOBLASTOMA (RB1) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| WO2006078414A2 (en) * | 2004-12-23 | 2006-07-27 | Ambion, Inc. | METHODS AND COMPOSITIONS CONCERNING siRNA'S AS MEDIATORS OF RNA INTERFERENCE |
| WO2006128141A2 (en) * | 2005-05-27 | 2006-11-30 | Sirna Therapeutics, Inc. | Rna interference mediated inhibition of stromal cell-derived factor-1 (sdf-1) gene expression using short interfering nucleic acid (sina) |
| EP1753292A2 (en) * | 2004-05-12 | 2007-02-21 | Kamel Khalili | COMPOSITIONS AND METHODS FOR siRNA INHIBITION OF PRIMATE POLYOMAVIRUS GENES |
| WO2007022369A2 (en) * | 2005-08-17 | 2007-02-22 | Sirna Therapeutics, Inc. | Chemically modified short interfering nucleic acid molecules that mediate rna interference |
| US7217807B2 (en) | 2002-11-26 | 2007-05-15 | Rosetta Genomics Ltd | Bioinformatically detectable group of novel HIV regulatory genes and uses thereof |
| EP1784498A2 (en) * | 2004-08-03 | 2007-05-16 | Duke University | Systems and methods for inhibiting metastasis |
| EP1786905A1 (en) * | 2004-08-18 | 2007-05-23 | GeneSense Technologies Inc. | Small interfering rna molecules against ribonucleotide reductase and uses thereof |
| WO2007092059A2 (en) * | 2005-10-03 | 2007-08-16 | Sirna Therapeutics, Inc. | Rna interference mediated inhibition of influenza virus gene expression using short interfering nucleic acid |
| WO2007107162A2 (en) * | 2006-03-23 | 2007-09-27 | Santaris Pharma A/S | Small internally segmented interfering rna |
| WO2007120527A2 (en) * | 2006-03-31 | 2007-10-25 | Sirna Therapeutics Inc. | Rna interference mediated inhibition of respiratory syncytial virus (rsv) expression using short interfering nucleic acid (sina) |
| WO2007147143A2 (en) * | 2006-06-16 | 2007-12-21 | Sirna Therapeutics, Inc. | Rna interference mediated inhibition of 11 beta-hydroxysteroid dehydrogenase-1 (11 beta-hsd-1) gene expression using short interfering nucleic acid (sina) |
| WO2008011467A2 (en) * | 2006-07-18 | 2008-01-24 | Sirna Therapeutics Inc. | Rna interference mediated inhibition of sterol regulatory element-binding protein 1 (srebp1) gene expression using short interfering nucleic acid (sina) |
| EP1884569A1 (en) * | 2006-07-31 | 2008-02-06 | Institut National De La Sante Et De La Recherche Medicale (Inserm) | Sensitization of cancer cells to therapy using siNA targeting genes from the 1p and 19q chromosomal regions |
| WO2008036933A2 (en) | 2006-09-21 | 2008-03-27 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inhibiting expression of the hamp gene |
| WO2008042973A2 (en) | 2006-10-03 | 2008-04-10 | Alnylam Pharmaceuticals, Inc. | Lipid containing formulations |
| EP1957648A2 (en) * | 2005-11-17 | 2008-08-20 | Board of Regents, The University of Texas System | Modulation of gene expression by oligomers targeted to chromosomal dna |
| WO2008109511A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting fcer2 gene expression and uses thereof |
| WO2008109494A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting stat3 gene expression and uses thereof |
| WO2008109381A2 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting hif1a gene expression and uses thereof |
| WO2008109354A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting il18 gene expression and uses thereof |
| WO2008109526A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting egr gene expression and uses thereof |
| WO2008109449A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna Inc. | Nucleic acid compounds for inhibiting bcl2 gene expression and uses thereof |
| WO2008109534A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting ezh2 gene expression and uses thereof |
| WO2008109373A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting erbb gene expression and uses thereof |
| WO2008109362A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting vegf gene expression and uses thereof |
| WO2008109375A2 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting pik3c gene expression and uses thereof |
| WO2008109551A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting tacstd1 gene expression and uses thereof |
| WO2008109492A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting igf1r gene expression and uses thereof |
| WO2008109357A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting apob gene expression and uses thereof |
| WO2008109547A2 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting tymp gene expression and uses thereof |
| WO2008109475A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting sirt2 gene expression and uses thereof |
| US20080279920A1 (en) * | 2004-11-05 | 2008-11-13 | Intradigm Corporation | Compositions For Treating Respiratory Viral Infections and Their Use |
| WO2008137867A2 (en) | 2007-05-03 | 2008-11-13 | Rosetta Inpharmatics Llc | Compositions comprising mir34 therapeutic agents for treating cancer |
| US7452987B2 (en) | 2002-08-05 | 2008-11-18 | Silence Therapeutics Aktiengesellschaft (Ag) | Interfering RNA molecules |
| EP2008274A2 (en) * | 2006-03-31 | 2008-12-31 | Alnylam Pharmaceuticals Inc. | Compositions and methods for inhibiting expression of eg5 gene |
| WO2009005095A1 (en) * | 2007-07-03 | 2009-01-08 | Kyorin Pharmaceutical Co., Ltd | Treatment of influenza |
| EP2018443A2 (en) * | 2006-05-22 | 2009-01-28 | Alnylam Pharmaceuticals Inc. | Compositions and methods for inhibiting expression of ikk-b gene |
| EP2021507A2 (en) * | 2006-05-11 | 2009-02-11 | Alnylam Pharmaceuticals Inc. | Compositions and methods for inhibiting expression of the pcsk9 gene |
| EP2021352A2 (en) * | 2006-05-19 | 2009-02-11 | The Scripps Research Institute | Treatment of protein misfolding |
| US7491805B2 (en) | 2001-05-18 | 2009-02-17 | Sirna Therapeutics, Inc. | Conjugates and compositions for cellular delivery |
| EP2023937A2 (en) * | 2006-05-19 | 2009-02-18 | Alnylam Pharmaceuticals Inc. | Rnai modulation of aha and therapeutic uses thereof |
| WO2009029293A2 (en) * | 2007-03-02 | 2009-03-05 | Mdrna, Inc. | Nucleic acid compounds for inhibiting myc gene expression and uses thereof |
| JP2009513716A (en) * | 2005-11-01 | 2009-04-02 | アルナイラム ファーマシューティカルズ インコーポレイテッド | Inhibition of influenza virus replication by RNAi |
| WO2009044392A2 (en) | 2007-10-03 | 2009-04-09 | Quark Pharmaceuticals, Inc. | Novel sirna structures |
| EP2072619A1 (en) | 2002-10-18 | 2009-06-24 | Silence Therapeutics Aktiengesellschaft | Factor involved in metastasis and uses thereof |
| WO2009118300A1 (en) | 2008-03-25 | 2009-10-01 | Novartis Forschungsstiftung Zweigniederlassung Friedrich Miescher Institute For Biomedical Research | Treating cancer by down-regulating frizzled-4 and/or frizzled-1 |
| US7618814B2 (en) | 2002-11-14 | 2009-11-17 | Rosetta Genomics Ltd. | Microrna-related nucleic acids and uses thereof |
| US7626015B2 (en) | 2006-06-09 | 2009-12-01 | Quark Pharmaceuticals, Inc. | Therapeutic uses of inhibitors of RTP801L |
| EP2135950A2 (en) | 2006-05-04 | 2009-12-23 | Novartis AG | Short interfering ribonucleic acid (siRNA) for oral administration |
| EP2143792A1 (en) * | 2007-05-09 | 2010-01-13 | Riken | Single-stranded cyclic rna, and method for production thereof |
| WO2010006342A2 (en) * | 2008-07-11 | 2010-01-14 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inhibiting expression of gsk-3 genes |
| US7655785B1 (en) | 2002-11-14 | 2010-02-02 | Rosetta Genomics Ltd. | Bioinformatically detectable group of novel regulatory oligonucleotides and uses thereof |
| US7687616B1 (en) | 2004-05-14 | 2010-03-30 | Rosetta Genomics Ltd | Small molecules modulating activity of micro RNA oligonucleotides and micro RNA targets and uses thereof |
| US7695902B2 (en) | 1996-06-06 | 2010-04-13 | Isis Pharmaceuticals, Inc. | Oligoribonucleotides and ribonucleases for cleaving RNA |
| US7709616B2 (en) | 2004-05-14 | 2010-05-04 | Rosetta Genomics Inc. | Micrornas and uses thereof |
| WO2010065756A2 (en) * | 2008-12-03 | 2010-06-10 | Mdrna, Inc. | Usirna complexes |
| US7741299B2 (en) | 2004-08-16 | 2010-06-22 | Quark Pharmaceuticals, Inc. | Therapeutic uses of inhibitors of RTP801 |
| WO2010100247A1 (en) | 2009-03-06 | 2010-09-10 | Novartis Forschungsstiftung, Zweigniederlassung, Friedrich Miescher Institute For Biomedical Research | Novel therapy for anxiety |
| US7795419B2 (en) | 2004-05-26 | 2010-09-14 | Rosetta Genomics Ltd. | Viral and viral associated miRNAs and uses thereof |
| EP2233573A1 (en) * | 2007-11-29 | 2010-09-29 | Suzhou Ribo Life Science Co., Ltd | A complex molecule interfering the expression of target genes and its preparing methods |
| US7807820B2 (en) | 2002-11-14 | 2010-10-05 | Dharmacon, Inc. | siRNA targeting beta secretase (BACE) |
| US7812149B2 (en) | 1996-06-06 | 2010-10-12 | Isis Pharmaceuticals, Inc. | 2′-Fluoro substituted oligomeric compounds and compositions for use in gene modulations |
| US7812002B2 (en) | 2007-03-21 | 2010-10-12 | Quark Pharmaceuticals, Inc. | Oligoribonucleotide inhibitors of NRF2 and methods of use thereof for treatment of cancer |
| EP2241323A1 (en) | 2009-04-14 | 2010-10-20 | Novartis Forschungsstiftung, Zweigniederlassung Friedrich Miescher Institute For Biomedical Research | Tenascin-W and brain cancers |
| US7825099B2 (en) | 2006-01-20 | 2010-11-02 | Quark Pharmaceuticals, Inc. | Treatment or prevention of oto-pathologies by inhibition of pro-apoptotic genes |
| US7842674B2 (en) | 2004-09-28 | 2010-11-30 | Quark Pharmaceuticals Inc. | Methods of treatment of acute renal failure |
| WO2010111490A3 (en) * | 2009-03-27 | 2010-12-29 | Merck Sharp & Dohme Corp. | RNA INTERFERENCE MEDIATED INHIBITION OF THE THYMIC STROMAL LYMPHOPOIETIN (TSLP) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US7872119B2 (en) | 2007-02-26 | 2011-01-18 | Quark Pharmaceuticals, Inc. | Inhibitors of RTP801 and their use in disease treatment |
| WO2011008730A2 (en) | 2009-07-13 | 2011-01-20 | Somagenics Inc. | Chemical modification of small hairpin rnas for inhibition of gene expression |
| US7884086B2 (en) | 2004-09-08 | 2011-02-08 | Isis Pharmaceuticals, Inc. | Conjugates for use in hepatocyte free uptake assays |
| US7888497B2 (en) | 2003-08-13 | 2011-02-15 | Rosetta Genomics Ltd. | Bioinformatically detectable group of novel regulatory oligonucleotides and uses thereof |
| WO2011019423A2 (en) | 2009-05-20 | 2011-02-17 | Schering Corporation | Modulation of pilr receptors to treat microbial infections |
| EP2292266A1 (en) | 2009-08-27 | 2011-03-09 | Novartis Forschungsstiftung, Zweigniederlassung | Treating cancer by modulating copine III |
| WO2011031600A1 (en) | 2009-09-10 | 2011-03-17 | Schering Corporation | Use of il-33 antagonists to treat fibrotic disease |
| US7910566B2 (en) | 2006-03-09 | 2011-03-22 | Quark Pharmaceuticals Inc. | Prevention and treatment of acute renal failure and other kidney diseases by inhibition of p53 by siRNA |
| US7915399B2 (en) | 2006-06-09 | 2011-03-29 | Protiva Biotherapeutics, Inc. | Modified siRNA molecules and uses thereof |
| AU2005313883B2 (en) * | 2004-12-09 | 2011-03-31 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inducing an immune response in a mammal and methods of avoiding an immune response to oligonucleotide agents such as short interfering RNAs |
| WO2011036118A1 (en) | 2009-09-22 | 2011-03-31 | Novartis Forschungsstiftung, Zweigniederlassung Friedrich Miescher Institute For Biomedical Research | Treating cancer by modulating mex-3 |
| US7919583B2 (en) | 2005-08-08 | 2011-04-05 | Discovery Genomics, Inc. | Integration-site directed vector systems |
| US7919473B2 (en) | 2004-03-12 | 2011-04-05 | Alnylam Pharmaceuticals, Inc. | IRNA agents targeting VEGF |
| US7923547B2 (en) | 2002-09-05 | 2011-04-12 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US7928217B2 (en) | 2004-05-27 | 2011-04-19 | Alnylam Pharmaceuticals, Inc. | Nuclease resistant double-stranded ribonucleic acid |
| WO2011045352A2 (en) | 2009-10-15 | 2011-04-21 | Novartis Forschungsstiftung | Spleen tyrosine kinase and brain cancers |
| EP2316943A1 (en) | 2007-07-05 | 2011-05-04 | Novartis AG | DSRNA for treating viral infection |
| WO2011084193A1 (en) | 2010-01-07 | 2011-07-14 | Quark Pharmaceuticals, Inc. | Oligonucleotide compounds comprising non-nucleotide overhangs |
| WO2011084357A1 (en) | 2009-12-17 | 2011-07-14 | Schering Corporation | Modulation of pilr to treat immune disorders |
| US7989612B2 (en) | 2002-02-20 | 2011-08-02 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| EP1423404B1 (en) * | 2002-02-20 | 2011-08-24 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED TREATMENT OF ALZHEIMER'S DISEASE USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| WO2011107586A1 (en) | 2010-03-05 | 2011-09-09 | Novartis Forschungsstiftung, Zweigniederlassung, Friedrich Miescher Institute For Biomedical Research, | Smoc1, tenascin-c and brain cancers |
| WO2011133868A2 (en) | 2010-04-22 | 2011-10-27 | Alnylam Pharmaceuticals, Inc. | Conformationally restricted dinucleotide monomers and oligonucleotides |
| WO2011133876A2 (en) | 2010-04-22 | 2011-10-27 | Alnylam Pharmaceuticals, Inc. | Oligonucleotides comprising acyclic and abasic nucleosides and analogs |
| WO2011131611A1 (en) | 2010-04-19 | 2011-10-27 | Novartis Forschungsstiftung, Zweigniederlassung Friedrich Miescher Institute For Biomedical Research | Modulating xrn1 |
| US8067570B2 (en) | 2006-01-20 | 2011-11-29 | Quark Pharmaceuticals, Inc. | Therapeutic uses of inhibitors of RTP801 |
| WO2011154485A1 (en) | 2010-06-10 | 2011-12-15 | Novartis Forschungsstiftung, Zweigniederlassung Friedrich Miescher Institute For Biomedical Research | Treating cancer by modulating mammalian sterile 20-like kinase 3 |
| US8101741B2 (en) | 2005-11-02 | 2012-01-24 | Protiva Biotherapeutics, Inc. | Modified siRNA molecules and uses thereof |
| EP2412724A1 (en) | 2010-07-29 | 2012-02-01 | Centre National de la Recherche Scientifique (C.N.R.S) | Regulation of Glypican 4 activity to modulate the fate of stem cells and uses thereof |
| EP2415869A1 (en) * | 2009-04-03 | 2012-02-08 | Biomics Biotechnologies Co., Ltd. | Modified oligo-nucleic acid molecule, preparation method and uses thereof |
| EP2441837A1 (en) | 2007-03-26 | 2012-04-18 | Alnylam Pharmaceuticals Inc. | dsRNA compositions and methods for treating HPV infections |
| US8163896B1 (en) | 2002-11-14 | 2012-04-24 | Rosetta Genomics Ltd. | Bioinformatically detectable group of novel regulatory genes and uses thereof |
| EP2446890A1 (en) | 2007-04-30 | 2012-05-02 | Allergan, Inc. | High viscosity macromolecular compositions for treating ocular conditions |
| CN102453066A (en) * | 2010-10-19 | 2012-05-16 | 南开大学 | Compound molecule, and preparation method and pharmaceutical composition thereof |
| US8222221B2 (en) | 2008-06-04 | 2012-07-17 | The Board Of Regents Of The University Of Texas System | Modulation of gene expression through endogenous small RNA targeting of gene promoters |
| EP1984382B1 (en) * | 2006-01-27 | 2012-08-15 | Santaris Pharma A/S | Lna modified phosphorothiolated oligonucleotides |
| US8273869B2 (en) | 2009-06-15 | 2012-09-25 | Alnylam Pharmaceuticals, Inc. | Lipid formulated dsRNA targeting the PCSK9 gene |
| US8278287B2 (en) | 2008-04-15 | 2012-10-02 | Quark Pharmaceuticals Inc. | siRNA compounds for inhibiting NRF2 |
| US8309530B2 (en) | 2009-02-04 | 2012-11-13 | Washington State University | Compositions and methods for modulating ghrelin-mediated conditions |
| US8309791B2 (en) | 2008-07-16 | 2012-11-13 | Recombinectics, Inc. | Method for producing a transgenic pig using a hyper-methylated transposon |
| US8329667B2 (en) | 2004-04-23 | 2012-12-11 | The Trustees Of Columbia University In The City Of New York | Inhibition of hairless protein mRNA |
| WO2012168259A1 (en) | 2011-06-06 | 2012-12-13 | Novartis Forschungsstiftung, Zweigniederlassung | Protein tyrosine phosphatase, non-receptor type 11 (ptpn11) and triple-negative breast cancer |
| EP2399575A3 (en) * | 2006-08-11 | 2012-12-19 | INSERM (Institut National de la Santé et de la Recherche Médicale) | Methods, uses and compositions for treatment of an infection by a virus of the family of flaviviridae through the farnesoid X receptor (FXR) inhibition |
| US8362229B2 (en) | 2006-02-08 | 2013-01-29 | Quark Pharmaceuticals, Inc. | Tandem siRNAS |
| AU2011203218B2 (en) * | 2004-12-09 | 2013-02-07 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inducing an immune response in a mammal and methods of avoiding an immune response to oligonucleotide agents such as short interfering RNAs |
| US8394947B2 (en) | 2004-06-03 | 2013-03-12 | Isis Pharmaceuticals, Inc. | Positionally modified siRNA constructs |
| US8431692B2 (en) | 2008-06-06 | 2013-04-30 | Quark Pharmaceuticals, Inc. | Compositions and methods for treatment of ear disorders |
| WO2013068431A1 (en) | 2011-11-08 | 2013-05-16 | Novartis Forschungsstiftung, Zweigniederlassung, Friedrich Miescher Institute For Biomedical Research | New treatment for neurodegenerative diseases |
| WO2013068432A1 (en) | 2011-11-08 | 2013-05-16 | Novartis Forschungsstiftung, Zweigniederlassung, Friedrich Miescher Institute For Biomedical Research | Early diagnostic of neurodegenerative diseases |
| WO2013072392A1 (en) | 2011-11-15 | 2013-05-23 | Novartis Forschungsstiftung, Zweigniederlassung Friedrich Miescher Institute For Biomedical Research | Combination of a phosphoinositide 3-kinase inhibitor and a modulator of the janus kinase 2-signal transducer and activator of transcription 5 pathway |
| WO2013102825A1 (en) | 2012-01-02 | 2013-07-11 | Novartis Ag | Cdcp1 and breast cancer |
| US8541568B2 (en) | 2008-05-24 | 2013-09-24 | Hai Yan | Compositions and methods using siRNA molecules for treatment of gliomas |
| WO2013144240A1 (en) | 2012-03-29 | 2013-10-03 | Friedrich Miescher Institute For Biomedical Research | Inhibition of interleukin- 8 and/or its receptor cxcrl in the treatment her2/her3 -overexpressing breast cancer |
| US8569474B2 (en) | 2004-03-09 | 2013-10-29 | Isis Pharmaceuticals, Inc. | Double stranded constructs comprising one or more short strands hybridized to a longer strand |
| US8575327B2 (en) | 2003-06-12 | 2013-11-05 | Alnylam Pharmaceuticals, Inc. | Conserved HBV and HCV sequences useful for gene silencing |
| US8604183B2 (en) | 2002-11-05 | 2013-12-10 | Isis Pharmaceuticals, Inc. | Compositions comprising alternating 2′-modified nucleosides for use in gene modulation |
| US8614311B2 (en) | 2007-12-12 | 2013-12-24 | Quark Pharmaceuticals, Inc. | RTP801L siRNA compounds and methods of use thereof |
| WO2014001482A1 (en) | 2012-06-29 | 2014-01-03 | Novartis Forschungsstiftung, Zweigniererlassung, Friedrich Miescher Institute For Biomedical Research | Treating diseases by modulating a specific isoform of mkl1 |
| WO2014006114A1 (en) | 2012-07-05 | 2014-01-09 | Novartis Forschungsstiftung, Zweigniederlassung Friedrich Miescher Institute For Biomedical Research | New treatment for neurodegenerative diseases |
| WO2014006115A1 (en) | 2012-07-06 | 2014-01-09 | Novartis Ag | Combination of a phosphoinositide 3-kinase inhibitor and an inhibitor of the il-8/cxcr interaction |
| US8653252B2 (en) | 2003-03-21 | 2014-02-18 | Santaris Pharma A/S | Short interfering RNA (siRNA) analogues |
| US8729339B2 (en) | 2003-10-09 | 2014-05-20 | E.I Du Pont De Nemours And Company | Gene silencing |
| US8729036B2 (en) | 2002-08-07 | 2014-05-20 | University Of Massachusetts | Compositions for RNA interference and methods of use thereof |
| US8735567B2 (en) | 2007-11-06 | 2014-05-27 | Patrick Y. Lu | Multi-targeted RNAi therapeutics for scarless wound healing of skin |
| US8785408B2 (en) | 2007-06-27 | 2014-07-22 | Quark Pharmaceuticals, Inc. | Compositions and methods for reducing or protecting against delayed graft function (DGF) |
| US8796235B2 (en) | 2003-02-21 | 2014-08-05 | University Of South Florida | Methods for attenuating dengue virus infection |
| US8796239B2 (en) | 2009-11-26 | 2014-08-05 | Quark Pharmaceuticals, Inc. | Sirna compounds comprising terminal substitutions |
| US8815586B2 (en) | 2009-04-24 | 2014-08-26 | The Board Of Regents Of The University Of Texas System | Modulation of gene expression using oligomers that target gene regions downstream of 3′ untranslated regions |
| US20140288158A1 (en) * | 2011-11-18 | 2014-09-25 | Alnylam Pharmaceuticals, Inc. | MODIFIED RNAi AGENTS |
| WO2014160871A2 (en) | 2013-03-27 | 2014-10-02 | The General Hospital Corporation | Methods and agents for treating alzheimer's disease |
| US8859516B2 (en) | 2009-09-15 | 2014-10-14 | Alnylam Pharmaceuticals, Inc. | Lipid formulated compositions and methods for inhibiting expression of Eg5 and VEGF genes |
| WO2014176259A1 (en) | 2013-04-22 | 2014-10-30 | Icahn School Of Medicine At Mount Sinai | Mutations in pdgfrb and notch3 as causes of autosomal dominant infantile myofibromatosis |
| US8889649B2 (en) | 2010-11-12 | 2014-11-18 | National University Corporation Ehime University | Composition containing antisense oligonucleotide to micro RNA |
| EP2810643A2 (en) | 2009-08-14 | 2014-12-10 | Alnylam Pharmaceuticals Inc. | Lipid formulated compositions and mehods for inhibiting expression of a gene from the ebola virus |
| WO2015019286A1 (en) | 2013-08-07 | 2015-02-12 | Friedrich Miescher Institute For Biomedical Research | New screening method for the treatment friedreich's ataxia |
| US8975471B2 (en) | 2004-10-12 | 2015-03-10 | The Rockefeller University | MicroRNAs |
| US8999943B2 (en) | 2005-03-14 | 2015-04-07 | Board Of Regents, The University Of Texas System | Antigene oligomers inhibit transcription |
| WO2015051366A2 (en) | 2013-10-04 | 2015-04-09 | Novartis Ag | Novel formats for organic compounds for use in rna interference |
| WO2015051045A2 (en) | 2013-10-04 | 2015-04-09 | Novartis Ag | 3'END CAPS FOR RNAi AGENTS FOR USE IN RNA INTERFERENCE |
| WO2015051044A2 (en) | 2013-10-04 | 2015-04-09 | Novartis Ag | Novel formats for organic compounds for use in rna interference |
| WO2015050871A2 (en) | 2013-10-04 | 2015-04-09 | Novartis Ag | Organic compounds to treat hepatitis b virus |
| US9006197B2 (en) | 2008-03-05 | 2015-04-14 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inhibiting expression of Eg5 and VEGF genes |
| US9012622B2 (en) | 2008-12-31 | 2015-04-21 | Patrick Y. Lu | Compositions and methods using siRNA molecules and siRNA cocktails for the treatment of breast cancer |
| US9051567B2 (en) | 2009-06-15 | 2015-06-09 | Tekmira Pharmaceuticals Corporation | Methods for increasing efficacy of lipid formulated siRNA |
| US9051570B2 (en) | 2007-05-22 | 2015-06-09 | Arcturus Therapeutics, Inc. | UNA oligomers for therapeutics |
| US9096636B2 (en) | 1996-06-06 | 2015-08-04 | Isis Pharmaceuticals, Inc. | Chimeric oligomeric compounds and their use in gene modulation |
| US9181551B2 (en) | 2002-02-20 | 2015-11-10 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US9187746B2 (en) | 2009-09-22 | 2015-11-17 | Alnylam Pharmaceuticals, Inc. | Dual targeting siRNA agents |
| US9200275B2 (en) | 2006-06-14 | 2015-12-01 | Merck Sharp & Dohme Corp. | Methods and compositions for regulating cell cycle progression |
| WO2015189816A1 (en) | 2014-06-13 | 2015-12-17 | Friedrich Miescher Institute For Biomedical Research | New treatment against influenza virus |
| WO2016001830A1 (en) | 2014-07-01 | 2016-01-07 | Friedrich Miescher Institute For Biomedical Research | Combination of a brafv600e inhibitor and mertk inhibitor to treat melanoma |
| US9243246B2 (en) | 2010-08-24 | 2016-01-26 | Sirna Therapeutics, Inc. | Single-stranded RNAi agents containing an internal, non-nucleic acid spacer |
| EP2980220A1 (en) | 2005-09-20 | 2016-02-03 | BASF Plant Science GmbH | Improved methods controlling gene expression |
| US9260471B2 (en) | 2010-10-29 | 2016-02-16 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using short interfering nucleic acids (siNA) |
| WO2016046768A1 (en) | 2014-09-24 | 2016-03-31 | Friedrich Miescher Institute For Biomedical Research | Lats and breast cancer |
| US9309313B2 (en) | 2008-01-09 | 2016-04-12 | The Schepens Eye Research Institute, Inc. | Therapeutic compositions for treatment of ocular inflammatory disorders |
| WO2016077620A1 (en) | 2014-11-12 | 2016-05-19 | The United States Of America, As Represented By The Secretary, Department Of Health And Human Services | Treatment of hormonal disorders of growth |
| US9446062B2 (en) | 2006-10-25 | 2016-09-20 | Quark Pharmaceuticals, Inc. | Methods of treating ischemia-reperfusion injury with siRNAs |
| US9506063B2 (en) | 2010-07-29 | 2016-11-29 | Sirnaomics, Inc. | SiRNA compositions and methods for treatment of HPV and other infections |
| WO2017072669A1 (en) | 2015-10-28 | 2017-05-04 | Friedrich Miescher Institute For Biomedical Research | Tenascin-w and biliary tract cancers |
| US9642873B2 (en) | 2010-05-04 | 2017-05-09 | Sirnaomics, Inc. | Combinations of TGFβ and COX-2 inhibitors and methods for their therapeutic application |
| WO2017079442A1 (en) | 2015-11-04 | 2017-05-11 | Icahn School Of Medicine At Mount Sinai | Methods of treating tumors and cancer, and identifying candidate subjects for such treatment |
| US9657294B2 (en) | 2002-02-20 | 2017-05-23 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US9719092B2 (en) | 2002-11-14 | 2017-08-01 | Thermo Fisher Scientific Inc. | RNAi targeting CNTD2 |
| US9719094B2 (en) | 2002-11-14 | 2017-08-01 | Thermo Fisher Scientific Inc. | RNAi targeting SEC61G |
| US9771586B2 (en) | 2002-11-14 | 2017-09-26 | Thermo Fisher Scientific Inc. | RNAi targeting ZNF205 |
| US9777275B2 (en) | 2002-02-01 | 2017-10-03 | Life Technologies Corporation | Oligonucleotide compositions with enhanced efficiency |
| US9777270B2 (en) | 2002-11-14 | 2017-10-03 | Thermo Fisher Scientific Inc. | Methods and compositions for selecting siRNA of improved functionality |
| WO2017189730A1 (en) | 2016-04-26 | 2017-11-02 | Icahn School Of Medicine At Mount Sinai | Treatment of hippo pathway mutant tumors and methods of identifying subjects as candidates for treatment |
| US9856475B2 (en) | 2014-03-25 | 2018-01-02 | Arcturus Therapeutics, Inc. | Formulations for treating amyloidosis |
| US9868952B2 (en) | 2012-07-08 | 2018-01-16 | Sirnaomics, Inc. | Compositions and methods for “resistance-proof” SiRNA therapeutics for influenza |
| US9885036B2 (en) | 2008-07-01 | 2018-02-06 | Daiichi Sankyo Company, Limited | Double-stranded polynucleotide |
| EP3129493A4 (en) * | 2014-04-09 | 2018-04-25 | The Scripps Research Institute | Import of unnatural or modified nucleoside triphosphates into cells via nucleic acid triphosphate transporters |
| US9982259B2 (en) | 2014-03-25 | 2018-05-29 | Arcturus Therapeutics, Inc. | Transthyretin allele selective UNA oligomers for gene silencing |
| US9994853B2 (en) | 2001-05-18 | 2018-06-12 | Sirna Therapeutics, Inc. | Chemically modified multifunctional short interfering nucleic acid molecules that mediate RNA interference |
| US10106793B2 (en) | 2002-02-01 | 2018-10-23 | Life Technologies Corporation | Double-stranded oligonucleotides |
| US10105441B2 (en) | 2007-08-16 | 2018-10-23 | The Schepens Eye Research Institute, Inc. | Method for inhibiting or reducing dry eye disease by IL-1Ra |
| WO2018191278A3 (en) * | 2017-04-11 | 2018-11-22 | Arbutus Biopharma Corporation | Targeted compositions |
| US10421964B2 (en) | 2015-07-23 | 2019-09-24 | Arcturus Therapeutics, Inc. | UNA oligomers and compositions for treating amyloidosis |
| US10478449B2 (en) * | 2002-11-05 | 2019-11-19 | Ionis Pharmaceuticals, Inc. | 2′-methoxy substituted oligomeric compounds and compositions for use in gene modulations |
| US10508277B2 (en) | 2004-05-24 | 2019-12-17 | Sirna Therapeutics, Inc. | Chemically modified multifunctional short interfering nucleic acid molecules that mediate RNA interference |
| US10513703B2 (en) | 2014-11-10 | 2019-12-24 | Alnylam Pharmaceuticals, Inc. | Hepatitis B virus (HBV) iRNA compositions and methods of use thereof |
| WO2019246112A1 (en) | 2018-06-18 | 2019-12-26 | University Of Rochester | Methods of treating schizophrenia and other neuropsychiatric disorders |
| WO2019246262A2 (en) | 2018-06-21 | 2019-12-26 | University Of Rochester | Methods of treating or inhibiting onset of huntington's disease |
| US10519447B2 (en) | 2015-04-01 | 2019-12-31 | Arcturus Therapeutics, Inc. | Therapeutic UNA oligomers and uses thereof |
| US10610571B2 (en) | 2017-08-03 | 2020-04-07 | Synthorx, Inc. | Cytokine conjugates for the treatment of proliferative and infectious diseases |
| WO2020093053A1 (en) * | 2018-11-02 | 2020-05-07 | Arbutus Biopharma Corporation | Bivalent targeted conjugates |
| US10683500B2 (en) | 2014-03-25 | 2020-06-16 | Arcturus Therapeutics, Inc. | UNA oligomers having reduced off-target effects in gene silencing |
| WO2020123663A1 (en) | 2018-12-11 | 2020-06-18 | University Of Rochester | Methods of treating schizophrenia and other neuropsychiatric disorders |
| WO2020167822A2 (en) | 2019-02-13 | 2020-08-20 | University Of Rochester | Gene networks that mediate remyelination of the human brain |
| WO2021011936A2 (en) | 2019-07-18 | 2021-01-21 | University Of Rochester | Cell-type selective immunoprotection of cells |
| US11077195B2 (en) | 2019-02-06 | 2021-08-03 | Synthorx, Inc. | IL-2 conjugates and methods of use thereof |
| EP3901263A1 (en) * | 2011-06-30 | 2021-10-27 | Arrowhead Pharmaceuticals, Inc. | Compositions and methods for inhibiting gene expression of hepatitis b virus |
| US11324820B2 (en) | 2017-04-18 | 2022-05-10 | Alnylam Pharmaceuticals, Inc. | Methods for the treatment of subjects having a hepatitis b virus (HBV) infection |
| US11492623B2 (en) | 2018-08-13 | 2022-11-08 | Alnylam Pharmaceuticals, Inc. | Hepatitis B virus (HBV) dsRNA agent compositions and methods of use thereof |
| US11517584B2 (en) | 2016-08-04 | 2022-12-06 | Arrowhead Pharmaceuticals, Inc. | RNAi agents for Hepatitis B virus infection |
| US11534453B2 (en) | 2015-08-07 | 2022-12-27 | Arrowhead Pharmaceuticals, Inc. | RNAi therapy for hepatitis B virus infection |
| WO2023150553A1 (en) | 2022-02-01 | 2023-08-10 | University Of Rochester | Gpr17 promoter-based targeting and transduction of glial progenitor cells |
| WO2023191630A1 (en) | 2022-03-30 | 2023-10-05 | Academisch Medisch Centrum | Antisense nucleic acids for use in the treatment for kcnq1 mutation carriers |
| WO2023191631A1 (en) | 2022-03-30 | 2023-10-05 | Academisch Medisch Centrum | Antisense nucleic acids for use in the treatment for lmna mutation carriers |
| US11834689B2 (en) | 2017-07-11 | 2023-12-05 | The Scripps Research Institute | Incorporation of unnatural nucleotides and methods thereof |
| US11896672B2 (en) | 2016-04-11 | 2024-02-13 | Arbutus Biopharma Corporation | Targeted nucleic acid conjugate compositions |
Families Citing this family (1)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US9035039B2 (en) | 2011-12-22 | 2015-05-19 | Protiva Biotherapeutics, Inc. | Compositions and methods for silencing SMAD4 |
Citations (13)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US5672695A (en) | 1990-10-12 | 1997-09-30 | Max-Planck-Gesellschaft Zur Forderung Der Wissenschaften E.V. | Modified ribozymes |
| US5889136A (en) | 1995-06-09 | 1999-03-30 | The Regents Of The University Of Colorado | Orthoester protecting groups in RNA synthesis |
| WO1999049029A1 (en) | 1998-03-20 | 1999-09-30 | Benitec Australia Ltd | Control of gene expression |
| US5998203A (en) | 1996-04-16 | 1999-12-07 | Ribozyme Pharmaceuticals, Inc. | Enzymatic nucleic acids containing 5'-and/or 3'-cap structures |
| CA2359180A1 (en) | 1999-01-30 | 2000-08-03 | Roland Kreutzer | Method and medicament for inhibiting the expression of a given gene |
| WO2000044914A1 (en) | 1999-01-28 | 2000-08-03 | Medical College Of Georgia Research Institute, Inc. | Composition and method for in vivo and in vitro attenuation of gene expression using double stranded rna |
| US6111086A (en) | 1998-02-27 | 2000-08-29 | Scaringe; Stephen A. | Orthoester protecting groups |
| WO2001068836A2 (en) | 2000-03-16 | 2001-09-20 | Genetica, Inc. | Methods and compositions for rna interference |
| WO2001070949A1 (en) | 2000-03-17 | 2001-09-27 | Benitec Australia Ltd | Genetic silencing |
| WO2002038805A2 (en) | 2000-11-09 | 2002-05-16 | Cenix Bioscience Gmbh | Eukaryotic cell division genes and their use in diagnosis and treatment of proliferative diseases |
| WO2002055693A2 (en) | 2001-01-09 | 2002-07-18 | Ribopharma Ag | Method for inhibiting the expression of a target gene |
| WO2002055692A2 (en) | 2001-01-09 | 2002-07-18 | Ribopharma Ag | Method for inhibiting the expression of a target gene and medicament for treating a tumor disease |
| US6506559B1 (en) | 1997-12-23 | 2003-01-14 | Carnegie Institute Of Washington | Genetic inhibition by double-stranded RNA |
Family Cites Families (3)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| JPH04507083A (en) * | 1989-05-19 | 1992-12-10 | ヘム・リサーチ・インコーポレーテッド | Short therapeutic dsRNA of defined structure |
| WO2000049035A1 (en) * | 1999-02-19 | 2000-08-24 | The General Hospital Corporation | Gene silencing |
| WO2001096584A2 (en) * | 2000-06-12 | 2001-12-20 | Akkadix Corporation | Materials and methods for the control of nematodes |
-
2003
- 2003-02-20 WO PCT/US2003/005346 patent/WO2003070918A2/en active Application Filing
Patent Citations (15)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US5672695A (en) | 1990-10-12 | 1997-09-30 | Max-Planck-Gesellschaft Zur Forderung Der Wissenschaften E.V. | Modified ribozymes |
| US5889136A (en) | 1995-06-09 | 1999-03-30 | The Regents Of The University Of Colorado | Orthoester protecting groups in RNA synthesis |
| US6008400A (en) | 1995-06-09 | 1999-12-28 | Scaringe; Stephen | Orthoester reagents for use as protecting groups in oligonucleotide synthesis |
| US5998203A (en) | 1996-04-16 | 1999-12-07 | Ribozyme Pharmaceuticals, Inc. | Enzymatic nucleic acids containing 5'-and/or 3'-cap structures |
| US6506559B1 (en) | 1997-12-23 | 2003-01-14 | Carnegie Institute Of Washington | Genetic inhibition by double-stranded RNA |
| US6111086A (en) | 1998-02-27 | 2000-08-29 | Scaringe; Stephen A. | Orthoester protecting groups |
| WO1999049029A1 (en) | 1998-03-20 | 1999-09-30 | Benitec Australia Ltd | Control of gene expression |
| WO2000044914A1 (en) | 1999-01-28 | 2000-08-03 | Medical College Of Georgia Research Institute, Inc. | Composition and method for in vivo and in vitro attenuation of gene expression using double stranded rna |
| EP1144623B1 (en) | 1999-01-30 | 2002-08-28 | Ribopharma AG | Method and medicament for inhibiting the expression of a defined gene |
| CA2359180A1 (en) | 1999-01-30 | 2000-08-03 | Roland Kreutzer | Method and medicament for inhibiting the expression of a given gene |
| WO2001068836A2 (en) | 2000-03-16 | 2001-09-20 | Genetica, Inc. | Methods and compositions for rna interference |
| WO2001070949A1 (en) | 2000-03-17 | 2001-09-27 | Benitec Australia Ltd | Genetic silencing |
| WO2002038805A2 (en) | 2000-11-09 | 2002-05-16 | Cenix Bioscience Gmbh | Eukaryotic cell division genes and their use in diagnosis and treatment of proliferative diseases |
| WO2002055693A2 (en) | 2001-01-09 | 2002-07-18 | Ribopharma Ag | Method for inhibiting the expression of a target gene |
| WO2002055692A2 (en) | 2001-01-09 | 2002-07-18 | Ribopharma Ag | Method for inhibiting the expression of a target gene and medicament for treating a tumor disease |
Non-Patent Citations (3)
| Title |
|---|
| BURGIN ET AL., BIOCHEMISTRY, vol. 35, 1996, pages 14090 |
| FIRE ET AL., NATURE, vol. 391, 1998, pages 806 |
| See also references of EP1458741A2 |
Cited By (455)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US9096636B2 (en) | 1996-06-06 | 2015-08-04 | Isis Pharmaceuticals, Inc. | Chimeric oligomeric compounds and their use in gene modulation |
| US7695902B2 (en) | 1996-06-06 | 2010-04-13 | Isis Pharmaceuticals, Inc. | Oligoribonucleotides and ribonucleases for cleaving RNA |
| US7812149B2 (en) | 1996-06-06 | 2010-10-12 | Isis Pharmaceuticals, Inc. | 2′-Fluoro substituted oligomeric compounds and compositions for use in gene modulations |
| US7491805B2 (en) | 2001-05-18 | 2009-02-17 | Sirna Therapeutics, Inc. | Conjugates and compositions for cellular delivery |
| US7964578B2 (en) | 2001-05-18 | 2011-06-21 | Sirna Therapeutics, Inc. | Conjugates and compositions for cellular delivery |
| US9994853B2 (en) | 2001-05-18 | 2018-06-12 | Sirna Therapeutics, Inc. | Chemically modified multifunctional short interfering nucleic acid molecules that mediate RNA interference |
| US10626398B2 (en) | 2002-02-01 | 2020-04-21 | Life Technologies Corporation | Oligonucleotide compositions with enhanced efficiency |
| US10196640B1 (en) | 2002-02-01 | 2019-02-05 | Life Technologies Corporation | Oligonucleotide compositions with enhanced efficiency |
| US10106793B2 (en) | 2002-02-01 | 2018-10-23 | Life Technologies Corporation | Double-stranded oligonucleotides |
| US10036025B2 (en) | 2002-02-01 | 2018-07-31 | Life Technologies Corporation | Oligonucleotide compositions with enhanced efficiency |
| US9796978B1 (en) | 2002-02-01 | 2017-10-24 | Life Technologies Corporation | Oligonucleotide compositions with enhanced efficiency |
| US9777275B2 (en) | 2002-02-01 | 2017-10-03 | Life Technologies Corporation | Oligonucleotide compositions with enhanced efficiency |
| EP1423406A2 (en) * | 2002-02-20 | 2004-06-02 | Sirna Therapeutics, Inc. | Rna interference mediated inhibition of gene expression using short interfering nucleic acid (sina) |
| US9657294B2 (en) | 2002-02-20 | 2017-05-23 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US9957517B2 (en) | 2002-02-20 | 2018-05-01 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| EP1572128B1 (en) * | 2002-02-20 | 2011-08-24 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF HIV GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US10000754B2 (en) | 2002-02-20 | 2018-06-19 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US9738899B2 (en) | 2002-02-20 | 2017-08-22 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US9732344B2 (en) | 2002-02-20 | 2017-08-15 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| WO2005045036A2 (en) * | 2002-02-20 | 2005-05-19 | Sirna Therapeutics, Inc. | Rna interference mediated inhibition of hairless (hr) gene expression using short interfering nucleic acid (sina) |
| EP1572128A2 (en) * | 2002-02-20 | 2005-09-14 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF HIV GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US10351852B2 (en) | 2002-02-20 | 2019-07-16 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US9181551B2 (en) | 2002-02-20 | 2015-11-10 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US9771588B2 (en) | 2002-02-20 | 2017-09-26 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| EP1423406B1 (en) * | 2002-02-20 | 2010-09-01 | Sirna Therapeutics, Inc. | Rna interference mediated inhibition of gene expression using short interfering nucleic acid (sina) |
| US10662428B2 (en) | 2002-02-20 | 2020-05-26 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US10889815B2 (en) | 2002-02-20 | 2021-01-12 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US8846894B2 (en) | 2002-02-20 | 2014-09-30 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| EP1430157A2 (en) * | 2002-02-20 | 2004-06-23 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF HEPATITIS C VIRUS (HCV) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US7989612B2 (en) | 2002-02-20 | 2011-08-02 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| WO2005045036A3 (en) * | 2002-02-20 | 2006-03-02 | Sirna Therapeutics Inc | Rna interference mediated inhibition of hairless (hr) gene expression using short interfering nucleic acid (sina) |
| EP1430157B1 (en) * | 2002-02-20 | 2011-08-10 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF HEPATITIS C VIRUS (HCV) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| EP1423404B1 (en) * | 2002-02-20 | 2011-08-24 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED TREATMENT OF ALZHEIMER'S DISEASE USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US9790501B2 (en) | 2002-08-05 | 2017-10-17 | Silence Therapeutics Gmbh | Interfering RNA molecules |
| US9790505B2 (en) | 2002-08-05 | 2017-10-17 | Silence Therapeutics Gmbh | Interfering RNA molecules |
| US7452987B2 (en) | 2002-08-05 | 2008-11-18 | Silence Therapeutics Aktiengesellschaft (Ag) | Interfering RNA molecules |
| US11578328B2 (en) | 2002-08-05 | 2023-02-14 | Silence Therapeutics Gmbh | Interfering RNA molecules |
| EP1857547B1 (en) | 2002-08-05 | 2018-01-17 | Silence Therapeutics GmbH | Further novel forms of interfering RNA molecules |
| US9222092B2 (en) | 2002-08-05 | 2015-12-29 | Silence Therapeutics Gmbh | Interfering RNA molecules |
| EP2258847B1 (en) | 2002-08-05 | 2017-03-15 | Silence Therapeutics GmbH | Futher novel forms of interfering RNA molecules |
| US10266829B2 (en) | 2002-08-05 | 2019-04-23 | Silence Therapeutics Gmbh | Interfering RNA molecules |
| US10323246B2 (en) | 2002-08-05 | 2019-06-18 | Silence Therapeutics Gmbh | Interfering RNA molecules |
| US10329568B2 (en) | 2002-08-05 | 2019-06-25 | Silence Therapeutics Gmbh | Interfering RNA molecules |
| US9695423B2 (en) | 2002-08-05 | 2017-07-04 | Silence Therapeutics Gmbh | Interfering RNA molecules |
| US8324370B2 (en) | 2002-08-05 | 2012-12-04 | Silence Therapeutics Aktiengesellschaft (Ag) | Interfering RNA molecules |
| US9758784B1 (en) | 2002-08-05 | 2017-09-12 | Silence Therapeutics Gmbh | Interfering RNA molecules |
| US9783802B2 (en) | 2002-08-05 | 2017-10-10 | Silence Therapeutics Gmbh | Interfering RNA molecules |
| US7893245B2 (en) | 2002-08-05 | 2011-02-22 | Silence Therapeutics Aktiengesellschaft (Ag) | Interfering RNA molecules |
| US8933215B2 (en) | 2002-08-05 | 2015-01-13 | Silence Therapeutics Aktiengesellschaft (Ag) | Interfering RNA molecules |
| US10774332B2 (en) | 2002-08-05 | 2020-09-15 | Silence Therapeutics Gmbh | Interfering RNA molecules |
| US9611472B2 (en) | 2002-08-07 | 2017-04-04 | University Of Massachusetts | Compositions for RNA interference and methods of use thereof |
| US8729036B2 (en) | 2002-08-07 | 2014-05-20 | University Of Massachusetts | Compositions for RNA interference and methods of use thereof |
| US7956176B2 (en) | 2002-09-05 | 2011-06-07 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| US7923547B2 (en) | 2002-09-05 | 2011-04-12 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) |
| EP2072619A1 (en) | 2002-10-18 | 2009-06-24 | Silence Therapeutics Aktiengesellschaft | Factor involved in metastasis and uses thereof |
| EP1580264A1 (en) * | 2002-10-29 | 2005-09-28 | Oriental Yeast Co., Ltd. | Construction of knockdown animal by transferring double-stranded rna expression vector |
| EP1580264A4 (en) * | 2002-10-29 | 2006-03-08 | Oriental Yeast Co Ltd | Construction of knockdown animal by transferring double-stranded rna expression vector |
| EP1563070A4 (en) * | 2002-11-05 | 2008-05-28 | Isis Pharmaceuticals Inc | 2'-substituted oligomeric compounds and compositions for use in gene modulations |
| US8604183B2 (en) | 2002-11-05 | 2013-12-10 | Isis Pharmaceuticals, Inc. | Compositions comprising alternating 2′-modified nucleosides for use in gene modulation |
| EP1563070A2 (en) * | 2002-11-05 | 2005-08-17 | Isis Pharmaceuticals, Inc. | 2'-substituted oligomeric compounds and compositions for use in gene modulations |
| US10478449B2 (en) * | 2002-11-05 | 2019-11-19 | Ionis Pharmaceuticals, Inc. | 2′-methoxy substituted oligomeric compounds and compositions for use in gene modulations |
| US10233449B2 (en) | 2002-11-14 | 2019-03-19 | Thermo Fisher Scientific Inc. | Methods and compositions for selecting siRNA of improved functionality |
| US7807820B2 (en) | 2002-11-14 | 2010-10-05 | Dharmacon, Inc. | siRNA targeting beta secretase (BACE) |
| US9719094B2 (en) | 2002-11-14 | 2017-08-01 | Thermo Fisher Scientific Inc. | RNAi targeting SEC61G |
| US7618814B2 (en) | 2002-11-14 | 2009-11-17 | Rosetta Genomics Ltd. | Microrna-related nucleic acids and uses thereof |
| US7655785B1 (en) | 2002-11-14 | 2010-02-02 | Rosetta Genomics Ltd. | Bioinformatically detectable group of novel regulatory oligonucleotides and uses thereof |
| US8084598B1 (en) | 2002-11-14 | 2011-12-27 | Rosetta Genomics Inc. | Bioionformality detectable group of novel regulatory oligonucleotides and uses thereof |
| US11198870B2 (en) | 2002-11-14 | 2021-12-14 | Thermo Fisher Scientific Inc. | Methods and compositions for selecting siRNA of improved functionality |
| US8163896B1 (en) | 2002-11-14 | 2012-04-24 | Rosetta Genomics Ltd. | Bioinformatically detectable group of novel regulatory genes and uses thereof |
| US9777270B2 (en) | 2002-11-14 | 2017-10-03 | Thermo Fisher Scientific Inc. | Methods and compositions for selecting siRNA of improved functionality |
| US9719092B2 (en) | 2002-11-14 | 2017-08-01 | Thermo Fisher Scientific Inc. | RNAi targeting CNTD2 |
| US9771586B2 (en) | 2002-11-14 | 2017-09-26 | Thermo Fisher Scientific Inc. | RNAi targeting ZNF205 |
| US7217807B2 (en) | 2002-11-26 | 2007-05-15 | Rosetta Genomics Ltd | Bioinformatically detectable group of novel HIV regulatory genes and uses thereof |
| US7777022B2 (en) | 2002-12-05 | 2010-08-17 | Rosetta Genomics, Ltd. | Bioinformatically detectable group of novel regulatory viral and viral associated oligonucleotides and uses thereof |
| US8796235B2 (en) | 2003-02-21 | 2014-08-05 | University Of South Florida | Methods for attenuating dengue virus infection |
| US9738894B2 (en) | 2003-03-21 | 2017-08-22 | Roche Innovation Center Copenhagen A/S | Short interfering RNA (siRNA) analogues |
| US9297010B2 (en) | 2003-03-21 | 2016-03-29 | Roche Innovation Center Copenhagen A/S | Short interfering RNA (siRNA) analogues |
| US8653252B2 (en) | 2003-03-21 | 2014-02-18 | Santaris Pharma A/S | Short interfering RNA (siRNA) analogues |
| US8575327B2 (en) | 2003-06-12 | 2013-11-05 | Alnylam Pharmaceuticals, Inc. | Conserved HBV and HCV sequences useful for gene silencing |
| WO2005014806A3 (en) * | 2003-06-12 | 2005-12-01 | Nucleonics Inc | Conserved hbv and hcv sequences useful for gene silencing |
| US9200281B2 (en) | 2003-06-12 | 2015-12-01 | Alnylam Pharmaceuticals, Inc. | Conserved HBV and HCV sequences useful for gene silencing |
| US8350021B2 (en) | 2003-06-12 | 2013-01-08 | Alnylam Pharmaceuticals, Inc. | Conserved HBV and HCV sequences useful for gene silencing |
| US9982263B2 (en) | 2003-06-12 | 2018-05-29 | Alnylam Pharmaceuticals, Inc. | Conserved HBV and HCV sequences useful for gene silencing |
| US10982212B2 (en) | 2003-06-12 | 2021-04-20 | Alnylam Pharmaceuticals, Inc. | Conserved HBV and HCV sequences useful for gene silencing |
| EP1636342A2 (en) * | 2003-06-20 | 2006-03-22 | Isis Pharmaceuticals, Inc. | Oligomeric compounds for use in gene modulation |
| EP1644475A4 (en) * | 2003-06-20 | 2009-06-03 | Isis Pharmaceuticals Inc | Double stranded compositions comprising a 3'-endo modified strand for use in gene modulation |
| EP1636342A4 (en) * | 2003-06-20 | 2008-10-08 | Isis Pharmaceuticals Inc | Oligomeric compounds for use in gene modulation |
| EP1644475A2 (en) * | 2003-06-20 | 2006-04-12 | Isis Pharmaceuticals, Inc. | DOUBLE STRANDED COMPOSITIONS COMPRISING A 3’-ENDO MODIFIED STRAND FOR USE IN GENE MODULATION |
| US7888497B2 (en) | 2003-08-13 | 2011-02-15 | Rosetta Genomics Ltd. | Bioinformatically detectable group of novel regulatory oligonucleotides and uses thereof |
| EP2338995A3 (en) * | 2003-08-28 | 2012-01-11 | Novartis AG | Interfering RNA duplex having blunt-ends and 3'-modifications |
| US8097716B2 (en) | 2003-08-28 | 2012-01-17 | Novartis Ag | Interfering RNA duplex having blunt-ends and 3′-modifications |
| WO2005021749A1 (en) * | 2003-08-28 | 2005-03-10 | Novartis Ag | Interfering rna duplex having blunt-ends and 3’-modifications |
| US8729339B2 (en) | 2003-10-09 | 2014-05-20 | E.I Du Pont De Nemours And Company | Gene silencing |
| WO2005045032A3 (en) * | 2003-10-20 | 2006-03-02 | Sirna Therapeutics Inc | RNA INTERFERENCE MEDIATED INHIBITION OF EARLY GROWTH RESPONSE GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| WO2005045032A2 (en) * | 2003-10-20 | 2005-05-19 | Sima Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF EARLY GROWTH RESPONSE GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| WO2005045041A3 (en) * | 2003-10-23 | 2005-08-18 | Sirna Therapeutics Inc | Rna interference mediated inhibition of cholesteryl ester transfer protein (cetp) gene expression using short interfering nucleic acid (sina) |
| JP2007522794A (en) * | 2003-10-23 | 2007-08-16 | サーナ・セラピューティクス・インコーポレイテッド | Inhibition of CETP gene expression via RNA interference using short interfering nucleic acids (siNA) |
| WO2005040379A3 (en) * | 2003-10-23 | 2006-03-02 | Sirna Therapeutics Inc | RNA INTERFERENCE MEDIATED INHIBITION OF RAS GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| WO2005045038A3 (en) * | 2003-10-23 | 2006-03-02 | Ivan Richards | RNA interference mediated inhibition of GPRA and AAA1 gene expression using short nucleic acid (siNA) |
| WO2005045034A3 (en) * | 2003-10-23 | 2005-08-11 | Sirna Therapeutics Inc | RNA INTERFERENCE MEDIATED TREATMENT OF PARKINSON DISEASE USING SHORT INTERERING NUCLEIC ACID (siNA) |
| WO2005040379A2 (en) * | 2003-10-23 | 2005-05-06 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF RAS GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| WO2005045040A3 (en) * | 2003-10-23 | 2006-02-23 | Sirna Therapeutics Inc | RNA INTERFERANCE MEDIATED INHIBITION OF CHOLINERGIC MUSCARINIC RECEPTOR (CHRM3) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID /siNA) |
| JP2007522793A (en) * | 2003-10-23 | 2007-08-16 | サーナ・セラピューティクス・インコーポレイテッド | Inhibition of NOGO and / or NOGO receptor gene expression via RNA interference using short interfering nucleic acids (siNA) |
| WO2005045034A2 (en) * | 2003-10-23 | 2005-05-19 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED TREATMENT OF PARKINSON DISEASE USING SHORT INTERERING NUCLEIC ACID (siNA) |
| WO2005045037A3 (en) * | 2003-10-23 | 2005-11-03 | Sirna Therapeutics Inc | RNA INTERFERENCE MEDIATED INHIBITION OF 5-ALPHA REDUCTASE AND ANDROGEN RECEPTOR GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| WO2005045040A2 (en) * | 2003-10-23 | 2005-05-19 | Sirna Therapeutics, Inc. | RNA INTERFERANCE MEDIATED INHIBITION OF CHOLINERGIC MUSCARINIC RECEPTOR (CHRM3) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID /siNA) |
| WO2005045038A2 (en) * | 2003-10-23 | 2005-05-19 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF GPRA AND AAA1 GENE EXPRESSION USING SHORT NUCLEIC ACID (siNA) |
| WO2005045041A2 (en) * | 2003-10-23 | 2005-05-19 | Sirna Therapeutics, Inc. | Rna interference mediated inhibition of cholesteryl ester transfer protein (cetp) gene expression using short interfering nucleic acid (sina) |
| WO2005045037A2 (en) * | 2003-10-23 | 2005-05-19 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF 5-ALPHA REDUCTASE AND ANDROGEN RECEPTOR GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| WO2005045035A2 (en) * | 2003-10-23 | 2005-05-19 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF NOGO AND NOGO RECEPTOR GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| WO2005045039A2 (en) * | 2003-10-23 | 2005-05-19 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF INTERCELLULAR ADHESION MOLECULE (ICAM) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| WO2005045035A3 (en) * | 2003-10-23 | 2005-12-08 | Sirna Therapeutics Inc | RNA INTERFERENCE MEDIATED INHIBITION OF NOGO AND NOGO RECEPTOR GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| WO2005045039A3 (en) * | 2003-10-23 | 2006-01-12 | Ivan Richards | RNA INTERFERENCE MEDIATED INHIBITION OF INTERCELLULAR ADHESION MOLECULE (ICAM) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| US9089590B2 (en) | 2003-12-04 | 2015-07-28 | University Of South Florida | Polynucleotides for reducing respiratory syncytial virus gene expression |
| WO2005056021A1 (en) * | 2003-12-04 | 2005-06-23 | University Of South Florida | Polynucleotides for reducing respiratory syncytial virus gene expression |
| WO2005059134A1 (en) * | 2003-12-17 | 2005-06-30 | Index Pharmaceuticals Ab | COMPOUNDS AND METHODS FOR RNA INTERFERENCE OF THE p65 SUBUNIT OF NF-KAPPA-B |
| WO2005073378A1 (en) * | 2004-01-30 | 2005-08-11 | Santaris Pharma A/S | MODIFIED SHORT INTERFERING RNA (MODIFIED siRNA) |
| US7858769B2 (en) | 2004-02-10 | 2010-12-28 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using multifunctional short interfering nucleic acid (multifunctional siNA) |
| WO2005078097A3 (en) * | 2004-02-10 | 2005-10-27 | Sirna Therapeutics Inc | RNA INTERFERENCE MEDIATED INHIBITION OF GENE EXPRESSION USING MULTIFUNCTIONAL SHORT INTERFERING NUCLEIC ACID (Multifunctional siNA) |
| WO2005080568A1 (en) * | 2004-02-20 | 2005-09-01 | Index Pharmaceuticals Ab | Methods and compositions for treatment or prevention of secondary ischemic injury |
| US8569474B2 (en) | 2004-03-09 | 2013-10-29 | Isis Pharmaceuticals, Inc. | Double stranded constructs comprising one or more short strands hybridized to a longer strand |
| US7919473B2 (en) | 2004-03-12 | 2011-04-05 | Alnylam Pharmaceuticals, Inc. | IRNA agents targeting VEGF |
| US8329667B2 (en) | 2004-04-23 | 2012-12-11 | The Trustees Of Columbia University In The City Of New York | Inhibition of hairless protein mRNA |
| US8946402B2 (en) | 2004-04-23 | 2015-02-03 | The Trustees Of Columbia University In The City Of New York | Inhibition of hairless protein mRNA |
| EP1753292A4 (en) * | 2004-05-12 | 2007-09-26 | Kamel Khalili | COMPOSITIONS AND METHODS FOR siRNA INHIBITION OF PRIMATE POLYOMAVIRUS GENES |
| EP1753292A2 (en) * | 2004-05-12 | 2007-02-21 | Kamel Khalili | COMPOSITIONS AND METHODS FOR siRNA INHIBITION OF PRIMATE POLYOMAVIRUS GENES |
| US7687616B1 (en) | 2004-05-14 | 2010-03-30 | Rosetta Genomics Ltd | Small molecules modulating activity of micro RNA oligonucleotides and micro RNA targets and uses thereof |
| US7709616B2 (en) | 2004-05-14 | 2010-05-04 | Rosetta Genomics Inc. | Micrornas and uses thereof |
| US10508277B2 (en) | 2004-05-24 | 2019-12-17 | Sirna Therapeutics, Inc. | Chemically modified multifunctional short interfering nucleic acid molecules that mediate RNA interference |
| US7795419B2 (en) | 2004-05-26 | 2010-09-14 | Rosetta Genomics Ltd. | Viral and viral associated miRNAs and uses thereof |
| US8455633B2 (en) | 2004-05-26 | 2013-06-04 | Rosetta Genomics Ltd. | Viral and viral associated mirnas and uses thereof |
| US8334373B2 (en) | 2004-05-27 | 2012-12-18 | Alnylam Pharmaceuticals, Inc. | Nuclease resistant double-stranded ribonucleic acid |
| US7928217B2 (en) | 2004-05-27 | 2011-04-19 | Alnylam Pharmaceuticals, Inc. | Nuclease resistant double-stranded ribonucleic acid |
| EP2399924A2 (en) | 2004-05-27 | 2011-12-28 | Alnylam Pharmaceuticals, Inc. | Nuclease resistant double-stranded ribonucleic acid |
| US8993746B2 (en) | 2004-05-27 | 2015-03-31 | Alnylam Pharmaceuticals, Inc. | Nuclease resistant double-stranded ribonucleic acid |
| US8394947B2 (en) | 2004-06-03 | 2013-03-12 | Isis Pharmaceuticals, Inc. | Positionally modified siRNA constructs |
| WO2005121370A3 (en) * | 2004-06-03 | 2006-05-26 | Isis Pharmaceuticals Inc | Oligomeric compounds that facilitate risc loading |
| WO2005121372A3 (en) * | 2004-06-03 | 2006-04-13 | Isis Pharmaceuticals Inc | Double strand compositions comprising differentially modified strands for use in gene modulation |
| EP1784498A2 (en) * | 2004-08-03 | 2007-05-16 | Duke University | Systems and methods for inhibiting metastasis |
| EP1784498A4 (en) * | 2004-08-03 | 2007-12-05 | Univ Duke | Systems and methods for inhibiting metastasis |
| US8642571B2 (en) | 2004-08-06 | 2014-02-04 | Quark Pharmaceuticals, Inc. | Therapeutic uses of inhibitors of RTP801 |
| US8168607B2 (en) | 2004-08-06 | 2012-05-01 | Quark Pharmaceuticals Inc. | Methods of treating eye diseases in diabetic patients |
| US7741299B2 (en) | 2004-08-16 | 2010-06-22 | Quark Pharmaceuticals, Inc. | Therapeutic uses of inhibitors of RTP801 |
| EP2319925A2 (en) | 2004-08-16 | 2011-05-11 | Quark Pharmaceuticals, Inc. | Therapeutic uses of inhibitors of RTP801 |
| US8309532B2 (en) | 2004-08-16 | 2012-11-13 | Quark Pharmaceuticals, Inc. | Therapeutic uses of inhibitors of RTP801 |
| EP1786905A1 (en) * | 2004-08-18 | 2007-05-23 | GeneSense Technologies Inc. | Small interfering rna molecules against ribonucleotide reductase and uses thereof |
| EP1786905A4 (en) * | 2004-08-18 | 2008-08-20 | Lorus Therapeutics Inc | Small interfering rna molecules against ribonucleotide reductase and uses thereof |
| WO2006025154A1 (en) * | 2004-08-30 | 2006-03-09 | Gifu University | Modified oligonucleotides |
| JPWO2006025154A1 (en) * | 2004-08-30 | 2008-05-08 | 国立大学法人岐阜大学 | Modified oligonucleotide |
| US7884086B2 (en) | 2004-09-08 | 2011-02-08 | Isis Pharmaceuticals, Inc. | Conjugates for use in hepatocyte free uptake assays |
| US8765699B2 (en) | 2004-09-28 | 2014-07-01 | Quark Pharmaceuticals, Inc. | Oligoribonucleotides and methods of use thereof for treatment of alopecia, acute renal failure and other diseases |
| US7842674B2 (en) | 2004-09-28 | 2010-11-30 | Quark Pharmaceuticals Inc. | Methods of treatment of acute renal failure |
| US8148342B2 (en) | 2004-09-28 | 2012-04-03 | Quark Pharmaceuticals Inc. | Oligoribonucleotides and methods of use thereof for treatment of alopecia, acute renal failure and other diseases |
| US8975471B2 (en) | 2004-10-12 | 2015-03-10 | The Rockefeller University | MicroRNAs |
| US20080279920A1 (en) * | 2004-11-05 | 2008-11-13 | Intradigm Corporation | Compositions For Treating Respiratory Viral Infections and Their Use |
| US8691781B2 (en) | 2004-11-05 | 2014-04-08 | Sirnaomics, Inc. | Compositions for treating respiratory viral infections and their use |
| WO2006060598A3 (en) * | 2004-12-01 | 2006-10-26 | Sirna Therapeutics Inc | RNA INTERFERENCE MEDIATED INHIBITION OF INTERLEUKIN AND INTERLEUKIN RECEPTOR GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| WO2006060598A2 (en) * | 2004-12-01 | 2006-06-08 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF INTERLEUKIN AND INTERLEUKIN RECEPTOR GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| AU2011203218B2 (en) * | 2004-12-09 | 2013-02-07 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inducing an immune response in a mammal and methods of avoiding an immune response to oligonucleotide agents such as short interfering RNAs |
| US8003619B2 (en) | 2004-12-09 | 2011-08-23 | Alnylam Pharmaceuticals, Inc. | Method of stimulating an immune response and inhibiting expression of a gene using an oligonucleotide |
| AU2005313883B2 (en) * | 2004-12-09 | 2011-03-31 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inducing an immune response in a mammal and methods of avoiding an immune response to oligonucleotide agents such as short interfering RNAs |
| WO2006078414A2 (en) * | 2004-12-23 | 2006-07-27 | Ambion, Inc. | METHODS AND COMPOSITIONS CONCERNING siRNA'S AS MEDIATORS OF RNA INTERFERENCE |
| WO2006078414A3 (en) * | 2004-12-23 | 2007-01-25 | Ambion Inc | METHODS AND COMPOSITIONS CONCERNING siRNA'S AS MEDIATORS OF RNA INTERFERENCE |
| WO2006078798A3 (en) * | 2005-01-18 | 2007-01-25 | Sirna Therapeutics Inc | RNA INTERFERENCE MEDIATED INHIBITION OF RETINOBLASTOMA (RB1) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| WO2006078798A2 (en) * | 2005-01-18 | 2006-07-27 | Sirna Therapeutics, Inc. | RNA INTERFERENCE MEDIATED INHIBITION OF RETINOBLASTOMA (RB1) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| EP1681347A1 (en) * | 2005-01-18 | 2006-07-19 | Metanomics GmbH & Co. KGaA | Improved methods for double-stranded RNA mediated gene silencing |
| US8999943B2 (en) | 2005-03-14 | 2015-04-07 | Board Of Regents, The University Of Texas System | Antigene oligomers inhibit transcription |
| WO2006128141A2 (en) * | 2005-05-27 | 2006-11-30 | Sirna Therapeutics, Inc. | Rna interference mediated inhibition of stromal cell-derived factor-1 (sdf-1) gene expression using short interfering nucleic acid (sina) |
| WO2006128141A3 (en) * | 2005-05-27 | 2007-07-12 | Sirna Therapeutics Inc | Rna interference mediated inhibition of stromal cell-derived factor-1 (sdf-1) gene expression using short interfering nucleic acid (sina) |
| US7919583B2 (en) | 2005-08-08 | 2011-04-05 | Discovery Genomics, Inc. | Integration-site directed vector systems |
| WO2007022369A2 (en) * | 2005-08-17 | 2007-02-22 | Sirna Therapeutics, Inc. | Chemically modified short interfering nucleic acid molecules that mediate rna interference |
| WO2007022369A3 (en) * | 2005-08-17 | 2007-07-26 | Sirna Therapeutics Inc | Chemically modified short interfering nucleic acid molecules that mediate rna interference |
| AU2006279454B2 (en) * | 2005-08-17 | 2011-12-15 | Sirna Therapeutics, Inc. | Chemically modified short interfering nucleic acid molecules that mediate rna interference |
| EP2980220A1 (en) | 2005-09-20 | 2016-02-03 | BASF Plant Science GmbH | Improved methods controlling gene expression |
| WO2007092059A3 (en) * | 2005-10-03 | 2007-12-13 | Sirna Therapeutics Inc | Rna interference mediated inhibition of influenza virus gene expression using short interfering nucleic acid |
| WO2007092059A2 (en) * | 2005-10-03 | 2007-08-16 | Sirna Therapeutics, Inc. | Rna interference mediated inhibition of influenza virus gene expression using short interfering nucleic acid |
| JP2009513716A (en) * | 2005-11-01 | 2009-04-02 | アルナイラム ファーマシューティカルズ インコーポレイテッド | Inhibition of influenza virus replication by RNAi |
| US8513403B2 (en) | 2005-11-02 | 2013-08-20 | Protiva Biotherapeutics, Inc. | Modified siRNA molecules and uses thereof |
| US8188263B2 (en) | 2005-11-02 | 2012-05-29 | Protiva Biotherapeutics, Inc. | Modified siRNA molecules and uses thereof |
| US8101741B2 (en) | 2005-11-02 | 2012-01-24 | Protiva Biotherapeutics, Inc. | Modified siRNA molecules and uses thereof |
| US9074208B2 (en) | 2005-11-02 | 2015-07-07 | Protiva Biotherapeutics, Inc. | Modified siRNA molecules and uses thereof |
| EP1957648A2 (en) * | 2005-11-17 | 2008-08-20 | Board of Regents, The University of Texas System | Modulation of gene expression by oligomers targeted to chromosomal dna |
| EP1957648A4 (en) * | 2005-11-17 | 2009-01-21 | Univ Texas | Modulation of gene expression by oligomers targeted to chromosomal dna |
| US8324181B2 (en) | 2005-11-17 | 2012-12-04 | Board Of Regents, The University Of Texas System | Modulation of gene expression by oligomers targeted to chromosomal DNA |
| US7709456B2 (en) | 2005-11-17 | 2010-05-04 | Board Of Regents, The University Of Texas System | Modulation of gene expression by oligomers targeted to chromosomal DNA |
| US7825099B2 (en) | 2006-01-20 | 2010-11-02 | Quark Pharmaceuticals, Inc. | Treatment or prevention of oto-pathologies by inhibition of pro-apoptotic genes |
| US8404654B2 (en) | 2006-01-20 | 2013-03-26 | Quark Pharmaceuticals, Inc. | Treatment or prevention of oto-pathologies by inhibition of pro-apoptotic genes |
| EP2402443A2 (en) | 2006-01-20 | 2012-01-04 | Quark Pharmaceuticals, Inc. | Therapeutic uses of inhibitors of rtp801 |
| US9056903B2 (en) | 2006-01-20 | 2015-06-16 | Quark Pharmaceuticals, Inc. | Therapeutic uses of inhibitors of RTP801 |
| US8067570B2 (en) | 2006-01-20 | 2011-11-29 | Quark Pharmaceuticals, Inc. | Therapeutic uses of inhibitors of RTP801 |
| EP1984382B1 (en) * | 2006-01-27 | 2012-08-15 | Santaris Pharma A/S | Lna modified phosphorothiolated oligonucleotides |
| US8362229B2 (en) | 2006-02-08 | 2013-01-29 | Quark Pharmaceuticals, Inc. | Tandem siRNAS |
| US7910566B2 (en) | 2006-03-09 | 2011-03-22 | Quark Pharmaceuticals Inc. | Prevention and treatment of acute renal failure and other kidney diseases by inhibition of p53 by siRNA |
| WO2007107162A2 (en) * | 2006-03-23 | 2007-09-27 | Santaris Pharma A/S | Small internally segmented interfering rna |
| US8329888B2 (en) | 2006-03-23 | 2012-12-11 | Santaris Pharma A/S | Small internally segmented interfering RNA |
| EA015563B1 (en) * | 2006-03-23 | 2011-08-30 | Сантарис Фарма А/С | Small internally segmented interfering rna |
| WO2007107162A3 (en) * | 2006-03-23 | 2008-05-08 | Santaris Pharma As | Small internally segmented interfering rna |
| EP2008274A2 (en) * | 2006-03-31 | 2008-12-31 | Alnylam Pharmaceuticals Inc. | Compositions and methods for inhibiting expression of eg5 gene |
| EP2008274A4 (en) * | 2006-03-31 | 2009-10-28 | Alnylam Pharmaceuticals Inc | Compositions and methods for inhibiting expression of eg5 gene |
| WO2007120527A3 (en) * | 2006-03-31 | 2008-07-03 | Sirna Therapeutics Inc | Rna interference mediated inhibition of respiratory syncytial virus (rsv) expression using short interfering nucleic acid (sina) |
| US7718629B2 (en) | 2006-03-31 | 2010-05-18 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inhibiting expression of Eg5 gene |
| EP2527354A1 (en) * | 2006-03-31 | 2012-11-28 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inhibiting expression of Eg5 gene |
| WO2007120527A2 (en) * | 2006-03-31 | 2007-10-25 | Sirna Therapeutics Inc. | Rna interference mediated inhibition of respiratory syncytial virus (rsv) expression using short interfering nucleic acid (sina) |
| US9493771B2 (en) | 2006-05-04 | 2016-11-15 | Novartis Ag | Short interfering ribonucleic acid (siRNA) for oral administration |
| US8404831B2 (en) | 2006-05-04 | 2013-03-26 | Novartis Ag | Short interfering ribonucleic acid (siRNA) for oral administration |
| EP3121278A1 (en) | 2006-05-04 | 2017-01-25 | Novartis Ag | Short interfering ribonucleic acid (sirna) |
| US8957041B2 (en) | 2006-05-04 | 2015-02-17 | Novartis Ag | Short interfering ribonucleic acid (siRNA) for oral administration |
| EP2322617A2 (en) | 2006-05-04 | 2011-05-18 | Novartis AG | Short interfering ribonucleic acid (siRNA) for oral administration |
| US8344128B2 (en) | 2006-05-04 | 2013-01-01 | Novartis Ag | Short interfering ribonucleic acid (siRNA) for oral administration |
| EP2135950A2 (en) | 2006-05-04 | 2009-12-23 | Novartis AG | Short interfering ribonucleic acid (siRNA) for oral administration |
| EP2765196A1 (en) | 2006-05-04 | 2014-08-13 | Novartis AG | Short interfering ribonucleic acid (siRNA) |
| US8084600B2 (en) | 2006-05-04 | 2011-12-27 | Novartis Ag | Short interfering ribonucleic acid (siRNA) with improved pharmacological properties |
| EP2759596A1 (en) | 2006-05-04 | 2014-07-30 | Novartis AG | Short interfering ribonucleic acid (siRNA) |
| US8404832B2 (en) | 2006-05-04 | 2013-03-26 | Novartis Ag | Short interfering ribonucleic acid (siRNA) for oral administration |
| EP2762568A1 (en) | 2006-05-04 | 2014-08-06 | Novartis AG | Short interfering ribonucleic acid (siRNA) |
| EP2584048A1 (en) | 2006-05-11 | 2013-04-24 | Alnylam Pharmaceuticals Inc. | Compositions and methods for inhibiting expression of the PCSK9 gene |
| US8222222B2 (en) | 2006-05-11 | 2012-07-17 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inhibiting expression of the PCSK9 gene |
| EP2021507A2 (en) * | 2006-05-11 | 2009-02-11 | Alnylam Pharmaceuticals Inc. | Compositions and methods for inhibiting expression of the pcsk9 gene |
| JP2009536827A (en) * | 2006-05-11 | 2009-10-22 | アルニラム ファーマシューティカルズ, インコーポレイテッド | Compositions and methods for inhibiting the expression of the PCSK9 gene |
| EP2584047A1 (en) | 2006-05-11 | 2013-04-24 | Alnylam Pharmaceuticals Inc. | Compositions and methods for inhibiting expression of the PCSK9 gene |
| US9822365B2 (en) | 2006-05-11 | 2017-11-21 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inhibiting expression of the PCSK9 gene |
| EP3249052A1 (en) | 2006-05-11 | 2017-11-29 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inhibiting expression of the pcsk9 gene |
| US10501742B2 (en) | 2006-05-11 | 2019-12-10 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inhibiting expression of the PCSK9 gene |
| EP3578656A1 (en) | 2006-05-11 | 2019-12-11 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inhibiting expression of the pcsk9 gene |
| EP2021507A4 (en) * | 2006-05-11 | 2009-10-28 | Alnylam Pharmaceuticals Inc | Compositions and methods for inhibiting expression of the pcsk9 gene |
| KR101320916B1 (en) * | 2006-05-11 | 2013-10-23 | 알닐람 파마슈티칼스 인코포레이티드 | Compositions and methods for inhibiting expression of the PCSK9 gene |
| EP2194128A1 (en) | 2006-05-11 | 2010-06-09 | Alnylam Pharmaceuticals Inc. | Compositions and methods for inhibiting expression of the PCSK9 gene |
| EP3872179A1 (en) | 2006-05-11 | 2021-09-01 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inhibiting expression of the pcsk9 gene |
| US9260718B2 (en) | 2006-05-11 | 2016-02-16 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inhibiting expression of the PCSK9 gene |
| EP2835429A1 (en) | 2006-05-11 | 2015-02-11 | Alnylam Pharmaceuticals Inc. | Compositions and methods for inhibiting expression of the PCSK9 gene |
| US8809292B2 (en) | 2006-05-11 | 2014-08-19 | Alnylam Pharmaceuticals, Inc | Compositions and methods for inhibiting expression of the PCSK9 gene |
| EP2023937A4 (en) * | 2006-05-19 | 2009-10-28 | Alnylam Pharmaceuticals Inc | Rnai modulation of aha and therapeutic uses thereof |
| EP2021352A4 (en) * | 2006-05-19 | 2009-10-28 | Scripps Research Inst | Treatment of protein misfolding |
| EP2021352A2 (en) * | 2006-05-19 | 2009-02-11 | The Scripps Research Institute | Treatment of protein misfolding |
| EP2023937A2 (en) * | 2006-05-19 | 2009-02-18 | Alnylam Pharmaceuticals Inc. | Rnai modulation of aha and therapeutic uses thereof |
| US7812150B2 (en) | 2006-05-19 | 2010-10-12 | Alnylam Pharmaceuticals, Inc. | RNAi modulation of Aha and therapeutic uses thereof |
| EP2392583A1 (en) * | 2006-05-19 | 2011-12-07 | Alnylam Europe AG. | RNAi modulation of Aha and therapeutic uses thereof |
| JP2009537153A (en) * | 2006-05-19 | 2009-10-29 | アルニラム ファーマシューティカルズ, インコーポレイテッド | Aha RNAi regulation and its therapeutic use |
| AU2007253803B2 (en) * | 2006-05-19 | 2012-05-24 | Alnylam Pharmaceuticals, Inc. | RNAi modulation of Aha and therapeutic uses thereof |
| EP2018443A4 (en) * | 2006-05-22 | 2009-11-11 | Alnylam Pharmaceuticals Inc | Compositions and methods for inhibiting expression of ikk-b gene |
| EP2584051A1 (en) * | 2006-05-22 | 2013-04-24 | Alnylam Pharmaceuticals Inc. | Compositions and methods for inhibiting expressions of IKK-B gene |
| EP2192200A1 (en) * | 2006-05-22 | 2010-06-02 | Alnylam Pharmaceuticals, Inc | Compositions and methods for inhibiting expression of IKK-B gene |
| EP2018443A2 (en) * | 2006-05-22 | 2009-01-28 | Alnylam Pharmaceuticals Inc. | Compositions and methods for inhibiting expression of ikk-b gene |
| US7888498B2 (en) | 2006-05-22 | 2011-02-15 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inhibiting expression of IKK-B gene |
| US7626015B2 (en) | 2006-06-09 | 2009-12-01 | Quark Pharmaceuticals, Inc. | Therapeutic uses of inhibitors of RTP801L |
| US8017764B2 (en) | 2006-06-09 | 2011-09-13 | Quark Pharmaceuticals Inc. | Therapeutic uses of inhibitors of RTP801L |
| US7915399B2 (en) | 2006-06-09 | 2011-03-29 | Protiva Biotherapeutics, Inc. | Modified siRNA molecules and uses thereof |
| US9200275B2 (en) | 2006-06-14 | 2015-12-01 | Merck Sharp & Dohme Corp. | Methods and compositions for regulating cell cycle progression |
| WO2007147143A3 (en) * | 2006-06-16 | 2008-03-06 | Sirna Therapeutics Inc | Rna interference mediated inhibition of 11 beta-hydroxysteroid dehydrogenase-1 (11 beta-hsd-1) gene expression using short interfering nucleic acid (sina) |
| WO2007147143A2 (en) * | 2006-06-16 | 2007-12-21 | Sirna Therapeutics, Inc. | Rna interference mediated inhibition of 11 beta-hydroxysteroid dehydrogenase-1 (11 beta-hsd-1) gene expression using short interfering nucleic acid (sina) |
| WO2008011467A2 (en) * | 2006-07-18 | 2008-01-24 | Sirna Therapeutics Inc. | Rna interference mediated inhibition of sterol regulatory element-binding protein 1 (srebp1) gene expression using short interfering nucleic acid (sina) |
| WO2008011467A3 (en) * | 2006-07-18 | 2008-08-07 | Sirna Therapeutics Inc | Rna interference mediated inhibition of sterol regulatory element-binding protein 1 (srebp1) gene expression using short interfering nucleic acid (sina) |
| US8293886B2 (en) | 2006-07-31 | 2012-10-23 | Universite Joseph Fourier | Sensizitation of cancer cells to therapy using sina targeting genes from the 1P and 19Q chromosomal regions |
| WO2008015577A2 (en) * | 2006-07-31 | 2008-02-07 | Universite Joseph Fourier (Grenoble 1) | Sensizitation of cancer cells to therapy using sina targeting genes from the 1p and 19q chromosomal regions |
| EP1884569A1 (en) * | 2006-07-31 | 2008-02-06 | Institut National De La Sante Et De La Recherche Medicale (Inserm) | Sensitization of cancer cells to therapy using siNA targeting genes from the 1p and 19q chromosomal regions |
| US7939653B2 (en) | 2006-07-31 | 2011-05-10 | Universite Joseph Fourier | Sensizitation of cancer cells to therapy using siNA targeting genes from the 1p and 19q chromosomal regions |
| WO2008015577A3 (en) * | 2006-07-31 | 2008-10-30 | Inst Nat Sante Rech Med | Sensizitation of cancer cells to therapy using sina targeting genes from the 1p and 19q chromosomal regions |
| EP2336320A1 (en) * | 2006-07-31 | 2011-06-22 | Universite Joseph Fourier (Grenoble 1) | Sensizitation of cancer cells to therapy using sina targeting genes from the 1p and 19q chromosomal regions |
| EP2399575A3 (en) * | 2006-08-11 | 2012-12-19 | INSERM (Institut National de la Santé et de la Recherche Médicale) | Methods, uses and compositions for treatment of an infection by a virus of the family of flaviviridae through the farnesoid X receptor (FXR) inhibition |
| WO2008036933A2 (en) | 2006-09-21 | 2008-03-27 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inhibiting expression of the hamp gene |
| WO2008042973A2 (en) | 2006-10-03 | 2008-04-10 | Alnylam Pharmaceuticals, Inc. | Lipid containing formulations |
| US9446062B2 (en) | 2006-10-25 | 2016-09-20 | Quark Pharmaceuticals, Inc. | Methods of treating ischemia-reperfusion injury with siRNAs |
| US7872119B2 (en) | 2007-02-26 | 2011-01-18 | Quark Pharmaceuticals, Inc. | Inhibitors of RTP801 and their use in disease treatment |
| WO2008109357A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting apob gene expression and uses thereof |
| WO2008109511A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting fcer2 gene expression and uses thereof |
| WO2008109526A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting egr gene expression and uses thereof |
| WO2008109449A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna Inc. | Nucleic acid compounds for inhibiting bcl2 gene expression and uses thereof |
| WO2008109534A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting ezh2 gene expression and uses thereof |
| WO2009029293A3 (en) * | 2007-03-02 | 2009-05-14 | Mdrna Inc | Nucleic acid compounds for inhibiting myc gene expression and uses thereof |
| WO2009029293A2 (en) * | 2007-03-02 | 2009-03-05 | Mdrna, Inc. | Nucleic acid compounds for inhibiting myc gene expression and uses thereof |
| WO2008109373A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting erbb gene expression and uses thereof |
| WO2008109375A3 (en) * | 2007-03-02 | 2009-03-05 | Mdrna Inc | Nucleic acid compounds for inhibiting pik3c gene expression and uses thereof |
| WO2008109362A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting vegf gene expression and uses thereof |
| WO2008109375A2 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting pik3c gene expression and uses thereof |
| WO2008109494A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting stat3 gene expression and uses thereof |
| WO2008109547A3 (en) * | 2007-03-02 | 2008-11-06 | Mdrna Inc | Nucleic acid compounds for inhibiting tymp gene expression and uses thereof |
| WO2008109381A2 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting hif1a gene expression and uses thereof |
| WO2008109381A3 (en) * | 2007-03-02 | 2008-10-30 | Mdrna Inc | Nucleic acid compounds for inhibiting hif1a gene expression and uses thereof |
| WO2008109475A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting sirt2 gene expression and uses thereof |
| WO2008109547A2 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting tymp gene expression and uses thereof |
| WO2008109354A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting il18 gene expression and uses thereof |
| WO2008109551A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting tacstd1 gene expression and uses thereof |
| WO2008109492A1 (en) * | 2007-03-02 | 2008-09-12 | Mdrna, Inc. | Nucleic acid compounds for inhibiting igf1r gene expression and uses thereof |
| US8410069B2 (en) | 2007-03-21 | 2013-04-02 | Quark Pharmaceuticals, Inc. | Oligoribonucleotide inhibitors of Nrf2 and methods of use thereof for treatment of cancer |
| US7812002B2 (en) | 2007-03-21 | 2010-10-12 | Quark Pharmaceuticals, Inc. | Oligoribonucleotide inhibitors of NRF2 and methods of use thereof for treatment of cancer |
| EP2441837A1 (en) | 2007-03-26 | 2012-04-18 | Alnylam Pharmaceuticals Inc. | dsRNA compositions and methods for treating HPV infections |
| EP2446890A1 (en) | 2007-04-30 | 2012-05-02 | Allergan, Inc. | High viscosity macromolecular compositions for treating ocular conditions |
| US11078262B2 (en) | 2007-04-30 | 2021-08-03 | Allergan, Inc. | High viscosity macromolecular compositions for treating ocular conditions |
| WO2008137867A2 (en) | 2007-05-03 | 2008-11-13 | Rosetta Inpharmatics Llc | Compositions comprising mir34 therapeutic agents for treating cancer |
| TWI468515B (en) * | 2007-05-09 | 2015-01-11 | Riken | Single-chain circular rna and method of producing the same |
| EP2143792A1 (en) * | 2007-05-09 | 2010-01-13 | Riken | Single-stranded cyclic rna, and method for production thereof |
| AU2008250075B2 (en) * | 2007-05-09 | 2014-06-19 | Otsuka Pharmaceutical Co., Ltd. | Single-chain circular RNA and method of producing the same |
| EP2143792A4 (en) * | 2007-05-09 | 2011-08-24 | Riken | Single-stranded cyclic rna, and method of production thereof |
| US9303260B2 (en) | 2007-05-22 | 2016-04-05 | Arcturus Therapeutics, Inc. | UNA duplex oligomers for therapeutics |
| US10457945B2 (en) | 2007-05-22 | 2019-10-29 | Arcturus Therapeutics, Inc. | UNA oligomers for therapeutics with prolonged stability |
| US9297009B2 (en) | 2007-05-22 | 2016-03-29 | Arcturus Therapeutics, Inc. | UNA oligomers targeting micro-RNA for therapeutics |
| US9051570B2 (en) | 2007-05-22 | 2015-06-09 | Arcturus Therapeutics, Inc. | UNA oligomers for therapeutics |
| US9944929B2 (en) | 2007-05-22 | 2018-04-17 | Arcturus Therapeutics, Inc. | UNA single stranded oligomers for therapeutics |
| US8785408B2 (en) | 2007-06-27 | 2014-07-22 | Quark Pharmaceuticals, Inc. | Compositions and methods for reducing or protecting against delayed graft function (DGF) |
| JP5298014B2 (en) * | 2007-07-03 | 2013-09-25 | 杏林製薬株式会社 | Influenza treatment |
| WO2009005095A1 (en) * | 2007-07-03 | 2009-01-08 | Kyorin Pharmaceutical Co., Ltd | Treatment of influenza |
| US8710024B2 (en) | 2007-07-03 | 2014-04-29 | Kyorin Pharmaceutical Co., Ltd. | Treatment of influenza |
| US8198256B2 (en) | 2007-07-03 | 2012-06-12 | Kyorin Pharmaceutical Co., Ltd. | Treatment of influenza |
| EP2319926A1 (en) | 2007-07-05 | 2011-05-11 | Novartis AG | DSRNA for treating viral infection |
| EP2316943A1 (en) | 2007-07-05 | 2011-05-04 | Novartis AG | DSRNA for treating viral infection |
| US10273482B2 (en) | 2007-07-05 | 2019-04-30 | Arrowhead Pharmaceuticals, Inc. | dsRNA for treating viral infection |
| US10105441B2 (en) | 2007-08-16 | 2018-10-23 | The Schepens Eye Research Institute, Inc. | Method for inhibiting or reducing dry eye disease by IL-1Ra |
| US8614309B2 (en) | 2007-10-03 | 2013-12-24 | Quark Pharmaceuticals, Inc. | Double-stranded RNA directed to CASP2 and methods of use thereof |
| WO2009044392A2 (en) | 2007-10-03 | 2009-04-09 | Quark Pharmaceuticals, Inc. | Novel sirna structures |
| US8735567B2 (en) | 2007-11-06 | 2014-05-27 | Patrick Y. Lu | Multi-targeted RNAi therapeutics for scarless wound healing of skin |
| USRE46873E1 (en) | 2007-11-06 | 2018-05-29 | Sirnaomics, Inc. | Multi-targeted RNAi therapeutics for scarless wound healing of skin |
| EP2233573A4 (en) * | 2007-11-29 | 2011-02-23 | Suzhou Ribo Life Science Co Ltd | A complex molecule interfering the expression of target genes and its preparing methods |
| EP2233573A1 (en) * | 2007-11-29 | 2010-09-29 | Suzhou Ribo Life Science Co., Ltd | A complex molecule interfering the expression of target genes and its preparing methods |
| CN101889087B (en) * | 2007-11-29 | 2013-04-03 | 苏州瑞博生物技术有限公司 | A complex molecule interfering the expression of target genes and preparing methods thereof |
| US8614311B2 (en) | 2007-12-12 | 2013-12-24 | Quark Pharmaceuticals, Inc. | RTP801L siRNA compounds and methods of use thereof |
| US9872901B2 (en) | 2008-01-09 | 2018-01-23 | The Schepens Eye Research Institute, Inc. | Therapeutic compositions for treatment of ocular inflammatory disorders |
| US9309313B2 (en) | 2008-01-09 | 2016-04-12 | The Schepens Eye Research Institute, Inc. | Therapeutic compositions for treatment of ocular inflammatory disorders |
| US11241497B2 (en) | 2008-01-09 | 2022-02-08 | The Schepens Eye Research Institute, Inc. | Therapeutic compositions for treatment of ocular inflammatory disorders |
| US9006197B2 (en) | 2008-03-05 | 2015-04-14 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inhibiting expression of Eg5 and VEGF genes |
| WO2009118300A1 (en) | 2008-03-25 | 2009-10-01 | Novartis Forschungsstiftung Zweigniederlassung Friedrich Miescher Institute For Biomedical Research | Treating cancer by down-regulating frizzled-4 and/or frizzled-1 |
| US8278287B2 (en) | 2008-04-15 | 2012-10-02 | Quark Pharmaceuticals Inc. | siRNA compounds for inhibiting NRF2 |
| US8541568B2 (en) | 2008-05-24 | 2013-09-24 | Hai Yan | Compositions and methods using siRNA molecules for treatment of gliomas |
| US8222221B2 (en) | 2008-06-04 | 2012-07-17 | The Board Of Regents Of The University Of Texas System | Modulation of gene expression through endogenous small RNA targeting of gene promoters |
| US8431692B2 (en) | 2008-06-06 | 2013-04-30 | Quark Pharmaceuticals, Inc. | Compositions and methods for treatment of ear disorders |
| US9089591B2 (en) | 2008-06-06 | 2015-07-28 | Quark Pharmaceuticals, Inc. | Compositions and methods for treatment of ear disorders |
| US9885036B2 (en) | 2008-07-01 | 2018-02-06 | Daiichi Sankyo Company, Limited | Double-stranded polynucleotide |
| WO2010006342A3 (en) * | 2008-07-11 | 2010-08-12 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inhibiting expression of gsk-3 genes |
| US9029525B2 (en) | 2008-07-11 | 2015-05-12 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inhibiting expression of GSK-3 genes |
| WO2010006342A2 (en) * | 2008-07-11 | 2010-01-14 | Alnylam Pharmaceuticals, Inc. | Compositions and methods for inhibiting expression of gsk-3 genes |
| US8785718B2 (en) | 2008-07-16 | 2014-07-22 | Recombinetics, Inc. | Methods for producing genetically modified animals using hypermethylated transposons |
| US8309791B2 (en) | 2008-07-16 | 2012-11-13 | Recombinectics, Inc. | Method for producing a transgenic pig using a hyper-methylated transposon |
| WO2010065756A3 (en) * | 2008-12-03 | 2010-11-25 | Mdrna, Inc. | Usirna complexes |
| US9340789B2 (en) | 2008-12-03 | 2016-05-17 | Arcturus Therapeutics, Inc. | UNA oligomer structures for therapeutic agents |
| WO2010065756A2 (en) * | 2008-12-03 | 2010-06-10 | Mdrna, Inc. | Usirna complexes |
| US9012622B2 (en) | 2008-12-31 | 2015-04-21 | Patrick Y. Lu | Compositions and methods using siRNA molecules and siRNA cocktails for the treatment of breast cancer |
| US8309530B2 (en) | 2009-02-04 | 2012-11-13 | Washington State University | Compositions and methods for modulating ghrelin-mediated conditions |
| WO2010100247A1 (en) | 2009-03-06 | 2010-09-10 | Novartis Forschungsstiftung, Zweigniederlassung, Friedrich Miescher Institute For Biomedical Research | Novel therapy for anxiety |
| WO2010111490A3 (en) * | 2009-03-27 | 2010-12-29 | Merck Sharp & Dohme Corp. | RNA INTERFERENCE MEDIATED INHIBITION OF THE THYMIC STROMAL LYMPHOPOIETIN (TSLP) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) |
| EP2415869A1 (en) * | 2009-04-03 | 2012-02-08 | Biomics Biotechnologies Co., Ltd. | Modified oligo-nucleic acid molecule, preparation method and uses thereof |
| US8563710B2 (en) | 2009-04-03 | 2013-10-22 | Biomics Biotechnologies Co., Ltd. | Modified oligonucleotide and its preparation and application |
| EP2415869A4 (en) * | 2009-04-03 | 2013-06-19 | Biomics Biotechnologies Co Ltd | Modified oligo-nucleic acid molecule, preparation method and uses thereof |
| EP2241323A1 (en) | 2009-04-14 | 2010-10-20 | Novartis Forschungsstiftung, Zweigniederlassung Friedrich Miescher Institute For Biomedical Research | Tenascin-W and brain cancers |
| US8815586B2 (en) | 2009-04-24 | 2014-08-26 | The Board Of Regents Of The University Of Texas System | Modulation of gene expression using oligomers that target gene regions downstream of 3′ untranslated regions |
| WO2011019423A2 (en) | 2009-05-20 | 2011-02-17 | Schering Corporation | Modulation of pilr receptors to treat microbial infections |
| US8273869B2 (en) | 2009-06-15 | 2012-09-25 | Alnylam Pharmaceuticals, Inc. | Lipid formulated dsRNA targeting the PCSK9 gene |
| US9051567B2 (en) | 2009-06-15 | 2015-06-09 | Tekmira Pharmaceuticals Corporation | Methods for increasing efficacy of lipid formulated siRNA |
| US10053689B2 (en) | 2009-06-15 | 2018-08-21 | Arbutus Biopharma Corporation | Methods for increasing efficacy of lipid formulated siRNA |
| US10870850B2 (en) | 2009-07-13 | 2020-12-22 | Somagenics, Inc. | Chemical modification of short small hairpin RNAs for inhibition of gene expression |
| EP2454371A2 (en) * | 2009-07-13 | 2012-05-23 | Somagenics, Inc. | Chemical modification of small hairpin rnas for inhibition of gene expression |
| US8871730B2 (en) | 2009-07-13 | 2014-10-28 | Somagenics Inc. | Chemical modification of short small hairpin RNAs for inhibition of gene expression |
| US9816091B2 (en) | 2009-07-13 | 2017-11-14 | Somagenics, Inc. | Chemical modification of short small hairpin RNAs for inhibition of gene expression |
| WO2011008730A2 (en) | 2009-07-13 | 2011-01-20 | Somagenics Inc. | Chemical modification of small hairpin rnas for inhibition of gene expression |
| EP2454371A4 (en) * | 2009-07-13 | 2013-10-09 | Somagenics Inc | Chemical modification of small hairpin rnas for inhibition of gene expression |
| EP2810643A2 (en) | 2009-08-14 | 2014-12-10 | Alnylam Pharmaceuticals Inc. | Lipid formulated compositions and mehods for inhibiting expression of a gene from the ebola virus |
| EP2292266A1 (en) | 2009-08-27 | 2011-03-09 | Novartis Forschungsstiftung, Zweigniederlassung | Treating cancer by modulating copine III |
| WO2011031600A1 (en) | 2009-09-10 | 2011-03-17 | Schering Corporation | Use of il-33 antagonists to treat fibrotic disease |
| US8859516B2 (en) | 2009-09-15 | 2014-10-14 | Alnylam Pharmaceuticals, Inc. | Lipid formulated compositions and methods for inhibiting expression of Eg5 and VEGF genes |
| US9187746B2 (en) | 2009-09-22 | 2015-11-17 | Alnylam Pharmaceuticals, Inc. | Dual targeting siRNA agents |
| WO2011036118A1 (en) | 2009-09-22 | 2011-03-31 | Novartis Forschungsstiftung, Zweigniederlassung Friedrich Miescher Institute For Biomedical Research | Treating cancer by modulating mex-3 |
| WO2011045352A2 (en) | 2009-10-15 | 2011-04-21 | Novartis Forschungsstiftung | Spleen tyrosine kinase and brain cancers |
| US8796239B2 (en) | 2009-11-26 | 2014-08-05 | Quark Pharmaceuticals, Inc. | Sirna compounds comprising terminal substitutions |
| US9701960B2 (en) | 2009-11-26 | 2017-07-11 | Quark Pharmaceuticals, Inc. | siRNA compounds comprising terminal substitutions |
| US10494631B2 (en) | 2009-11-26 | 2019-12-03 | Quark Pharmaceuticals, Inc. | siRNA compounds comprising terminal substitutions |
| WO2011084357A1 (en) | 2009-12-17 | 2011-07-14 | Schering Corporation | Modulation of pilr to treat immune disorders |
| US8859751B2 (en) | 2010-01-07 | 2014-10-14 | Quark Pharmaceuticals, Inc. | Oligonucleotide compounds comprising non-nucleotide overhangs |
| WO2011084193A1 (en) | 2010-01-07 | 2011-07-14 | Quark Pharmaceuticals, Inc. | Oligonucleotide compounds comprising non-nucleotide overhangs |
| US9249414B2 (en) | 2010-01-07 | 2016-02-02 | Quark Pharmaceuticals, Inc. | Oligonucleotide compounds comprising non-nucleotide overhangs |
| WO2011107586A1 (en) | 2010-03-05 | 2011-09-09 | Novartis Forschungsstiftung, Zweigniederlassung, Friedrich Miescher Institute For Biomedical Research, | Smoc1, tenascin-c and brain cancers |
| WO2011131611A1 (en) | 2010-04-19 | 2011-10-27 | Novartis Forschungsstiftung, Zweigniederlassung Friedrich Miescher Institute For Biomedical Research | Modulating xrn1 |
| WO2011133876A2 (en) | 2010-04-22 | 2011-10-27 | Alnylam Pharmaceuticals, Inc. | Oligonucleotides comprising acyclic and abasic nucleosides and analogs |
| WO2011133868A2 (en) | 2010-04-22 | 2011-10-27 | Alnylam Pharmaceuticals, Inc. | Conformationally restricted dinucleotide monomers and oligonucleotides |
| US9642873B2 (en) | 2010-05-04 | 2017-05-09 | Sirnaomics, Inc. | Combinations of TGFβ and COX-2 inhibitors and methods for their therapeutic application |
| WO2011154485A1 (en) | 2010-06-10 | 2011-12-15 | Novartis Forschungsstiftung, Zweigniederlassung Friedrich Miescher Institute For Biomedical Research | Treating cancer by modulating mammalian sterile 20-like kinase 3 |
| US9506063B2 (en) | 2010-07-29 | 2016-11-29 | Sirnaomics, Inc. | SiRNA compositions and methods for treatment of HPV and other infections |
| EP2412724A1 (en) | 2010-07-29 | 2012-02-01 | Centre National de la Recherche Scientifique (C.N.R.S) | Regulation of Glypican 4 activity to modulate the fate of stem cells and uses thereof |
| WO2012013784A1 (en) | 2010-07-29 | 2012-02-02 | Centre National De La Recherche Scientifique (C.N.R.S) | Regulation of glypican 4 activity to modulate the fate of stem cells and uses thereof |
| US9845466B2 (en) | 2010-08-24 | 2017-12-19 | Sirna Therapeutics, Inc. | Single-stranded RNAi agents containing an internal, non-nucleic acid spacer |
| US10584335B2 (en) | 2010-08-24 | 2020-03-10 | Sirna Therapeutics, Inc. | Single-stranded RNAi agents containing an internal, non-nucleic acid spacer |
| US9243246B2 (en) | 2010-08-24 | 2016-01-26 | Sirna Therapeutics, Inc. | Single-stranded RNAi agents containing an internal, non-nucleic acid spacer |
| CN102453066A (en) * | 2010-10-19 | 2012-05-16 | 南开大学 | Compound molecule, and preparation method and pharmaceutical composition thereof |
| CN102453066B (en) * | 2010-10-19 | 2016-07-06 | 南开大学 | A kind of compound molecule and preparation method thereof and pharmaceutical composition |
| US9260471B2 (en) | 2010-10-29 | 2016-02-16 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using short interfering nucleic acids (siNA) |
| US11193126B2 (en) | 2010-10-29 | 2021-12-07 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using short interfering nucleic acids (siNA) |
| US11932854B2 (en) | 2010-10-29 | 2024-03-19 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using short interfering nucleic acids (siNA) |
| US9970005B2 (en) | 2010-10-29 | 2018-05-15 | Sirna Therapeutics, Inc. | RNA interference mediated inhibition of gene expression using short interfering nucleic acids (siNA) |
| US8889649B2 (en) | 2010-11-12 | 2014-11-18 | National University Corporation Ehime University | Composition containing antisense oligonucleotide to micro RNA |
| WO2012168259A1 (en) | 2011-06-06 | 2012-12-13 | Novartis Forschungsstiftung, Zweigniederlassung | Protein tyrosine phosphatase, non-receptor type 11 (ptpn11) and triple-negative breast cancer |
| US9181553B2 (en) | 2011-06-06 | 2015-11-10 | Novartis Forschungsstiftung Zweigniederlassung Friedrich Miescher Institute For Biomedical Research | Method of treatment of breast cancers over-expressing the SHP2 signature genes |
| EP3901263A1 (en) * | 2011-06-30 | 2021-10-27 | Arrowhead Pharmaceuticals, Inc. | Compositions and methods for inhibiting gene expression of hepatitis b virus |
| WO2013068432A1 (en) | 2011-11-08 | 2013-05-16 | Novartis Forschungsstiftung, Zweigniederlassung, Friedrich Miescher Institute For Biomedical Research | Early diagnostic of neurodegenerative diseases |
| WO2013068431A1 (en) | 2011-11-08 | 2013-05-16 | Novartis Forschungsstiftung, Zweigniederlassung, Friedrich Miescher Institute For Biomedical Research | New treatment for neurodegenerative diseases |
| WO2013072392A1 (en) | 2011-11-15 | 2013-05-23 | Novartis Forschungsstiftung, Zweigniederlassung Friedrich Miescher Institute For Biomedical Research | Combination of a phosphoinositide 3-kinase inhibitor and a modulator of the janus kinase 2-signal transducer and activator of transcription 5 pathway |
| US11406716B2 (en) | 2011-11-18 | 2022-08-09 | Alnylam Pharmaceuticals, Inc. | Modified RNAi agents |
| US9796974B2 (en) * | 2011-11-18 | 2017-10-24 | Alnylam Pharmaceuticals, Inc. | Modified RNAi agents |
| US20140288158A1 (en) * | 2011-11-18 | 2014-09-25 | Alnylam Pharmaceuticals, Inc. | MODIFIED RNAi AGENTS |
| US10668170B2 (en) | 2011-11-18 | 2020-06-02 | Alnylam Pharmaceuticals, Inc. | Modified RNAi agents |
| WO2013102825A1 (en) | 2012-01-02 | 2013-07-11 | Novartis Ag | Cdcp1 and breast cancer |
| WO2013144240A1 (en) | 2012-03-29 | 2013-10-03 | Friedrich Miescher Institute For Biomedical Research | Inhibition of interleukin- 8 and/or its receptor cxcrl in the treatment her2/her3 -overexpressing breast cancer |
| WO2014001482A1 (en) | 2012-06-29 | 2014-01-03 | Novartis Forschungsstiftung, Zweigniererlassung, Friedrich Miescher Institute For Biomedical Research | Treating diseases by modulating a specific isoform of mkl1 |
| WO2014006114A1 (en) | 2012-07-05 | 2014-01-09 | Novartis Forschungsstiftung, Zweigniederlassung Friedrich Miescher Institute For Biomedical Research | New treatment for neurodegenerative diseases |
| WO2014006115A1 (en) | 2012-07-06 | 2014-01-09 | Novartis Ag | Combination of a phosphoinositide 3-kinase inhibitor and an inhibitor of the il-8/cxcr interaction |
| US9868952B2 (en) | 2012-07-08 | 2018-01-16 | Sirnaomics, Inc. | Compositions and methods for “resistance-proof” SiRNA therapeutics for influenza |
| EP3708184A1 (en) | 2013-03-27 | 2020-09-16 | The General Hospital Corporation | Methods and agents for treating alzheimer s disease |
| WO2014160871A2 (en) | 2013-03-27 | 2014-10-02 | The General Hospital Corporation | Methods and agents for treating alzheimer's disease |
| WO2014176259A1 (en) | 2013-04-22 | 2014-10-30 | Icahn School Of Medicine At Mount Sinai | Mutations in pdgfrb and notch3 as causes of autosomal dominant infantile myofibromatosis |
| WO2015019286A1 (en) | 2013-08-07 | 2015-02-12 | Friedrich Miescher Institute For Biomedical Research | New screening method for the treatment friedreich's ataxia |
| US10227588B2 (en) | 2013-10-04 | 2019-03-12 | Novartis Ag | 3′end caps for RNAi agents for use in RNA interference |
| WO2015050871A2 (en) | 2013-10-04 | 2015-04-09 | Novartis Ag | Organic compounds to treat hepatitis b virus |
| WO2015051044A2 (en) | 2013-10-04 | 2015-04-09 | Novartis Ag | Novel formats for organic compounds for use in rna interference |
| WO2015051045A2 (en) | 2013-10-04 | 2015-04-09 | Novartis Ag | 3'END CAPS FOR RNAi AGENTS FOR USE IN RNA INTERFERENCE |
| WO2015051366A2 (en) | 2013-10-04 | 2015-04-09 | Novartis Ag | Novel formats for organic compounds for use in rna interference |
| US11008570B2 (en) | 2013-10-04 | 2021-05-18 | Novartis Ag | 3′ end caps for RNAi agents for use in RNA interference |
| US10519446B2 (en) | 2013-10-04 | 2019-12-31 | Novartis Ag | Organic compounds to treat hepatitis B virus |
| US9988627B2 (en) | 2013-10-04 | 2018-06-05 | Novartis Ag | Formats for organic compounds for use in RNA interference |
| EP3722277A2 (en) | 2013-10-04 | 2020-10-14 | Novartis AG | 3'end caps for rna-interferring agents for use in rna |
| US9982259B2 (en) | 2014-03-25 | 2018-05-29 | Arcturus Therapeutics, Inc. | Transthyretin allele selective UNA oligomers for gene silencing |
| US9856475B2 (en) | 2014-03-25 | 2018-01-02 | Arcturus Therapeutics, Inc. | Formulations for treating amyloidosis |
| US10683500B2 (en) | 2014-03-25 | 2020-06-16 | Arcturus Therapeutics, Inc. | UNA oligomers having reduced off-target effects in gene silencing |
| US10604758B2 (en) | 2014-03-25 | 2020-03-31 | Arcturus Therapeutics, Inc. | Therapeutic oligomers for treating amyloidosis |
| EP3129493A4 (en) * | 2014-04-09 | 2018-04-25 | The Scripps Research Institute | Import of unnatural or modified nucleoside triphosphates into cells via nucleic acid triphosphate transporters |
| US11466279B2 (en) | 2014-04-09 | 2022-10-11 | The Scripps Research Institute | Import of unnatural or modified nucleoside triphosphates into cells via nucleic acid triphosphate transporters |
| US10513706B2 (en) | 2014-04-09 | 2019-12-24 | The Scripps Research Institute | Import of unnatural or modified nucleoside triphosphates into cells via nucleic acid triphosphate transporters |
| WO2015189816A1 (en) | 2014-06-13 | 2015-12-17 | Friedrich Miescher Institute For Biomedical Research | New treatment against influenza virus |
| WO2016001830A1 (en) | 2014-07-01 | 2016-01-07 | Friedrich Miescher Institute For Biomedical Research | Combination of a brafv600e inhibitor and mertk inhibitor to treat melanoma |
| WO2016046768A1 (en) | 2014-09-24 | 2016-03-31 | Friedrich Miescher Institute For Biomedical Research | Lats and breast cancer |
| US10513703B2 (en) | 2014-11-10 | 2019-12-24 | Alnylam Pharmaceuticals, Inc. | Hepatitis B virus (HBV) iRNA compositions and methods of use thereof |
| US11060091B2 (en) | 2014-11-10 | 2021-07-13 | Alnylam Pharmaceuticals, Inc. | Hepatitis B virus (HBV) iRNA compositions and methods of use thereof |
| WO2016077620A1 (en) | 2014-11-12 | 2016-05-19 | The United States Of America, As Represented By The Secretary, Department Of Health And Human Services | Treatment of hormonal disorders of growth |
| US10519447B2 (en) | 2015-04-01 | 2019-12-31 | Arcturus Therapeutics, Inc. | Therapeutic UNA oligomers and uses thereof |
| US10421964B2 (en) | 2015-07-23 | 2019-09-24 | Arcturus Therapeutics, Inc. | UNA oligomers and compositions for treating amyloidosis |
| US11534453B2 (en) | 2015-08-07 | 2022-12-27 | Arrowhead Pharmaceuticals, Inc. | RNAi therapy for hepatitis B virus infection |
| WO2017072669A1 (en) | 2015-10-28 | 2017-05-04 | Friedrich Miescher Institute For Biomedical Research | Tenascin-w and biliary tract cancers |
| WO2017079442A1 (en) | 2015-11-04 | 2017-05-11 | Icahn School Of Medicine At Mount Sinai | Methods of treating tumors and cancer, and identifying candidate subjects for such treatment |
| US11896672B2 (en) | 2016-04-11 | 2024-02-13 | Arbutus Biopharma Corporation | Targeted nucleic acid conjugate compositions |
| WO2017189730A1 (en) | 2016-04-26 | 2017-11-02 | Icahn School Of Medicine At Mount Sinai | Treatment of hippo pathway mutant tumors and methods of identifying subjects as candidates for treatment |
| US11590156B2 (en) | 2016-08-04 | 2023-02-28 | Arrowhead Pharmaceuticals, Inc. | RNAi agents for hepatitis B virus infection |
| US11517584B2 (en) | 2016-08-04 | 2022-12-06 | Arrowhead Pharmaceuticals, Inc. | RNAi agents for Hepatitis B virus infection |
| US11427823B2 (en) | 2017-04-11 | 2022-08-30 | Arbutus Biopharma Corporation | Targeted compositions |
| WO2018191278A3 (en) * | 2017-04-11 | 2018-11-22 | Arbutus Biopharma Corporation | Targeted compositions |
| US11324820B2 (en) | 2017-04-18 | 2022-05-10 | Alnylam Pharmaceuticals, Inc. | Methods for the treatment of subjects having a hepatitis b virus (HBV) infection |
| US11834689B2 (en) | 2017-07-11 | 2023-12-05 | The Scripps Research Institute | Incorporation of unnatural nucleotides and methods thereof |
| US10610571B2 (en) | 2017-08-03 | 2020-04-07 | Synthorx, Inc. | Cytokine conjugates for the treatment of proliferative and infectious diseases |
| US11701407B2 (en) | 2017-08-03 | 2023-07-18 | Synthorx, Inc. | Cytokine conjugates for the treatment of proliferative and infectious diseases |
| US11622993B2 (en) | 2017-08-03 | 2023-04-11 | Synthorx, Inc. | Cytokine conjugates for the treatment of autoimmune diseases |
| WO2019246112A1 (en) | 2018-06-18 | 2019-12-26 | University Of Rochester | Methods of treating schizophrenia and other neuropsychiatric disorders |
| WO2019246262A2 (en) | 2018-06-21 | 2019-12-26 | University Of Rochester | Methods of treating or inhibiting onset of huntington's disease |
| US11492623B2 (en) | 2018-08-13 | 2022-11-08 | Alnylam Pharmaceuticals, Inc. | Hepatitis B virus (HBV) dsRNA agent compositions and methods of use thereof |
| WO2020093053A1 (en) * | 2018-11-02 | 2020-05-07 | Arbutus Biopharma Corporation | Bivalent targeted conjugates |
| WO2020123663A1 (en) | 2018-12-11 | 2020-06-18 | University Of Rochester | Methods of treating schizophrenia and other neuropsychiatric disorders |
| US11077195B2 (en) | 2019-02-06 | 2021-08-03 | Synthorx, Inc. | IL-2 conjugates and methods of use thereof |
| WO2020167822A2 (en) | 2019-02-13 | 2020-08-20 | University Of Rochester | Gene networks that mediate remyelination of the human brain |
| WO2021011936A2 (en) | 2019-07-18 | 2021-01-21 | University Of Rochester | Cell-type selective immunoprotection of cells |
| WO2023150553A1 (en) | 2022-02-01 | 2023-08-10 | University Of Rochester | Gpr17 promoter-based targeting and transduction of glial progenitor cells |
| WO2023191630A1 (en) | 2022-03-30 | 2023-10-05 | Academisch Medisch Centrum | Antisense nucleic acids for use in the treatment for kcnq1 mutation carriers |
| WO2023191631A1 (en) | 2022-03-30 | 2023-10-05 | Academisch Medisch Centrum | Antisense nucleic acids for use in the treatment for lmna mutation carriers |
Also Published As
| Publication number | Publication date |
|---|---|
| WO2003070918A3 (en) | 2004-07-08 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| US10662428B2 (en) | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (siNA) | |
| AU2003221258B2 (en) | RNA interference by modified short interfering nucleic acid | |
| EP1713915B1 (en) | RNA INTERFERENCE MEDIATED INHIBITION OF GENE EXPRESSION USING MULTIFUNCTIONAL SHORT INTERFERING NUCLEIC ACID (MULTIFUNCTIONAL siNA) | |
| US8273866B2 (en) | RNA interference mediated inhibition of gene expression using chemically modified short interfering nucleic acid (SINA) | |
| WO2003070918A2 (en) | Rna interference by modified short interfering nucleic acid | |
| US20050106726A1 (en) | RNA interference mediated inhibition of platelet-derived endothelial cell growth factor (ECGF1) gene expression using short interfering nucleic acid (siNA) | |
| AU2003216323B2 (en) | Inhibition of vascular endothelial growth factor (vegf) and vegf receptor gene expression using short interfereing nucleic acid (sina) | |
| WO2003070881A2 (en) | RNA INTERFERENCE MEDIATED INHIBITION OF PROTEIN TYPROSINE PHOSPHATASE-1B (PTP-1B) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) | |
| US20050233329A1 (en) | Inhibition of gene expression using duplex forming oligonucleotides | |
| GB2406568A (en) | RNA interfernece mediated inhibition of gene expression by chemically modified short interfering nucleic acids | |
| EP1710307A2 (en) | RNA interference mediated inhibition of gene expression using short interfering nucleic acid (siNA) | |
| EP1495041A1 (en) | RNA INTERFERENCE MEDIATED INHIBITION OF G72 AND D-AMINO ACID OXIDASE (DAAO) GENE EXPRESSION USING SHORT INTERFERING NUCLEIC ACID (siNA) | |
| AU2006203725B2 (en) | RNA interference by modified short interfering nucleic acid | |
| GB2413557A (en) | RNA interference mediated by short nucleic acids containing modified nucleotides |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| WWE | Wipo information: entry into national phase |
Ref document number: 10424339 Country of ref document: US |
|
| AK | Designated states |
Kind code of ref document: A2 Designated state(s): AE AG AL AM AT AU AZ BA BB BG BR BY BZ CA CH CN CO CR CU CZ DE DK DM DZ EC EE ES FI GB GD GE GH GM HR HU ID IL IN IS JP KE KG KP KR KZ LC LK LR LS LT LU LV MA MD MG MK MN MW MX MZ NO NZ OM PH PL PT RO RU SC SD SE SG SK SL TJ TM TN TR TT TZ UA UG US UZ VC VN YU ZA ZM ZW |
|
| AL | Designated countries for regional patents |
Kind code of ref document: A2 Designated state(s): GH GM KE LS MW MZ SD SL SZ TZ UG ZM ZW AM AZ BY KG KZ MD RU TJ TM AT BE BG CH CY CZ DE DK EE ES FI FR GB GR HU IE IT LU MC NL PT SE SI SK TR BF BJ CF CG CI CM GA GN GQ GW ML MR NE SN TD TG |
|
| ENP | Entry into the national phase |
Ref document number: 0406022 Country of ref document: GB Kind code of ref document: A Free format text: PCT FILING DATE = 20030220 |
|
| 121 | Ep: the epo has been informed by wipo that ep was designated in this application | ||
| WWE | Wipo information: entry into national phase |
Ref document number: 10693059 Country of ref document: US |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 2003221258 Country of ref document: AU |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 2459532 Country of ref document: CA |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 2003716126 Country of ref document: EP |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 2003569811 Country of ref document: JP |
|
| WWP | Wipo information: published in national office |
Ref document number: 2003716126 Country of ref document: EP |
|
| WWP | Wipo information: published in national office |
Ref document number: 10424339 Country of ref document: US |