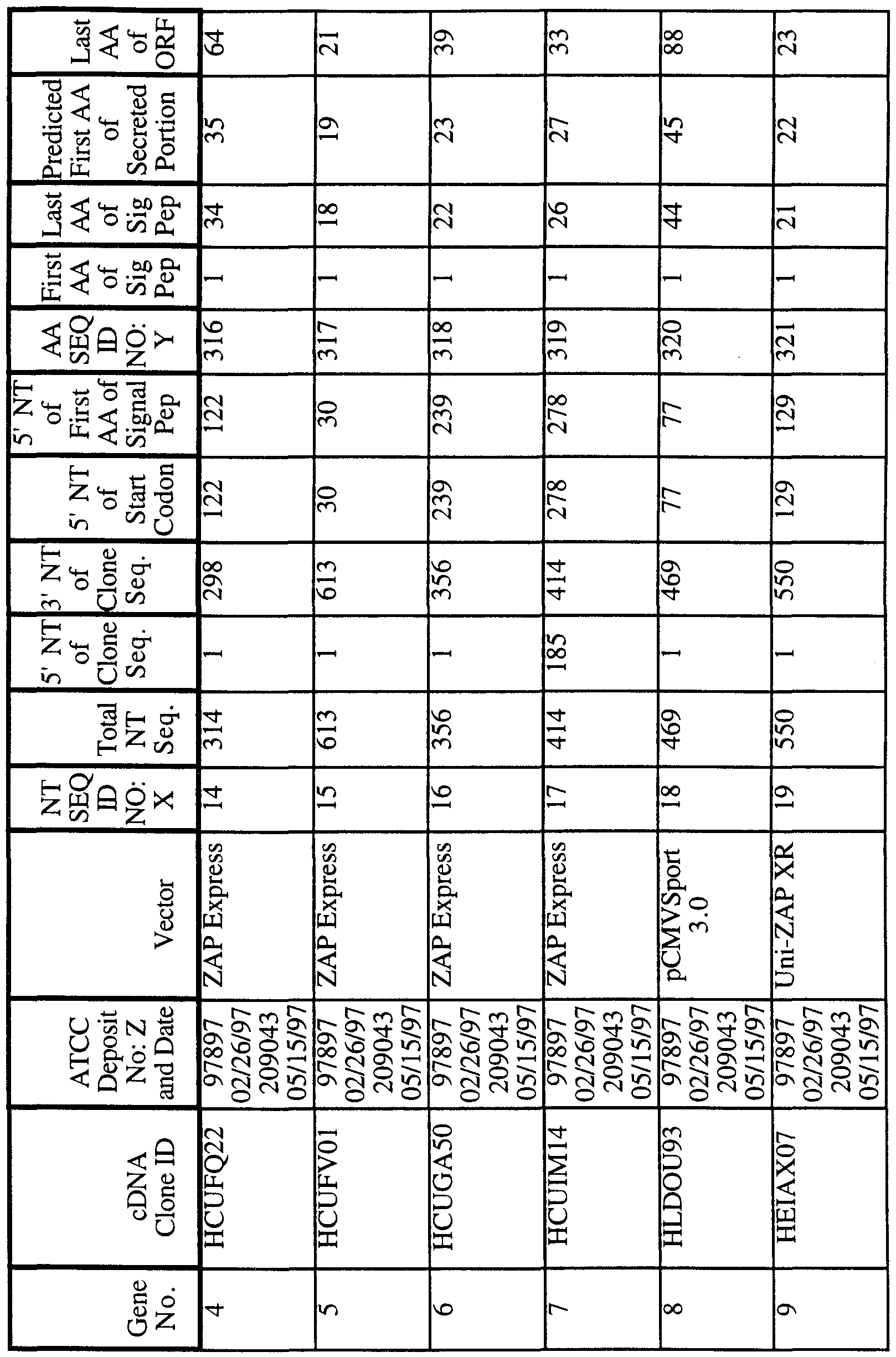
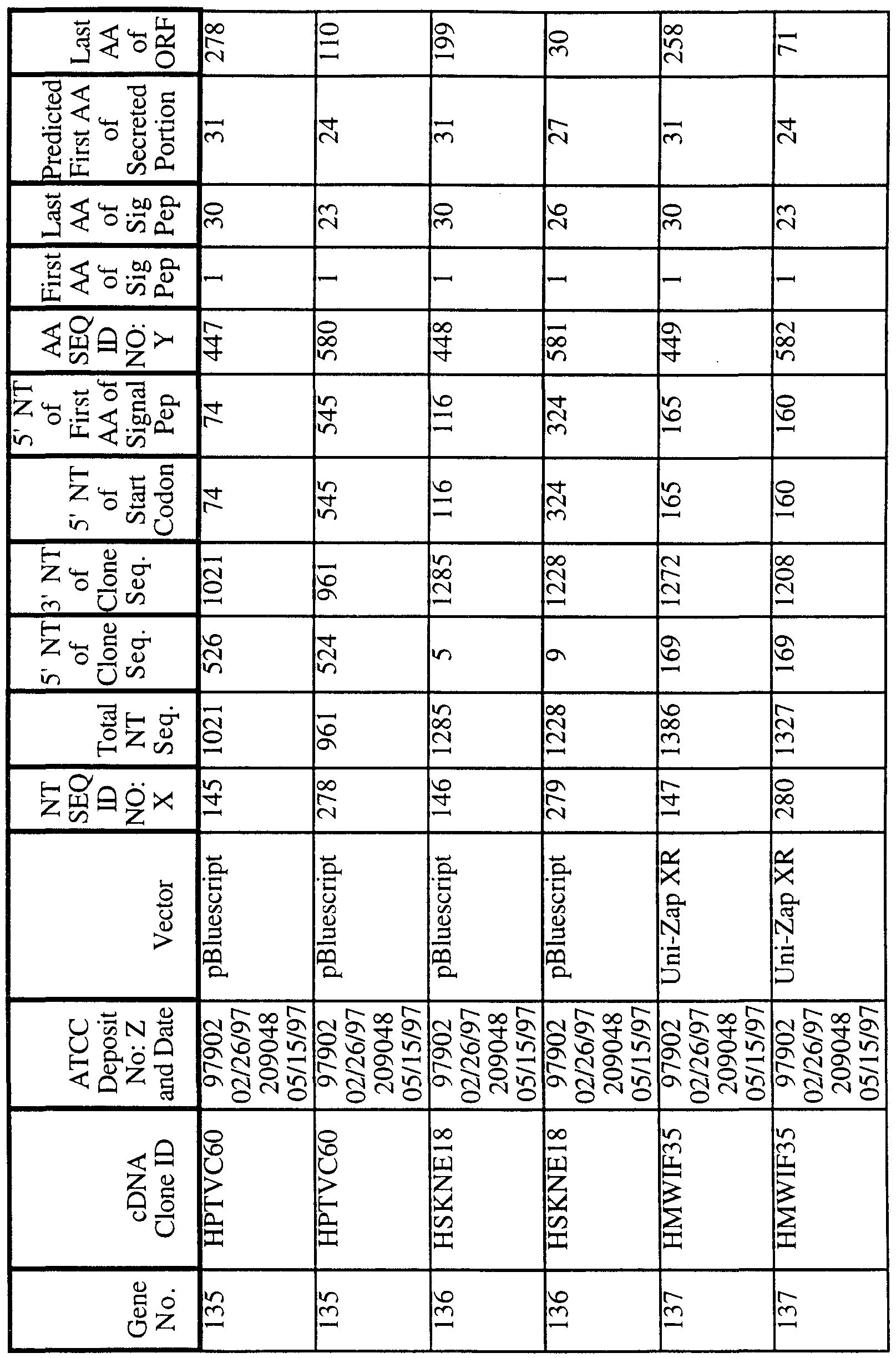
186 Human Secreted Proteins
Field of the Invention
This invention relates to newly identified polynucleotides and the polypeptides encoded by these polynucleotides, uses of such polynucleotides and polypeptides, and their production.
Background of the Invention
Unlike bacterium, which exist as a single compartment surrounded by a membrane, human cells and other eucaryotes are subdivided by membranes into many functionally distinct compartments. Each membrane -bounded compartment, or organelle, contains different proteins essential for the function of the organelle. The cell uses "sorting signals," which are amino acid motifs located within the protein, to target proteins to particular cellular organelles.
One type of sorting signal, called a signal sequence, a signal peptide, or a leader sequence, directs a class of proteins to an organelle called the endoplasmic reticulum (ER). The ER separates the membrane-bounded proteins from all other types of proteins. Once localized to the ER, both groups of proteins can be further directed to another organelle called the Golgi apparatus. Here, the Golgi distributes the proteins to vesicles, including secretory vesicles, the cell membrane, lysosomes, and the other organelles. Proteins targeted to the ER by a signal sequence can be released into the extracellular space as a secreted protein. For example, vesicles containing secreted proteins can fuse with the cell membrane and release their contents into the extracellular space - a process called exocytosis. Exocytosis can occur constitutively or after receipt of a triggering signal. In the latter case, the proteins are stored in secretory vesicles (or secretory granules) until exocytosis is triggered. Similarly, proteins residing on the cell membrane can also be secreted into the extracellular space by proteolytic cleavage of a "linker" holding the protein to the membrane.
Despite the great progress made in recent years, only a small number of genes encoding human secreted proteins have been identified. These secreted proteins include the commercially valuable human insulin, interferon, Factor VIII, human growth hormone, tissue plasminogen activator, and erythropoeitin. Thus, in light of the pervasive role of secreted proteins in human physiology, a need exists for identifying and characterizing novel human secreted proteins and the genes that encode them. This knowledge will allow one to detect, to treat, and to prevent medical disorders by using secreted proteins or the genes that encode them.
Summary of the Invention
The present invention relates to novel polynucleotides and the encoded polypeptides. Moreover, the present invention relates to vectors, host cells, antibodies, and recombinant methods for producing the polypeptides and polynucleotides. Also provided are diagnostic methods for detecting disorders related to the polypeptides, and therapeutic methods for treating such disorders. The invention further relates to screening methods for identifying binding partners of the polypeptides.
Detailed Description
Definitions
The following definitions are provided to facilitate understanding of certain terms used throughout this specification.
In the present invention, "isolated" refers to material removed from its original environment (e.g., the natural environment if it is naturally occurring), and thus is altered "by the hand of man" from its natural state. For example, an isolated polynucleotide could be part of a vector or a composition of matter, or could be contained within a cell, and still be "isolated" because that vector, composition of matter, or particular cell is not the original environment of the polynucleotide. In the present invention, a "secreted" protein refers to those proteins capable of being directed to the ER, secretory vesicles, or the extracellular space as a result of a signal sequence, as well as those proteins released into the extracellular space without necessarily containing a signal sequence. If the secreted protein is released into the extracellular space, the secreted protein can undergo extracellular processing to produce a "mature" protein. Release into the extracellular space can occur by many mechanisms, including exocytosis and proteolytic cleavage.
As used herein , a "polynucleotide" refers to a molecule having a nucleic acid sequence contained in SEQ ID NO:X or the cDNA contained within the clone deposited with the ATCC. For example, the polynucleotide can contain the nucleotide sequence of the full length cDNA sequence, including the 5' and 3' untranslated sequences, the coding region, with or without the signal sequence, the secreted protein coding region, as well as fragments, epitopes, domains, and variants of the nucleic acid sequence. Moreover, as used herein, a "polypeptide" refers to a molecule having the translated amino acid sequence generated from the polynucleotide as broadly defined. In the present invention, the full length sequence identified as SEQ ID NO:X was often generated by overlapping sequences contained in multiple clones (contig
analysis). A representative clone containing all or most of the sequence for SEQ ID NO:X was deposited with the American Type Culture Collection ("ATCC"). As shown in Table 1, each clone is identified by a cDNA Clone ID (Identifier) and the ATCC Deposit Number. The ATCC is located at 12301 Park Lawn Drive, Rockville, Maryland 20852, USA. The ATCC deposit was made pursuant to the terms of the Budapest Treaty on the international recognition of the deposit of microorganisms for purposes of patent procedure.
A "polynucleotide" of the present invention also includes those polynucleotides capable of hybridizing, under stringent hybridization conditions, to sequences contained in SEQ ID NO:X, the complement thereof, or the cDNA contained within the clone deposited with the ATCC. "Stringent hybridization conditions" refers to an overnight incubation at 42° C in a solution comprising 50% formamide, 5x SSC (750 mM NaCl,
75 mM sodium citrate), 50 mM sodium phosphate (pH 7.6), 5x Denhardt's solution, 10% dextran sulfate, and 20 μg/ml denatured, sheared salmon sperm DNA, followed by washing the filters in 0. lx SSC at about 65°C.
Also contemplated are nucleic acid molecules that hybridize to the polynucleotides of the present invention at lower stringency hybridization conditions. Changes in the stringency of hybridization and signal detection are primarily accomplished through the manipulation of formamide concentration (lower percentages of formamide result in lowered stringency); salt conditions, or temperature. For example, lower stringency conditions include an overnight incubation at 37°C in a solution comprising 6X SSPE (20X SSPE = 3M NaCl; 0.2M NaH2PO4; 0.02M EDTA, pH 7.4), 0.5% SDS, 30% formamide, 100 ug/ml salmon sperm blocking DNA; followed by washes at 50°C with 1XSSPE, 0.1% SDS. In addition, to achieve even lower stringency, washes performed following stringent hybridization can be done at higher salt concentrations (e.g. 5X SSC).
Note that variations in the above conditions may be accomplished through the inclusion and/or substitution of alternate blocking reagents used to suppress background in hybridization experiments. Typical blocking reagents include Denhardt's reagent, BLOTTO, heparin, denatured salmon sperm DNA, and commercially available proprietary formulations. The inclusion of specific blocking reagents may require modification of the hybridization conditions described above, due to problems with compatibility.
Of course, a polynucleotide which hybridizes only to polyA+ sequences (such as any 3' terminal polyA+ tract of a cDNA shown in the sequence listing), or to a
complementary stretch of T (or U) residues, would not be included in the definition of "polynucleotide," since such a polynucleotide would hybridize to any nucleic acid molecule containing a poly (A) stretch or the complement thereof (e.g., practically any double-stranded cDNA clone). The polynucleotide of the present invention can be composed of any polyribonucleotide or polydeoxribonucleotide, which may be unmodified RNA or DNA or modified RNA or DNA. For example, polynucleotides can be composed of single- and double-stranded DNA, DNA that is a mixture of single- and double-stranded regions, single- and double-stranded RNA, and RNA that is mixture of single- and double-stranded regions, hybrid molecules comprising DNA and RNA that may be single-stranded or, more typically, double-stranded or a mixture of single- and double- stranded regions. In addition, the polynucleotide can be composed of triple-stranded regions comprising RNA or DNA or both RNA and DNA. A polynucleotide may also contain one or more modified bases or DNA or RNA backbones modified for stability or for other reasons. "Modified" bases include, for example, tritylated bases and unusual bases such as inosine. A variety of modifications can be made to DNA and RNA; thus, "polynucleotide" embraces chemically, enzymatically, or metabolically modified forms.
The polypeptide of the present invention can be composed of amino acids joined to each other by peptide bonds or modified peptide bonds, i.e., peptide isosteres, and may contain amino acids other than the 20 gene-encoded amino acids. The polypeptides may be modified by either natural processes, such as posttranslational processing, or by chemical modification techniques which are well known in the art. Such modifications are well described in basic texts and in more detailed monographs, as well as in a voluminous research literature. Modifications can occur anywhere in a polypeptide, including the peptide backbone, the amino acid side-chains and the amino or carboxyl termini. It will be appreciated that the same type of modification may be present in the same or varying degrees at several sites in a given polypeptide. Also, a given polypeptide may contain many types of modifications. Polypeptides may be branched , for example, as a result of ubiquitination, and they may be cyclic, with or without branching. Cyclic, branched, and branched cyclic polypeptides may result from posttranslation natural processes or may be made by synthetic methods. Modifications include acetylation, acylation, ADP-ribosylation, amidation, covalent attachment of flavin, covalent attachment of a heme moiety, covalent attachment of a nucleotide or nucleotide derivative, covalent attachment of a lipid or lipid derivative, covalent attachment of phosphotidylinositol, cross-linking, cyclization, disulfide bond formation, demethylation, formation of covalent cross-links, formation of cysteine,
formation of pyroglutamate, formylation, gamma-carboxylation, glycosylation, GPI anchor formation, hydroxylation, iodination, methylation, myristoylation, oxidation, pegylation, proteolytic processing, phosphorylation, prenylation, racemization, selenoylation, sulfation, transfer-RNA mediated addition of amino acids to proteins such as arginylation, and ubiquitination. (See, for instance, PROTEINS -
STRUCTURE AND MOLECULAR PROPERTIES, 2nd Ed., T. E. Creighton, W. H. Freeman and Company, New York (1993); POSTTRANSLATIONAL COVALENT MODIFICATION OF PROTEINS, B. C. Johnson, Ed., Academic Press, New York, pgs. 1-12 (1983); Seifter et al., Meth Enzymol 182:626-646 (1990); Rattan et al., Ann NY Acad Sci 663:48-62 (1992).)
"SEQ ID NO:X" refers to a polynucleotide sequence while "SEQ ID NO:Y" refers to a polypeptide sequence, both sequences identified by an integer specified in Table 1.
"A polypeptide having biological activity" refers to polypeptides exhibiting activity similar, but not necessarily identical to, an activity of a polypeptide of the present invention, including mature forms, as measured in a particular biological assay, with or without dose dependency. In the case where dose dependency does exist, it need not be identical to that of the polypeptide, but rather substantially similar to the dose-dependence in a given activity as compared to the polypeptide of the present invention (i.e., the candidate polypeptide will exhibit greater activity or not more than about 25-fold less and, preferably, not more than about tenfold less activity, and most preferably, not more than about three-fold less activity relative to the polypeptide of the present invention.)
Polynucleotides and Polypeptides of the Invention
FEATURES OF PROTEIN ENCODED BY GENE NO: 1
This gene is expressed primarily in testes tumor and to a lesser extent in fetal brain. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions which include, but are not limited to, cancer particularly of the testes, and defects of the central nervous system such as seizure and neurodegenerative disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly cancer of the testes and central nervous system,
expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., testes and other reproductive tissue, brain and other tissue of the nervous system, and blood cells, and spleen, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment/diagnosis of testicular cancer and treatment of central nervous system disorders since this gene is primarily expressed in the testes tumor and developing brain.
FEATURES OF PROTEIN ENCODED BY GENE NO: 2
This gene is expressed primarily in cancer tissues, such as breast cancer and Wilm's tumor, and to a lesser extent in fetal tissues.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions which include, but are not limited to, and/or tumors, particularly, those found in the breast, and developmental abnormalities or disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the glandular tissues, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., mammary tissue, and fetal tissue and, cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. Preferred epitopes include those comprising a sequence shown in SEQ ID NO. 314 as residues: Pro-1 1 to Thr-18, Leu-43 to Pro-50, Gly-64 to Leu-72, and Leu-81 to Lys-86.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment/diagnosis of cancers and/or tumors, particularly, those found in the breast since expression is mainly in cancer/tumor tissues. May serve as therapeutic proteins for proliferation/differentiation of fetal tissues.
FEATURES OF PROTEIN ENCODED BY GENE NO: 3
This gene is expressed primarily in CD34 depleted buffy coat and to a lesser extent in spleen, chronic lymphocytic leukemia. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: blood disorders or leukemias, diseases of the immune system. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, and spleen, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment/diagnosis of blood disorders or leukemias, diseases of the immune system since expression is in tissues related to immune function.
FEATURES OF PROTEIN ENCODED BY GENE NO: 4
This gene is expressed primarily in CD34 depleted buffy coat. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: blood disorders or lymphocytic diseases. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment/diagnosis of blood disorders since expression is in tissues related to immune function.
FEATURES OF PROTEIN ENCODED BY GENE NO: 5
This gene is expressed primarily in CD34 depleted buffy coat. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: blood or immune diseases. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. Preferred epitopes include those comprising a sequence shown in SEQ ID NO. 317 as residues: Pro- 13 to Lys-21.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment/diagnosis of blood disorders since expression is in tissues related to immune function.
FEATURES OF PROTEIN ENCODED BY GENE NO: 6
This gene is expressed primarily in CD34 depleted buffy coat. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: blood or immune diseases. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., and blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level
in healthy tissue or bodily fluid from an individual not having the disorder. Preferred epitopes include those comprising a sequence shown in SEQ ID NO. 318 as residues: Lys-31 to Lys-39.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment/diagnosis of blood diseases since it is expressed in tissues related to immune function.
FEATURES OF PROTEIN ENCODED BY GENE NO: 7
This gene is expressed primarily in CD34 depleted buffy coat and to a lesser extent in pineal gland.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: diseases of the immune system and brain associated diseases. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, and pineal gland, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment/diagnosis of blood disorders, immune diseases or brain associated diseases (specifically of the pineal gland) since expression is in tissues related to immune function.
FEATURES OF PROTEIN ENCODED BY GENE NO: 8 The translation product of this gene shares sequence homology with an organic cation transporter which is thought to be important in organic cation uptake in the kidney and liver. (See Accession No. 2343059.) Preferred polypeptide fragments comprise the amino acid sequence ITIAIQMICLVNXELYPTFVRNXGVMVCSSLCDIGGIITP FIVFRLREVWQALPLILFAVLGLLAAGVTLLLPETKGVALPETMKDAENLGRKAKPKENTIYLK VQTSEPSGT (SEQ ID NO: 615) or TMKDAENLGRKAKPKENT (SEQ ID NO: 616) as well as N-terminal and C-terminal deletions of these fragments. Also preferred are polynucleotide fragments encoding these polypeptide fragments.
This gene is expressed primarily in liver.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: hepatic and renal diseases where drug elimination/cation exchange (organic cation uptake) in the liver and kidney are problematic. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the hepatic or renal system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., kidney and liver, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. Preferred epitopes include those comprising a sequence shown in SEQ ID NO. 320 as residues: Asn-64 to Asn-74, and Gln-81 to Gly-87.
The tissue distribution and homology to organic cation transporter indicate that polynucleotides and polypeptides corresponding to this gene are useful as a polyspecific transporter that is important for drug elimination in the liver (and possibly kidney) since expression is found in the liver.
FEATURES OF PROTEIN ENCODED BY GENE NO: 9
This gene is expressed primarily in eosinophil induced with EL- 5 and to a lesser extent in fetal liver and spleen. This gene also maps to chromosome 15, and therefore can be used in linkage analysis as a marker for chromosome 15.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: diseases of the immune system, particularly allergies or asthma. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, liver, and spleen, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the
standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treating/diagnosis of diseases involving esosinphil reactions since expression seems to be concentrated in eosinophils and other tissues involved in immunity such as the liver and spleen.
FEATURES OF PROTEIN ENCODED BY GENE NO: 10
This gene is expressed primarily in tissues of hematopoietic lineage and to a lesser extent in Hodgkins lymphoma. Any frame shifts in this sequence can easily be clarified using known molecular biology techniques.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions which include, but are not limited to, and immune deficiency or dysfunction. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., hematopoietic cells, lymphoid and reticuloendothelial tissues, and cancerous tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment/ diagnosis for lymphomas or immune dysfuction or as a therapeutic protein useful in immune modulation based on expression in anergic T-cells and lymphomas.
FEATURES OF PROTEIN ENCODED BY GENE NO: 11
This gene is expressed primarily in neutrophils and to a lesser extent in activated lymphoid cells.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the cell type present in a biological sample and for diagnosis of diseases and conditions: inflamation. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders
of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells and lymphoid tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. Preferred epitopes include those comprising a sequence shown in SEQ ID NO. 323 as residues: Glu-40 to Lys-46. The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for modulation of an immune reaction or as a growth factor for the differentiation or proliferation of neutrphils for the treatment of neutropenia.
FEATURES OF PROTEIN ENCODED BY GENE NO: 12 This gene is expressed primarily in brain and to a lesser extent in activated T- cells. It is likely that the open reading frame containing the predicted signal peptide continues in the 5' direction. Preferred polypeptide fragments comprise the amino acid sequence PRVRNSPEDLGLSLTGDSCKL (SEQ ID NO:617).
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: neurodegenerative disorders including ischemic shock, alzheimers and cognitive disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the central nervous system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, and brain, and other tissue of the nervous system and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. Preferred epitopes include those comprising a sequence shown in SEQ ID NO. 324 as residues: Ser-5 to Glu- 14, Ile-21 to Pro-35, Ser-65 to Asp-81, Cys-89 to Val-96, Lys-136 to Ser-145, Ile-152 to Met-169, and Arg-189 to Lys-196. The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnostic/treatment for cancers of the given tissue or in the treatment of neurological disorders of the CNS.
FEATURES OF PROTEIN ENCODED BY GENE NO: 13
This gene was also recently cloned by other groups, naming this calcium-activated potassium channel gene, hKCa4. (See Accession No. AF033021, see also, Accession No. 2584866.) This gene is mapped to human chromosome 19ql3.2. A second signal sequence likely exists upstream from the predicted signal sequence as described in Table 1. Preferred polypeptide fragments comprise: QADDLQATVAALCVLRGGGPWAG SWLSPKTPGAMGGDLVLGLGALRRRKRLL (SEQ NO: 618); or EQEKSLAGWALVLAXXGIGL MVLHAEMLWFGGCSAVNATGHLSDTLWLIPITFLΗGYGDVVPGTMWGKIVCLCTGVMGVCC TALLV AVVARKLEFNKAEKHVHNFMMDIQ YTKEMKES AARVLQEA WMFYKHTRRKESHAAR XHQRXLLAAINAFRQVRLKHRKLREQλ^SMVDISKMHMILYDLQQNLSSSHRALEKQIDTLAG
KLDALTELLSTALGPRQLPEPSQQSK (SEQ ID NO: 619), as well as N-terminal and C- terminal deletions. Also preferred are polynulcleotide fragments encoding these polypeptide fragments. This gene is expressed primarily in breast lymph node and T-cells, and to a lesser extent in placenta.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: hematologic and immune disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., lymphoid tissue, blood cells and placenta, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. Preferred epitopes include those comprising a sequence shown in SEQ ID NO. 325 as residues: Arg- 13 to Lys-23.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for the treatment/diagnosis of hematologic and diseases involving immune modulation based or distribution in the lymph node and T- cells.
FEATURES OF PROTEIN ENCODED BY GENE NO: 14
This gene was recently cloned by another group, calling it PAPS synethase. (See Accession No. el204135.) Preferred polypeptide fragments comprise the amino acid sequence YQAHHVSRNKRGQVVGTRGGFRGCTVWLTGLSGAGK (SEQ ID NO: 620). Also preferred are the polynucleotide fragments encoding this polypeptide fragment. It has been discovered that this gene is expressed primarily in benign prostate hyperplasia, Human Umbilical Vein Endothelial Cells and to a lesser extent in smooth muscle and Human endome trial stromal cells-treated with estradiol.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: inflamation, ischemia, and restenosis, based on endothelial cell and smooth muscle cell expression, and prostate diseases such as benign prostate hyperplasia or prostate cancer. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the prostate or vessels of the circulatory system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., prostate, endothelial cells, smooth muscle, and endometrium, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. Preferred epitopes include those comprising a sequence shown in SEQ ID NO. 326 as residues: Arg-21 to Asp-26, Lys-35 to Lys-44, Glu-49 to Asn-58.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treating/diagnosing diseases or conditions where the endothelial cell lining of the veins and arteries of underlying smooth muscle are involved.
FEATURES OF PROTEIN ENCODED BY GENE NO: 15
This gene is expressed primarily in human 6 week embryo and to a lesser extent in placenta.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: developmental anomalies or fetal deficiencies. Similarly, polypeptides and antibodies directed to these
polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly developmental in nature, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., embryonic tissue, and placenta, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. Preferred epitopes include those comprising a sequence shown in SEQ ID NO. 327 as residues Lys-50 to Glu-57.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for detection of developmental abnormalities.
FEATURES OF PROTEIN ENCODED BY GENE NO: 16 This gene is expressed primarily in kidney and amygdala and to a lesser extent in fetal tissues. This gene is mapped to chromosome 14, and therefore is useful in linkage analysis as a marker for chromosome 14.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) present in a biological sample and for diagnosis of diseases and conditions: kidney diseases, neurological disorders and developmental abnormalities. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s). For a number of disorders of the above tissues, particularly of the renal system or developing fetal tissues, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., kidney, amygdala, and fetal tissues, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment or diagnosis of conditions affecting the brain, kidneys and fetal development.
FEATURES OF PROTEIN ENCODED BY GENE NO: 17
This gene is expressed primarily in ovarian cancer.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: solid tumors similar to ovarian cancer Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the reproductive system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., ovarian and other reproductive tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. Preferred epitopes include those comprising a sequence shown in SEQ ID NO. 329 as residues Ser-51 to Val-56. The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for the treatment of solid tumors of the reproductive system such as ovarian cancer.
FEATURES OF PROTEIN ENCODED BY GENE NO: 18
This gene is expressed primarily in brain medulloblastoma. Preferred polypeptide fragments comprise the amino acid sequence: IRHEQHPNFSLEMHSKGSSLLLFLPQL ILILPVCAHLHEELNC (SEQ ID NO: 643) and SFHSEEKGHLLLQAERHPWVAGALVGVSG GLTLTTCSGPTEKPATKNYFLKRLLQEMHIRAN (SEQ ID NO: 644), as well as N-terminal and C-terminal deletions. Also preferred are polynucleotide fragments encoding these polypeptide fragments. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: tumors particularly of the CNS or Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the Central nervous system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene
expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treating medulloblastoma or similar tumors.
FEATURES OF PROTEIN ENCODED BY GENE NO: 19 This gene is expressed primarily in adipocytes.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: obesity. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the adipose tissues expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., adipocytes and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treating obesity by regulating the function and number of adipocytes
FEATURES OF PROTEIN ENCODED BY GENE NO: 20 This gene is expressed primarily in B cell lymphoma.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions which include, but are not limited to, of the immune system with an emphasis on B cell lymphoma. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the tumors of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, and lymphoid tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e.,
the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of B cell derived tumors based on its expression in b cell lymphomas
FEATURES OF PROTEIN ENCODED BY GENE NO: 21
This gene is expressed primarily in immune cells and to a lesser extent in fetal tissues Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: inflammatory diseases Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., cells of the immune system, and fetal tissues, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. Preferred epitopes include those comprising a sequence shown in SEQ ID NO:333 as residues Asp- 10 to Pro- 19, Ser-74 to Tyr-79, Glu-95 to Lys- 110. The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment of diseases involving alterations in T cell activity.
FEATURES OF PROTEIN ENCODED BY GENE NO: 22
It has been discovered that this gene is expressed primarily in ovarian tumor. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: tumors particularly of the ovary. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of tumors of the reproductive organs, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., ovarian
and other reproductive tissue and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. Preferred epitopes include those comprising a sequence shown in SEQ ID NO. 334 as residues: Leu-22 to Gln-27.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of ovarian tumors as it has only been identified in ovarian tumors.
FEATURES OF PROTEIN ENCODED BY GENE NO: 23
It has been discovered that this gene is expressed primarily in fetal tissues and to a lesser extent in osteoclastoma cell line
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: osteoporosis or arthritis Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the skeletal expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., bone cells, and fetal tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment of conditions of abnormal bone remodeling due to enhanced activity of osteoclasts. This may be useful as a specific marker for malignancies derived from osteoclasts or their precursors.
FEATURES OF PROTEIN ENCODED BY GENE NO: 24
The translation product of this gene shares sequence homology with a periplasmic ribonuclease which is thought to be important in degrading extracellular polynucleotides It has been discovered that this gene is expressed primarily in serum treated smooth muscle cells
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: vascular disease such as restenosis. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the vasculature expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., smooth muscle, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. Preferred epitopes include those comprising a sequence shown in SEQ ID NO: 336 as residues: Gln-30 to Lys-36, and Pro-41 to Arg-48. The tissue distribution and homology to ribonucleases indicate that polynucleotides and polypeptides corresponding to this gene are useful for treatment of pathological conditions of smooth muscle associated with bacterial or viral infiltration
FEATURES OF PROTEIN ENCODED BY GENE NO: 25 This gene is expressed primarily in Early Stage Human Brain.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: human brain development and related diseases. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the human brain development and related diseases, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to this gene indicate that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of diseases affecting human brain development and related diseases.
FEATURES OF PROTEIN ENCODED BY GENE NO: 26
It has been discovered that this gene is expressed primarily in human brain tissue. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: human brain diseases and other diseases related to brain diseases, which may be caused by brain diseases. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the human brain diseases, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to the gene indicate that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of human brain diseases and other diseases related.
FEATURES OF PROTEIN ENCODED BY GENE NO: 27
It has been discovered that this gene is expressed primarily in Anergic T-cells. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: immune diseases, inflammatory diseases and diseases related to T lymph cells. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune diseases, inflammatory diseases and diseases related to T lymph cells, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene
expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to the gene indicate that polynucleotides and polypeptides corresponding to this gene are useful for immune diseases, inflammatory diseases and diseases related to T lymph cells.
FEATURES OF PROTEIN ENCODED BY GENE NO: 28
The translation product of this gene shares sequence homology with Shigella flexneri positive transcriptional regulator CriR (criR) gene which is thought to be important in regulation of gene expression.
This gene is expressed primarily in human synovial sarcoma and normal human brain tissues.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: human brain diseases particularly sarcomas of the synovium. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the human brain and synovium and other related human brain diseases, expression of this gene at significantly higher or lower levels may be routinely detected in certain (e.g., synovial tissue, and brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of human synovial sarcoma and other related human brain diseases.
FEATURES OF PROTEIN ENCODED BY GENE NO: 29
This gene is expressed in bone marrow, infant brain, fetal liver and spleen, prostate and to a lesser extent in pineal gland, adipose tissue, kidney, adrenal gland, umbilical vein endothelial cells, and T cells. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: diseases related to bone marrow or
hematoplastic tissues, prostate, kidney, adrenal gland, and cardiovascular tissue or organs. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the diseases related to hematoplastic tissues, immune system, prostate, kidney, adrenal gland, and cardiovascular tissue or organs, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., bone marrow, hematopoietic cells, pineal gland, adipose tissue, kidney, adrenal gland, endothelial cells, and blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to the gene indicate that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of diseases related to hematoplastic tissues, immune system, prostate, kidney, adrenal gland, and cardiovascular tissue or organs.
FEATURES OF PROTEIN ENCODED BY GENE NO: 30 This gene is expressed primarily in meningea and to a lesser extent in breast and adult brain.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: Diseases of the meningea and related brain diseases. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the meningea and related brain diseases, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., miningea, mammary tissue, and brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of diseases of the meningea and related brain diseases.
FEATURES OF PROTEIN ENCODED BY GENE NO: 31
This gene is expressed in meningea, fetal spleen, osteoblast and to a lesser extent in activated T-cells, endometrial stromal cells, fetal lung, HL-60, thymus, testis and endothelial cells.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: meningeal disease, osteoporosis, immune diseases, and hematoplastic diseases. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the meningeal diseases, osteoporosis, immune diseases, and hematoplastic diseases, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, endometrium, lung, thymus, testis, and endothelial cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to gene indicate that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of meningeal, osteoporosis, immune diseases, hematoplastic diseases, testis diseases and lung diseases.
FEATURES OF PROTEIN ENCODED BY GENE NO: 32
This gene is expressed primarily in human thymus and to a much lesser extent in infant brain, T-cells, smooth muscle, endothelial cells, bone marrow, human ovarian tumor and keratinocytes testes, osteoclastoma, breast, and tonsils.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: Diseases involving the thymus, particularly thymic cancer and diseases involving T-cell maturation. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a
number of disorders of the above tissues or cells, particularly of the thymus, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., thymus, brain, and other tissue of the nervous system, blood cells, bone marrow, ovaries, and testes, and other reproductive tissue, mammary tissue, tonsils, melanocytes and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution and homology to gene indicate that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of diseases of the thymus particularly thymic cancer and diseases involving T-cell maturation.
FEATURES OF PROTEIN ENCODED BY GENE NO: 33
This gene is expressed primarily in human tonsils, and placenta, and to a lesser extent in adipocytes, melanocyte, and infant brain.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: inflammatory diseases, immune diseases, and obesity. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the inflammatory diseases, immune diseases, and obesity, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., tonsils, placenta, adipocytes, melanocytes, and brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to this gene indicate that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of diseases such as inflammation, immune diseases, and obesity.
FEATURES OF PROTEIN ENCODED BY GENE NO: 34
This gene is expressed in activated T cells, and to a lesser extent in pituitary, testis, and breast lymph node.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: diseases relating to T cells. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the disorders of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., pituitary, testes and other reproductive tissue, mammary tissue, and lymphoid tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment of immune disorders.
FEATURES OF PROTEIN ENCODED BY GENE NO: 35
This gene is expressed primarily in infant brain.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: neurological disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the diseases relating to neurological disorders, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain, and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of neurological disorders.
FEATURES OF PROTEIN ENCODED BY GENE NO: 36
This gene is expressed primarily in infant brain.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: neurological disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the diseases relating to neurological disorders, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of neurological disorders.
FEATURES OF PROTEIN ENCODED BY GENE NO: 37 This gene is expressed primarily in human ovary.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: ovarian cancer. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the ovarian disorders such as those involving germ cells, ovarian follicles, stromal cells, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., ovary and other reproductive tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of ovariopathy.
FEATURES OF PROTEIN ENCODED BY GENE NO: 38
This gene is expressed primarily in lymph node breast cancer.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: breast cancer. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the breast cancer, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., mammary tissue and lymphoid tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for used as a diagnostic marker for breast cancer.
FEATURES OF PROTEIN ENCODED BY GENE NO: 39 This gene is expressed primarily in brain and to a lesser extent in other tissues.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: neuronal disorders such as trauma, brain degeneration, and brain tumor. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the brain, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and therapeutic treatment of neuronal disorders.
FEATURES OF PROTEIN ENCODED BY GENE NO: 40
This gene is expressed in early stage human embryo, adrenal gland tumor, and immune tissues such as fetal liver, fetal spleen, T-cell, and myoloid progenitor cell line and to a lesser extent in ovary, colon cancer, and a few orther tissues.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: tumorigenesis including adrenal gland tumor, colon cancer and various other tumors, developmental and immune disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the cancer tissues, early stage human tissues, and immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., liver, spleen, blood cells, bone marrow, ovary and other reproductive tissue, and colon, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and therapeutic treatment of immune and developmental disorders, and tumorigenesis.
FEATURES OF PROTEIN ENCODED BY GENE NO: 41
This gene is expressed primarily in fetal lung, endothelial cells, liver, thymus and a few other immune tissues.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: immune disorders such as immune deficiency and autoimmune diseases, pulmonary diseases, liver diseases, and tumor matasis. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the fetal lung, liver, endothelial cells, and immune tissues, expression of this gene at significantly higher or lower levels may be routinely detected in certain
tissues and cell types (e.g., lung, endothelial cells, liver, thymus, and other tissue of the immune system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis of immune disorders and pulmonary and hepatic diseases. Its promoter may also be used for immune system and lung- specific gene therapies. The expression of this gene in endothelial cells indicates that it may also involve in angiogenesis which therefore may play role in tumor matasis.
FEATURES OF PROTEIN ENCODED BY GENE NO: 42
This gene is expressed primarily in liver, thyroid, parathyroid and to a lesser extent in fetal lung, stomach and early embryos.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: metabolic regulation, obesity, heptic failure, heptacellular tumors or thyroiditis and thyroid tumors. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the digestive/endocrine system expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., liver, thyroid, parathyroid, lung, stomach, and embryonic tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution and the extracellular locations indicates that polynucleotides and polypeptides corresponding to this gene are useful for the detection and treatment of digestive/endocrine disorders, including metabolic regulation, heptic failure, malabsortion, gastritis and neoplasms.
FEATURES OF PROTEIN ENCODED BY GENE NO: 43
This gene is expressed primarily in Schizophrenic adult brain, pituitary, front cortex, hypothalmus and to a lesser extent in retina, adipose and stomach cancer and placenta. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: schizophrenia and other neurological disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the central nerve system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., retinal tissue, adipose, stomach, and placenta, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful in treatment/detection of disorders in the nerve system, including schizophrenia, neurodegeneration, and neoplasia. Additionally, a secreted protein in brain may serve as an endocrine.
FEATURES OF PROTEIN ENCODED BY GENE NO: 44
The translation product of this gene shares sequence homology with GTP binding proteins which are thought to be important in signal transduction and protein transport.
This gene is expressed primarily in umbilical vein and microvascular endothelial cells, GM-CSF treated macrophage, anergic T cells, osteoblast, osteoclast, CD34+ cells and to a lesser extent in gall bladder. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: bone formation and growth, osteonecrosis, osteoporosis, angiogenesis and/or hematopoeisis. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the skeletal and hematopoeisis systems, expression of this gene at significantly higher or lower levels
may be routinely detected in certain tissues and cell types (e.g., endothelial cells, blood cells, bone, and gall bladder, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to GTP binding proteins indicate that polynucleotides and polypeptides corresponding to this gene are useful for treatment/detection of bone formation and growth, osteonecrosis, osteoporosis, and/or hematopoeisis because its involvement in the growth signaling or angiogenesis.
FEATURES OF PROTEIN ENCODED BY GENE NO: 45
The translation product of this gene shares sequence homology with signal sequence receptor gamma subunit which is thought to be important in protein translocation on endoplasmic reticulum.
This gene is expressed primarily in adrenal gland, salivary gland, prostate, and to a lesser extent in endothelial cells and smooth muscle.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: protein secretion. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the secretory organs, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., adrenal gland, salivary gland, prostate, endothelial cells, and smooth muscle, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to SSR gamma subunit indicate that polynucleotides and polypeptides corresponding to this gene are useful for endocrine disorders, prostate cancer, xerostomia or sialorrhea.
FEATURES OF PROTEIN ENCODED BY GENE NO: 46
This gene is expressed primarily in osteoclastoma cells and to a lesser extent in melanocyte, amygdala, brain, and stomach.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: ossification, osteoporosis, fracture, osteonecrosis, osteosarcoma. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the skeletal systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., melanocytes, amygdala, brain and other tissue of the nervous system, and stomach, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful in intervention of ossification, osteoporosis, fracture, osteonecrosis and osteosarcoma.
FEATURES OF PROTEIN ENCODED BY GENE NO: 48 The translation product of this gene shares sequence homology with proline rich proteins which is thought to be important in protein-protein interaction. This gene is expressed primarily in brain.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: neurological and psychological disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the central nerve system and endocrine system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to proline-rich proteins indicate that polynucleotides and polypeptides corresponding to this gene are useful in intervention
and detection of neurological diseases, including trauma, neoplasia, degenerative or metabolic conditions in the central nerve system. Additionally, the gene product may be a secreted by the brain as an endocrine.
FEATURES OF PROTEIN ENCODED BY GENE NO: 49
The translation product of this gene shares sequence homology with the AOCB gene from Aspergillus nidulans which is important in asexual development.
This gene is expressed primarily in infant brain and to a lesser extent in the developing embryo, trachea tumors, B-cell lymphoma and synovial sarcoma. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: neurodegenerative diseases, leukemia and sarcoma's. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the brain and immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., embryonic tissue, blood cells, trachea, and synovial tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution in infant brain and sarcoma's and homology to a gene involved in a key step of eukaryotivc development (fungal spore formation) indicates that the protein product of this clone could play a role in neurological diseases such as schizophrenia, particularly in infants. The existence of the gene in a B-cell lymphoma indicates the gene may be used in the treatment and detection of leukemia.
FEATURES OF PROTEIN ENCODED BY GENE NO: 50 This gene is expressed primarily in fetal lung.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: pulmonary disorders including lung cancer. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the pulmonary system, expression of this gene at significantly higher or
lower levels may be routinely detected in certain tissues and cell types (e.g., lung, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution of this gene only in fetal lung indicates that it plays a key role in development of the pulmonary system. This would suggest that misregulation of the expression of this protein product in the adult could lead to lymphoma or sarcoma formation, particularly in the lung. It may also be involved in predisposition to certain pulmonary defects such as pulmonary edema and embolism, bronchitis and cystic fibrosis.
FEATURES OF PROTEIN ENCODED BY GENE NO: 51
This gene is expressed primarily in hematopoietic cell types and fetal cells and to a lesser extent in all tissue types.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: defects in the immune system and hematopoeisis. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune and hematopoietic systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., hematopoietic cells, and fetal tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution of this gene predominantly in hematopoeitic cells and in the developing embryo indicates that polynucleotides and polypeptides corresponding to this gene are useful for detection and treatment of lymphomas and disease states affecting the immune system or hematopoeisis disorders such as leukemia, AIDS, arthritis and asthma..
FEATURES OF PROTEIN ENCODED BY GENE NO: 52
This gene is expressed primarily in prostate and to a lesser extent in fetal spleen, fetal liver, infant brain and T cell leukemias.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: prostate disorders, prostate cancer, leukemia. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, and/or prostate gland expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., thymus, spleen, liver, brain and other tissue of the nervous system, and blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution of this gene in prostate indicates that polynucleotides and polypeptides corresponding to this gene are useful for detection or treatment of prostate disorders or prostate cancer. Its distribution in fetal liver and fetal spleen indicates it may play a role in the immune system and its misregulation could lead to immune disorders such as leukemia, arthritis and asthma.
FEATURES OF PROTEIN ENCODED BY GENE NO: 53
The translation product of this gene shares sequence homology with dynein.
This gene is expressed primarily in brain.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: neuro-degenerative diseases of the brain. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly neuro-degenerative diseases expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The predominant tissue distribution in the brain and homology to dynein, a microtubule motor protein involved in the positioning of cellular organelles and molecules indicates that polynucleotides and polypeptides corresponding to this gene are useful for detection/treatment of neurodegenerative diseases, such as Alzheimers, Huntigtons, Parkinsons diseases and shizophrenia.
FEATURES OF PROTEIN ENCODED BY GENE NO: 54
The translation product of this gene shares sequence homology with ubiquitin- conjugation protein, an enzyme which is thought to be important in the processing of the Huntingtons Disease causing gene.
This gene is expressed primarily in brain and to a lesser extent in activated macrophages.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: neurodegenerative disease states including Huntington's disease. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of brain tissues. For a number of disorders of the above tissues or cells, particularly of the neurological systems expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, and blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The predominant tissue distribution of this gene in the brain and its homology to a Huntington interacting protein indicates that polynucleotides and polypeptides corresponding to this gene are useful for the regulation of the expression of the Huntington disease gene and other neurodegenerative diseases including spinocerebuUar ataxia types I and III, dentatorubropallidoluysian and spinal bulbar muscular atrophy. In addition, the existence of elevated levels of free ubiquitin pools in Alzheimer's disease, Parkinson's disease and amylotrophic lateral sclerosis indicates that the ubiquitin pathway of protein degradation plays a role in these disease states. Thus, considering the gene described here is homologous to a ubiquitin-conjugation protein it may play a general role in neurodegenarative conditions.
FEATURES OF PROTEIN ENCODED BY GENE NO: 56
This gene is expressed primarily in T-cells (anergic T-cells, resting T-Cells, apoptotic T-cells) and lymph node (breast), as well as brain (hypothalamus, hippocampus, pituitary, infant brain, early-stage brain). Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: immune (e.g. immunodeficiencies, autoimmunities, inflammation, leukemias & lymphomas) and neurological (e.g. Alzheimer's disease, dementia, schizophrenia) disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the central nervous, hematopoietic and immune systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, lymphoid tissue, and brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful in the intervention or detection of pathologies associated with the hematopoietic and immune systems, such as anemias (leukemias). In addition, the expression in brain (including fetal) might suggest a role in developmental brain defects, neuro-degenerative diseases or behavioral abnomalities (e.g. schizophrenia, Alzheimer's, dementia, depression, etc.).
FEATURES OF PROTEIN ENCODED BY GENE NO: 57
This gene is expressed primarily in lung, and to a lesser extent in a variety of other hematological cell types (e.g. Raji cells, bone marrow cell line, activated monocytes).
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: pulmonary and/or hematological disfunction. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the vasculo-pulmonary and hematopoietic systems, expression of this
gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., lung and blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful in the intervention and detection of pathologies associated with the vasculo-pulmonary system. In addition the expression of this gene in a variety of leukocytic cell types and a bone marrow cell line might suggest a role in hematopoietic and immune system disorders, such as leukemias & lymphomas, inflammation, immunodeficiencies and autoimmunities.
FEATURES OF PROTEIN ENCODED BY GENE NO: 58 The translation product of this gene shares sequence homology with adenylate kinase isozyme 3 (gil 163528 GTP:AMP phosphotransferase (EC 2.7.4.10) [Bos taurus]), which is thought to be important in catalyzing the phosphorylation of AMP to ADP in the presence of ATP or inorganic triphosphate.
This gene is expressed primarily in fetal liver, heart and placenta, and to a lesser extent in many other tissues.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: hepatic, cardiovascular or reproductive disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the hepatic, cardiovascular and reproductive systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., liver, heart, and placenta, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for the treatment and diagnosis of conditions related to hepatic function and pathogenesis, in particular, those dealing with liver development and the differentiation of hepatocyte progenitor cells.
FEATURES OF PROTEIN ENCODED BY GENE NO: 59
This gene is expressed primarily in CD34 positive cells (Cord Blood). Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: hematopoietic differentiation and immune disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of hematopoietic and immune systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., hematopoietic cells, and blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful in the detection and treatment of conditions associated with CD34-positive cells, and therefore as a marker for cell differentiation in hematapoiesis, as well as immunological disorders.
FEATURES OF PROTEIN ENCODED BY GENE NO: 60
The translation product of the predicted open reading frame of this contig has sequence identity to the murine gene designated Insulin-Like Growth Factor-Binding Protein (IGFBP)-l as described by Lee and colleagues (Hepatology 19 (3), 656-665 ( 1994)).
This gene is expressed exclusively in hemangiopericytoma. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of hemangiopericytoma and other pericyte or endothelial cell proliferative disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the circulatory and immune systems, expression of this gene at significantly higher or lower levels may routinely be detected in certain tissues and cell types (e.g., pericyte or endothelial cells, and liver, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or
another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
Polynucleotides and polypeptides corresponding to this gene are useful as cell growth regulators since IGFBP-1-like molecules function as modulators of insulin-like growth factor activity. In addition, since IGFBP-1 is expressed at high levels following hepatectomy and during fetal liver development, polynucleotides of the present invention may also be used for the diagnosis of developmental disorders. Further, polypeptides of the present invention may be used therapeutically to treat developmental liver disorders as well as to regulate hepatocyte and supporting cell growth following hepatectomy or to treat liver disorders.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of hemangiopericytoma and liver disorders.
FEATURES OF PROTEIN ENCODED BY GENE NO: 61
This gene is expressed primarily in schizophrenic frontal cortex. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: nervous system and cognitive disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the frontal cortex and CNS expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for study, treatment and diagnosis of frontal cortex, neuro-degenerative and CNS disorders
FEATURES OF PROTEIN ENCODED BY GENE NO: 62
This gene is expressed primarily in human adrenal gland tumor, and to a lesser extent in human kidney, medulla and adult pulmonary tissue.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: metabolic, endocrine disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the endocrine and nervous system disorders and neoplasia, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., adrenal gland, kidney, brain and other tissue of the nervous system, pulmonary tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for study, treatment and diagnosis of neurological and endocrine disorders including neoplasia.
FEATURES OF PROTEIN ENCODED BY GENE NO: 63 This gene is expressed primarily in human adipocytes, and to a lesser extent in spleen, 12-week old human, and testes.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: immune, metabolic and growth disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., adipocytes, spleen, and testes and other reproductive tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for study, diagnosis and treatment of immune, developmental and metabolic disorders.
FEATURES OF PROTEIN ENCODED BY GENE NO: 64
One translated product of this clone is homologous to the mouse zinc finger protein PZF. (See Accession No. 453376; see also Gene 152 (2), 233-238 (1995).) Preferred polypeptide fragments correspond to the highly conserved domains shared between mouse and man. For example, preferred polypeptide fragments comprise the amino acid sequence: LQCEICGFTCRQKASLNWHMKKHDADSFYQFSCNICGKKFEKKDSVVAHKAKSH PEV (SEQ ID NO: 621 ); ITSTDILGTNPESLTQPSD (SEQ ID NO: 622); NSTSGECLLLEAEGM SKSY (SEQ ID NO: 623); CSGTERVSLMADGKIFVGSGSSGGTEGLVMNSDILGATTEVLIEDSD SAGP (SEQ ID NO: 624); IQYVRCEMEGCGTVLAHPRYLQHHIKYQHLLKKKYVCPHPSCGRLF RLQKQLLRHAKHHT (SEQ ID NO: 625); DQRDYICEYCARAFKSSHNLAVHRMIHTGEK (SEQ ID NO: 626); RSSRTSVSRHRDTENTRSSRSKTGSLQLICKSEPNTDQLDY (SEQ ID NO: 627); PFKDDPRDETYKPHLERETPKPRRKSG (SEQ ID NO: 630); QYVRCEMEGCGTVLAHPRYLQ HHIKYQHLLKKKYVCPHPSCGRLFRLQKQLLRHAKHHTD (SEQ ID NO: 629); or residues 151 - 182 of QRD YICEYCARAFKSSHNLAVHRMIHTGEKHY (SEQ ID NO: 628). Also preferred are polynucleotide fragments encoding these polypeptide fragments.
This gene is expressed primarily in Rhabdomyosarcoma, melanocyte and colon cancer tissue and to a lesser extent in smooth muscle, pancreatic tumor, and apoptotic T-cells. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions which include, but are not limited to,. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune and hemopoetic, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., striated muscle, melanocytes, colon, smooth muscle, pancreas, and blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for study, diagnosis and treatment of cancer and hemopoetic disorders.
FEATURES OF PROTEIN ENCODED BY GENE NO: 65
This gene is expressed primarily in human adipose and salivary gland tissue and to a lesser extent in human bone marrow and fetal kidney.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: metabolic and immune disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the metabolic and hemopoetic systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., adipose, salivary gland, bone marrow, and kidney, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for study, diagnosis of metabolic and immune disorders.
FEATURES OF PROTEIN ENCODED BY GENE NO: 66 This translated product of this gene was recently identified as oxytocinase splice variant 1. (See Accession Nos. 2209276 and dl010078.) Preferred polypeptide fragments comprise the amino acid sequence: EMFDSLSYFKGSSLLLMLKTYLSEDVFQHAVVLYLHN HSYASIQSDDLWDSFNEVTNQTLDVKRMMKTWTLQKGFPLVTVQKKGKELπQQERFFLNMK PEIQPSDTRYM (SEQ ID NO: 631). Also preferred are polynucleotide fragments encoding this polypeptide fragment.
FEATURES OF PROTEIN ENCODED BY GENE NO: 67 This gene is expressed primarily in hemopoetic cells, particularly apoptotic T- cells, and to lesser extent in primary dendritic cells and adipose tissue.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of apoptotic T-cells, primary denritic cells, and adipose tissue present in a biological sample and for diagnosis of diseases and conditions: hemopoetic diseases including cancer and general immune disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell
type(s). For a number of disorders of the above tissues or cells, particularly of the oral and intestinal mucosa as well as hemopoetic and immune systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., hematopoietic cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment of diseases of the immune system, including cancer, hemopoetic and infectious diseases.
FEATURES OF PROTEIN ENCODED BY GENE NO: 68
This gene is expressed primarily in kidney cortex and to a lesser extent in infant brain, heart, uterus, and blood.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of kidney tissue present in a biological sample and for diagnosis of diseases and conditions: soft tissue cancer, inflammation, kidney fibrosis. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the nervous and endocrines systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., kidney, brain, and other nervous tissue, heart, uterus, and blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for study and treatment of cancer and fibroses.
FEATURES OF PROTEIN ENCODED BY GENE NO: 69
The translation product of this gene shares strong sequence homology with vertebrate and invertebrate protein tyrosine phosphatases. This gene is expressed primarily in endometrial tumors, melanocytes, myeloid progenitors and to a lesser extent in infant brain, adipocytes, and several hematopoietic stem cells.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of transformed hematopoietic and epithelial cells present in a biological sample and for diagnosis of diseases and conditions which include, but are not limited to, of skin and endometrium, leukemia. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the nervous and hemopoietic systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., endometrium, melanocytes, bone marrow, adipocytes, hematopoietic cells, and brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and sequence similarity with tyrosine phosphatases indicate that polynucleotides and polypeptides corresponding to this gene are useful for study and treatment of cancer and hematopoietic disorders.
FEATURES OF PROTEIN ENCODED BY GENE NO: 70
This gene is expressed primarily in osteoclastoma, breast, and infant brain and to a lesser extent in various fetal and transformed bone, ovarian, and neuronal cells.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: degenerative conditions of the brain and skeleton. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the nervous and skeletal system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., bone, mammary tissue, and brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for study and treatment of degenerative, neurological and skeletal disorders.
FEATURES OF PROTEIN ENCODED BY GENE NO: 71
This gene was originally cloned from tumor cell lines. Recently another group has also cloned this gene, calling it the human malignant melanoma metastasis- suppressor (KiSS-1) gene. (See Accession No. U43527.) Preferred polypeptide fragments comprise the amino acid sequence: LEKVASVGNSRPTGQQLESLGLLA (SEQ ID NO: 632); VHREEASCYCQAEPSGDL (SEQ ID NO: 633); RPALRQAGGGTREPRQKRWAGL (SEQ ID NO: 634); and AVNFRPQRSQSM (SEQ ID NO: 635). Any frame shifts can easily be resolved using known molecular biology techniques.
This gene is expressed primarily in many types of carcinomas and to a lesser extent in many normal organs. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissues(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions which include, but are not limited to, cancer particularly melanomas, and other hyperproliferative disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of transformed organ tissue, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. As a tumor suppressor gene, increase amounts of the polypeptide can be used to treat patients having a particular cancer. The tissue distribution indicates that this gene and the translated product is useful for diagnosing and study of cancer.
FEATURES OF PROTEIN ENCODED BY GENE NO: 72
This gene is expressed primarily in striatum and to a lesser extent in adipocytes and hemangioperiocytoma. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of striatal cells present in a biological sample and for diagnosis of diseases and conditions: neurological, fat and lysosomal storage
diseases. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the nervous and immune systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., striatal tissue, adipocytes, and vascular tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis, study and treatment of neurodegenerative and growth disorders.
FEATURES OF PROTEIN ENCODED BY GENE NO: 73
This gene is expressed primarily in bone marrow stromal cells and to a lesser extent in smooth muscle, testes, endothelium, and brain.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of bone marrow present in a biological sample and for diagnosis of diseases and conditions: connective tissue and hematopoietic diseases. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the skeletal and hematopoietic systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., bone marrow, stromal cells, smooth muscle, testes and other reproductive tissue, endothelium, brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for study, diagnosis, and treatment of connective tissue and blood diseases.
FEATURES OF PROTEIN ENCODED BY GENE NO: 74
This gene is expressed primarily in brain, fetal liver and lung and to a lesser extent in retina, spinal chord, activated T-cells and endothelial cells.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of brain and regenerating liver present in a biological sample and for diagnosis of diseases and conditions: CNS and spinal chord injuries, immune disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the nervous and immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, liver, pulmonary tissue, blood cells, and endothelial cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for study and treatment of hematopoietic and neurological conditions.
FEATURES OF PROTEIN ENCODED BY GENE NO: 75
The translation product of this gene shares sequence homology with GTP binding proteins (intracellular). This gene is expressed primarily in bone marrow, brain, and melanocytes and to a lesser extent in various endocrine and hematopoietic tissues.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: hematopietic and nervous system conditions. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the nervous and immune, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., bone marrow, melanocytes, brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder,
relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to nucleotide binding factors indicate that polynucleotides and polypeptides corresponding to this gene are useful for study, diagnosis, and treatment of brain degenerative, skin and blood diseases.
FEATURES OF PROTEIN ENCODED BY GENE NO: 76
This gene is expressed primarily in activated T-cells and to a lesser extent in retina, brain, and fetal bone. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of activated T-cells and developing brain present in a biological sample and for diagnosis of diseases and conditions: immune deficiencies and skeletal and neuronal growth disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the nervous, immune, and skeletomuscular sustems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, brain and other tissue of the nervous system, retinal tissue, and bone, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis, study and treatment of cancer, urogenital, and brain degenerative diseases.
FEATURES OF PROTEIN ENCODED BY GENE NO: 77
This gene is expressed primarily in fetal liver, activated monocytes, osteoblasts and to a lesser extent in synovial, brain, and lymphoid tissues.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of myeloid and lymphoid present in a biological sample and for diagnosis of diseases and conditions: inflammation, immune deficiencies, cancer. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system and skeleton, expression of this gene at significantly
higher or lower levels may be routinely detected in certain tissues and cell types (e.g., liver, blood cells, bone, synovial tissue, brain and other tissue of the nervous system, and lymphoid tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for study, diagnosis, and treatment of lymphoid and mesenchymal cancers and nervous system diseases.
FEATURES OF PROTEIN ENCODED BY GENE NO: 78
The translation product of this gene shares sequence homology with polymerase polyprotein precursor which is thought to be important in DNA repair and replication This gene is expressed primarily in infant brain and to a lesser extent in tumors and tumor cell lines
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions which include, but are not limited to, especially of the neural system and developing organs. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the neural system expression of this gene at significantly higher or lower levels may be routinely detected in certain (e.g., brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution and homology to polymerase polyprotein precursor indicate that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of cancers especially of the neural system and developing organs
FEATURES OF PROTEIN ENCODED BY GENE NO: 79
This gene is expressed primarily in muscle and endothelial cells and to a lesser extent in brain.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: vascular diseases. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the vascular system, expression of this gene at significantly higher or lower levels may be routinely detected in certain (e.g., muscle, endothelial cells, brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment and diagnosis of disorders of the vascular and neural system including cardiovascular and endothelial.
FEATURES OF PROTEIN ENCODED BY GENE NO: 80
This gene is expressed primarily in placenta and to a lesser extent in fetal liver Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: developmental disorders and disorder of the haemopoietic system, fetal liver and placenta. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of developmental disorders and disorder of the haemopoietic system, fetal liver and placenta, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., placenta and liver, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of developmental disorders and disorders of the haemopoietic system, fetal liver and placenta.
FEATURES OF PROTEIN ENCODED BY GENE NO: 81
This gene is expressed primarily in bone marrow, placenta and tissues and organs of the hematopoietic system. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: disorders of the bone and haemopoietic system. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune, bone and hematopoietic system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., bone marrow, placenta, and hematopoietic cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of disorders of the immune, bone and hematopoietic system
FEATURES OF PROTEIN ENCODED BY GENE NO: 82
The translation product of this gene shares sequence homology with secretory carrier membrane protein which is thought to be important in protein transport and export. Any frame shifts in coding sequence can be easily resolved using standard molecular biology techniques. Another group recently cloned this gene, calling it SCAMP. (See Accession No. 2232243.)
This gene is expressed primarily in prostate, breast and spleen, and to a lesser extent in several other tissues and organs. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: disorders of the breast prostate and spleen. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly disorders of the breast prostate and spleen, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell
types (e.g., prostate, mammary tissue, and spleen, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to secretory carrier membrane protein indicate that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of disorders of the breast, prostate and spleen.
FEATURES OF PROTEIN ENCODED BY GENE NO: 83
This gene is expressed primarily in developing organs and tissue like placenta and infant brain and to a lesser extent in developed organs and tissue like cerebellum and heart.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: neurological diseases. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the neural system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., placenta, heart, brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment and diagnosis of diseases of the neural system including neurological disorders and cancer.
FEATURES OF PROTEIN ENCODED BY GENE NO: 84
The translation product of this gene shares sequence homology with ATPase 6 in Trypanosoma brucei which is thought to be important in metabolism.
This gene is expressed primarily in tumor and fetal tissues and to a lesser extent in melanocytes, kidney cortex, monocytes and ovary.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a
biological sample and for diagnosis of diseases and conditions: metabolism disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the fetal systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., fetal tissues, melanocytes, kidney, blood cells, ovary and other tissue of the reproductive system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to ATPase indicate that polynucleotides and polypeptides corresponding to this gene are useful for treatment and diagnosis of metabolism disorders, especially in fetal and tumor tissue growth.
FEATURES OF PROTEIN ENCODED BY GENE NO: 85
The translation product of this gene shares sequence homology with the immunoglobulin superfamily of proteins which are known to be important in immune response and immunity. This gene is expressed primarily in stromal cells, colon cancer, lung, amygdala, melanocyte and to a lesser extent in a variety of other tissues.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: defects of stromal cell development and cancer. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the stromal cells, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., stromal cells, colon, lung, amygdala, and melanocytes, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution and homology to immunoglobulin indicate that polynucleotides and polypeptides corresponding to this gene are useful for treatment and diagnosis of immune system disorders.
FEATURES OF PROTEIN ENCODED BY GENE NO: 86
The translation product of this gene shares sequence homology with transcription iniation factor eIF-4 gamma which is thought to be important in gene transcription.
This gene is expressed primarily in tumor tissues.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: tumorigenesis. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly in tumor tissues, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., endometrium and lung, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to transcription iniation factor eIF-4 gamma indicate that polynucleotides and polypeptides corresponding to this gene are useful for gene regulation in tumorigenesis.
FEATURES OF PROTEIN ENCODED BY GENE NO: 87
The translation product of this gene shares sequence homology at low level in prolines with secreted basic proline-rich peptide II-2 which is thought to be important in protein structure or inhibiting hydroxyapatite formation in vitro.
This gene is expressed primarily in endometrial tumor and fetal lung. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: endometrial tumors. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the muscular/skeletal and reproductive systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., endometrium, and lung, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample
taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to secreted basic proline-rich peptide II-2 indicate that polynucleotides and polypeptides corresponding to this gene are useful for inhibiting hydroxyapatite formation or establishing cell/tissue structure.
FEATURES OF PROTEIN ENCODED BY GENE NO: 88
This gene is expressed primarily in: amniotic cells inducted with TNF in culture; and to a lesser extent in colon tissue from a patient with Crohn's Disease; parathyroid tumor; activated T-cells; cells of the human Caco-2 cell line; adenocarcinoma; colon; corpus colosum; fetal kidney; pancreas tumor; fetal brain; early stage brain, and anergic T-cells.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: tumors. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system; e.g., tumors, expression of this gene at significantly higher or lower levels may be routinely detected in certain (e.g., amniotic cells, colon, kidney, pancreas, parathyroid, brain and other tissue of the nervous system, blood cells, hematopoietic cells, liver, spleen, bone, testes and other reproductive tissue, brain and other tissue of the nervous system, and epithelial cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that the protein product of this clone is useful for modulating tumorigenesis and other immune system conditions such as disorders in immune response.
FEATURES OF PROTEIN ENCODED BY GENE NO: 89
This gene is expressed primarily in fetal liver/spleen and hematopoietic cells and to a lesser extent in brain, osteosarcoma, and testis tumor.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a
biological sample and for diagnosis of diseases and conditions: leukemia and hematopoietic disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the hematopoietic and immune systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., hematopoietic cells, liver, spleen, bone, testes, and other reproductive tissue, brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of hematopoietic and immune disorders.
FEATURES OF PROTEIN ENCODED BY GENE NO: 90
The translation product of this gene shares weak sequence homology with mouse Gcap 1 protein which is developmentally regulated in brain. This gene is expressed primarily in infant and adult brain and fetal liver/spleen and to a lesser extent in smooth muscle, T cells, and a variety of other tissues.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: neurological or hematopoietic disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the nervous, hematopoietic, immune, and endocrine systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, blood cells, liver, spleen ,and smooth muscle, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and its homology to Gcapl protein indicate that polynucleotides and polypeptides corresponding to this gene are useful for treatubg and diagnosis of disorders in neuronal, hematopoietic, immune, and endocrine systems.
FEATURES OF PROTEIN ENCODED BY GENE NO: 91
This gene is expressed primarily in brain and hematopoietic cells and to a lesser extent in tumor tissues.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: disorder in nervous, hematopoietic, immune systems and tumorigenesis. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the in nervous, hematopoietic, immune systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that the protein product of this clone is useful for diagnosis and treatment of disorders in the nervous, hematopoietic, and immune systems.
FEATURES OF PROTEIN ENCODED BY GENE NO: 92
The translation product of this gene shares sequence homology with neuroendocrine-specific protein A which is thought to be important in neurologic systems. This gene is expressed primarily in brain tissues.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: neural disorders and degeneration disease. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the central or peripheral nervous systems, expression of this gene at
significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., hematopoietic cells, and brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to neuroendocrine-specific protein A indicate that polynucleotides and polypeptides corresponding to this gene are useful for treatment or diagnosis of neural disorders and degeneration disease.
FEATURES OF PROTEIN ENCODED BY GENE NO: 93
The translation product of this gene shares sequence homology with collagenlike protein and prolin-rich protein which are thought to be important in connective tissue function and tissue structure. This gene is expressed primarily in fetal liver/spleen and brain tissues.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: neuronal or hematopoietic disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the nervous and hematopoietic systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., liver, spleen, and brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to collagen-like protein and proline-rich proteins indicate that polynucleotides and polypeptides corresponding to this gene are useful for supporting brain and hematopoietic tissue function and diagnosis and treatment of disorders in these functions.
FEATURES OF PROTEIN ENCODED BY GENE NO: 94 This gene is expressed primarily in embryonic tissues and tumor tissues.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a
biological sample and for diagnosis of diseases and conditions which include, but are not limited to,. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system (e.g., tumors), expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., embryonic tissue and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of cancer.
FEATURES OF PROTEIN ENCODED BY GENE NO: 95
This gene is expressed primarily in brain tumor, placenta, and melanoma. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: brain tumor or melanoma. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the brain or melanocytes, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, placenta, and melanocytes, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution indicates that the translation product of this gene is useful in the diagnosis and treatment of brain tumors and melanoma.
FEATURES OF PROTEIN ENCODED BY GENE NO: 96
The translation product of this gene shares sequence homology with a yeast membrane protein, SUR4, which encodes for APAl that acts on a glucose-signaling pathway that controls the expression of several genes that are transcriptionally regulated by glucose.
This gene is expressed primarily in fetal liver, and to a lesser extent in placenta and breast tissue.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: defects of fetal liver or defects of glucose-regulated ATPase activities in tissues. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the fetal immune/hematopoietic system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., liver, placenta, and mammary tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to yeast SUR4 membrane protein indicate that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of defects of fetal liver or defects of glucose-regulated ATPase activities.
FEATURES OF PROTEIN ENCODED BY GENE NO: 97
This gene is expressed primarily in fetal liver, brain, and amniotic fluid. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: defects of the fetal immune system and adult brain. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the fetal immune system and adult brain, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., liver, and brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., amniotic fluid, serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that the protein product of this clone is useful for detecting defects of the fetal immune and hematopoietic systems since fetal liver is
the predominant organ responsible for hematopoiesis in the fetus. In addition, the gene product of this gene is thought to be useful for detecting certain neurological defects of the brain.
FEATURES OF PROTEIN ENCODED BY GENE NO: 98
The translation product of this gene shares sequence homology with an yolk protein precursor, Vitellogenin which is thought to be important in binding lipids such as phosvitin.
This gene is expressed primarily in amniotic cells and fetal liver. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: defects in amniotic cells, fetal liver development and the fetal immune system. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the [insert system where a related disease state is likely, e.g., immune], expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., amniotic cells, and liver, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to vitellogenin indicate that the protein product of this clone is useful for treatment and diagnosis of defects in amniotic cells, fetal liver development and the fetal immune system.
FEATURES OF PROTEIN ENCODED BY GENE NO: 99
This gene is expressed primarily in placenta, endometrial tumor, osteosarcoma and stromal cells.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: tumor of the endometrium or bone, and osteosarcoma. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the obstetric system (e.g. placenta,
endometrium) and the bones, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., placenta, endometrium, bone, and stromal cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of tumors and abnormalities of the endometrium, and the bones because of its abundance in the aforementioned tissues..
FEATURES OF PROTEIN ENCODED BY GENE NO: 100
This gene is expressed primarily in hepatocellular tumor. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: hepatocellular tumor. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the liver, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., liver, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that the protein product of this clone is useful for diagnosis and treatment of hepatocellular cancer because of its abundant expression in this tissue.
FEATURES OF PROTEIN ENCODED BY GENE NO: 101
This gene is expressed primarily in Corpus Colosum, fetal lung and infant brain.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: defects of the Corpus Colosum or defects of the fetal lung. Similarly, polypeptides and antibodies directed to
these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the Corpus Colosum and brain in general, and fetal lung, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., lung, and brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that the protein product of this clone is useful for diagnosis and treatment of defects of the Corpus Colosum and brain in general, and defects of fetal lung.
FEATURES OF PROTEIN ENCODED BY GENE NO: 102
This gene is expressed primarily in T cells and stromal cells, and to a lesser extent in adrenal gland.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: defects of T cell immunity and stromal cell development. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, stromal cells, and adrenal gland, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that the protein product of this clone is useful for diagnosis and treatment of defects of T cell immunity and stromal cell development because of its abundant expression in these tissues.
FEATURES OF PROTEIN ENCODED BY GENE NO: 103
This gene is expressed primarily in infant brain and placenta.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: defects of the brain and nervous system. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the nervous system, especially brain, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, and placenta, cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that the protein product of this clone is useful for detecting defects of the brain, especially in young children.
FEATURES OF PROTEIN ENCODED BY GENE NO: 105
This gene is expressed primarily in human osteoclastoma and to a lesser extent in human pancreas tumor. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions which include, but are not limited to, cancer particularly osteoclastoma and pancreatic tumor. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly in transformed tissues, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., bone and pancreas, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that the protein product of this clone is useful for diagnosis and treatment of some types of tumors, particularly pancreatic cancer and osteoclastoma.
FEATURES OF PROTEIN ENCODED BY GENE NO: 106
This gene is expressed primarily in fetal liver/spleen, and to a lesser extent in activated T-Cells.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: immune disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., liver, spleen, and blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis or treatment of immune disorders.
FEATURES OF PROTEIN ENCODED BY GENE NO: 107
This gene is expressed primarily in human embryo and to a lesser extent in spleen and chronic lymphocytic leukemia.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: leukemia. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune or hemopoietic systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., embryonic tissue, spleen, and blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that the protein product of this clone is useful for the diagnosis and treatment of leukemia.
FEATURES OF PROTEIN ENCODED BY GENE NO: 108
This gene is expressed primarily in placenta, and to a lesser extent in early stage human brain and in lung. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: fetal developmental abnormalities. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly in fetal and amniotic tissue, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., placenta, brain and other tissue of the nervous system, and lung, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that the protein product of this is useful for production of growth factor(s) associated with fetal development. Preferred polypeptides comprise the full-length polypeptide shown in the sequence listing, truncated however, at the amino terminus and beginning with QTIE.
FEATURES OF PROTEIN ENCODED BY GENE NO: 109
This gene is expressed primarily in fetal spleen, and to a lesser extent in B-Cell lymphoma and T-Cell lymphoma.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: lymphoma. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., spleen and blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that the protein product of this clone is useful for the treatment and diagnosis of human lymphomas.
FEATURES OF PROTEIN ENCODED BY GENE NO: 110 The translation product of this gene shares sequence homology with sarcoma amplified sequence (SAS), a tetraspan receptor which is thought to be important in malignant fibrous histiocytoma and liposarcoma.
This gene is expressed primarily in human osteoclastoma, and to a lesser extent in pineal gland and infant brain. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: malignant fibrous histiocytoma and liposarcoma. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., bone, pineal gland, and brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to sarcoma amplified sequence (SAS) indicate that the protein product of this clone is useful for treatment of, osteosarcoma, malignant fibrous histiocytoma and liposarcoma and related cancers, particularly sarcomas.
FEATURES OF PROTEIN ENCODED BY GENE NO: 111
The translation product of this gene shares sequence homology with 6.8K proteolipid protein, mitochondrial - bovine.
This gene is expressed primarily in Wilm's tumor and to a lesser extent in cerebellum and placenta.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: Wilm's tumor.
Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell
type(s). For a number of disorders of the above tissues or cells, particularly of the immune or renal systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, and placenta, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to 6.8K proteolipid protein indicate that the protein product of this clone is useful for diagnostic and therapeutics associated with tumors, particularly Wilm's tumor disease.
FEATURES OF PROTEIN ENCODED BY GENE NO: 112
This gene is expressed primarily in embryonic tissue and to a lesser extent in osteoblasts, endothelial cells, macrophages (GM-CSF treated), and bone marrow. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: immune disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., embryonic tissue, bone, endothelial cells, blood cells and bone marrow, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment or diagnosis of immune disorders. Preferred polypeptides encoded by this gene comprise the following amino acid sequence: MITDVQLAIFANMLGVSLFLLVVLYHYVAVNNPKKQE (SEQ ID NO: 636).
FEATURES OF PROTEIN ENCODED BY GENE NO: 113 This gene is expressed primarily in hepatocellular tumor, and to a lesser extent in fetal liver/spleen.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: tumors, particularly hepatocellular tumors. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the hepatic system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., liver, and spleen, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that the protein product of this clone is useful for diagnosis and treatment of tumors, particularly hepatocellular tumors.
FEATURES OF PROTEIN ENCODED BY GENE NO: 114
The translation product of this gene exhibits a very high degree of sequence identity with the human Pig8 gene which is thought to be important in p53 mediated apoptosis. The sequence of this gene has since been published by Polyak and colleagues (Nature 389, 300-306 (1997)). In addition, the predicted translation product of this contig exhibits very high sequence homology with a murine gene denoted as EI24 which is also thought to be important in p53 mediated apoptosis.
This gene is expressed primarily in infant brain and activated T-cells and to a lesser extent in bone marrow, fetal liver, and prostate.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions which include, but are not limited to, and tissue damage by radiation and anti-cancer drugs. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the nervous and immune systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, blood cells, bone marrow, liver, and prostate, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder,
relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to human Pig8 and murine EI24 genes indicate that polynucleotides and polypeptides corresponding to this gene are useful for preventing apoptosis in patients being treated with anti-oncogenic drugs such as etoposide, hydroperoxycyclophosphamide, and X-irradiation, since this protein product is upregulated in cells undergoing such treatment where p53 was overexpressed. It may also be useful in the treatment of hematopoietic disorders and in boosting numbers of hematopoietic stem cells by interfering with the apoptosis of progenitor cells. The mature polypeptide is predicted to comprise the following amino acid sequence:
EEMADSVKTFLQDLARGIKDSIWGICΗSKLDARIQQKREEQRRRRASSVLAQRRAQSIERKQES EPRIVSRIFQCCAWNGGVFWFSLLLFYRVFIPVLQSVTARIIGDPSLHGDVWSWLEFFLTSIFSA LVv^LPLFVLSKVVNAIWFQDIADLAFEVSGRKPHPFPSVSKIIADMLFNLLLQALFLIQGMFVSL FPIHLVGQLVSLLHMSLLYSLYCFEYRWFNKGIEMHQRLSNIERNWPYYFGFGLPLAFLTAMQ SS YIISGCLFSILFPLFIIS ANEAKTPGKAYLFQLRLFSLVVFLSNRLFHKTVYLQSALSSSTSAEK FPSPHPSPAKLKATAGH (SEQ ID NO: 637). Accordingly, polypeptides comprising the foregoing amino acid sequence are provided as are polynucleotides encoded such polypeptides.
FEATURES OF PROTEIN ENCODED BY GENE NO: 115
This gene is expressed primarily in stromal cells and to a lesser extent in multiple sclerosis.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: affecting the nervous system. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the nervous system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., stromal cells and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment and diagnosis of multiple sclerosis and other autoimmune diseases.
FEATURES OF PROTEIN ENCODED BY GENE NO: 116
This gene is expressed primarily in the gall bladder Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: gall stones or infection of the digestive system . Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the digestive system or renal system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., gall bladder and tissue of the digestive system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for possible prevention of digestive disorders where there may be a lack of digestive enzymes produced or in the detection and possible prevention of gall stones.
FEATURES OF PROTEIN ENCODED BY GENE NO: 117
The translation product of this gene shares sequence homology with dystrophin gene which is thought to be important in building and maintenance of muscles. This gene is expressed primarily in placenta and to a lesser extent in fetal brain and fetal liver, and spleen.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: muscular dystropy, Duchenne and Becker's muscular dystropies. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the skeletal muscle system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., placenta, brain and other tissue of the nervous system, muscle, liver, and spleen, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from
an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to the dystrophin gene indicate that polynucleotides and polypeptides corresponding to this gene are useful for diseases related the degenerative myopathies that are characterized by the weakness and atrophy of muscles without neural degradation; such as Duchenne and Becker's muscular dystropies.
FEATURES OF PROTEIN ENCODED BY GENE NO: 118
This gene is expressed primarily in olfactory tissue and to a lesser extent in cartilage.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: connective tissue diseases; chondrosarcoma. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the connective tissue, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., olfactory tissue and cartilage, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for tumors of connective tissues, osteoarthritis and the treatment and diagnosis of chondrosarcoma.
FEATURES OF PROTEIN ENCODED BY GENE NO: 119
This gene is expressed primarily in Activated Neutrophils and to a lesser extent in fetal spleen, and CD34 positive cells from cord blood.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: allergies, defects in hematopoiesis and inflammation. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential
identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system and hematopoiesis system the, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, and spleen, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for reducing the allergic effects felt by allergy suffers by neutralizing the activity of the immune system, especially since neutrophils are abundant in persons suffering from allergies and other inflammatory conditions.
FEATURES OF PROTEIN ENCODED BY GENE NO: 120 The translation product of this gene shares sequence homology with poly A binding protein II which is thought to be important in RNA binding for transcription of RNA to DNA
This gene is expressed primarily in colon and to a lesser extent in brain and immune system. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: colon cancer. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune and digestive system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., colon, tissue and cells of the immune system, and brain or other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to poly A binding protein II indicate that polynucleotides and polypeptides corresponding to this gene are useful for detection and treatment of colon cancer and other disorders of the digestive system..
FEATURES OF PROTEIN ENCODED BY GENE NO: 121
The translation product of this gene shares sequence homology with thymidine diphosphoglucose 4.6 dehydrase which is thought to be important in the metabolism of sugar. This gene is expressed primarily in fetal liver and spleen and to a lesser extent in infant brain.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: diabetes. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the endocrine system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., liver, spleen, and brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution and homology to thymidine diphospoglucose 4.6 dehydrase indicate that polynucleotides and polypeptides corresponding to this gene are useful for treatment of persons with diabetes since it appears that this protein is needed in the metabolism of sugar in to its more basic components.
FEATURES OF PROTEIN ENCODED BY GENE NO: 122
The translation product of this gene shares sequence homology with ceruloplasmin which is thought to be important in the metabolism and transport of iron and copper. Ceruloplasmin also contains domains with homology to clotting factors V and Vπi. Defects in the circulating levels of ceruloplasmin (aceruloplasminemia) have been associated with certain disease conditions such as Wilson disease, and the accompanying hepatolenticular degeneration.
This gene is expressed primarily in brain and retina and to a lesser extent in endothelial cells.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: diseases marked by defects in iron metabolism; aceruloplasminemia not characterized by defects in the
known ceruloplasmin gene locus; nonclassical Wilson disease; movement disorders; and tumors derived from a brain tissue origin. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the brain, retina, and nervous system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, retinal tissue, and endothelial cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to ceruloplasmin indicate that polynucleotides and polypeptides corresponding to this gene are useful for treatment of patients with aceruloplasminemia, or other defects in iron and/or copper metabolism. Mutations in this locus could also be diagnostic for patients currently experiencing or predicted to experience aceruloplasminemia.
FEATURES OF PROTEIN ENCODED BY GENE NO: 123 This gene is expressed primarily in brain and B cell lymphoma and to a lesser extent in fetal liver and spleen.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: B cell lymphoma; tumors and diseases of the brain and/or spleen; hematopoietic defects. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the brain and hematopoietic system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, blood cells, liver, and spleen, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment of disorders in neuronal,
hematopoietic, and immune systems. It could potentially be useful for neurodegenerative disorders and neuronal and/or hematopoietic cell survival or proliferation.
FEATURES OF PROTEIN ENCODED BY GENE NO: 124
This gene is expressed primarily in osteoclastoma, dermatofibrosarcoma, and B cell lymphoma and to a lesser extent in endothelial cells.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions which include, but are not limited to, cancer in particular osteoclastoma, dermatofibrosarcoma, and B cell lymphoma. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the bone, immune, and circulatory system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., bone, epidermis, blood cells, and endothelial cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of cancers and lymphoma; osteoporosis; and the control of cell proliferation and/or differentiation.
FEATURES OF PROTEIN ENCODED BY GENE NO: 125
This gene is expressed primarily in immune tissues and hematopoietic cells, particularly in activated T cells and neutrophils, spleen, and fetal liver, and to a lesser extent in infant adrenal gland. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: defects in T cell activation; hematopoietic disorders; tumors of a hematopoietic and/or adrenal gland origin. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the hematopoietic and/or endocrine systems, expression of this gene at significantly higher
or lower levels may be routinely detected in certain tissues and cell types (e.g., cells and tissues of the immune system, hematopoietic cells, blood cells, liver, and adrenal gland, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for immune and/or hematopoietic disorders; diseases related to proliferation and/or differentiation of hematopoietic cells; defects in T cell and neutrophil activation and responsiveness; and endocrine and/or metabolic disorders, particularly of early childhood.
FEATURES OF PROTEIN ENCODED BY GENE NO: 126 This gene is expressed primarily in placenta and endothelial cells and to a lesser extent in melanocytes and embryonic tissues.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: tumors of an endothelial cell origin; angiogenesis associated with tumor development and metastasis. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the vascular system and developing embryo, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., placenta, endothelial cells, melanocytes, and embryonic tissues, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment of developmental disorders; inhibition of angiogenesis; and vascular patterning.
FEATURES OF PROTEIN ENCODED BY GENE NO: 127
This gene is expressed primarily in endothelial cells and hematopoietic tissues, including spleen, tonsils, leukocytes, and both B- and T-cell lymphomas.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: tumors of an endothelial cell and/or hematopoietic origin; leukemias and lymphomas. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune and vascular systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., endothelial cells, hematopoietic cells, spleen, tonsils, and blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for the manipulation of angiogenesis; the differentiation and morphogenesis of endothelial cells; the proliferation and/or differentiation of hematopoietic cells; and the commitment of hematopoietic cells to distinct cell lineages.
FEATURES OF PROTEIN ENCODED BY GENE NO: 128
This gene is expressed primarily in kidney medulla and to a lesser extent in spleen from chronic myelogenous leukemia patients, prostate cancer, and some other tissues. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: tumors of a kidney origin; chromic myelogenous leukemia; prostate cancer. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the kidney and spleen, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., kidney, spleen, and prostate, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for the diagnosis and treatment of kidney disorders and cancer, particularly chromic myelogenous leukemia and prostate cancer. It may also be useful for the enhancement of kidney tubule regeneration in the treatment of acute renal failure.
FEATURES OF PROTEIN ENCODED BY GENE NO: 129
This gene is expressed primarily in adult and infant brain and to a lesser extent in mesenchymal or fibroblast cells, as well as tissues with a mesenchymal origin. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: tumors of a brain and/or mesenchymal origin; neurodegenerative disorders; cancer; fibrosis. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the brain and of mesenchymal cells and tissues, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for the diagnosis of tumors of a brain and/or mesenchymal origin; neurodegenerative disorders; cancer; and fibrosis, based upon the expression of this gene within those tissues. Fibrosis is considered as mesenchymal cells and fibroblasts are the primary cellular targets involved in this pathological condition.
FEATURES OF PROTEIN ENCODED BY GENE NO: 130
This gene is expressed primarily in hepatocellular cancer and to a lesser extent in fetal tissues as well as testes tumor.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: liver cancer. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing
immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the digestive system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., liver, fetal tissue, and testes and other reproductive tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of liver cancer.
FEATURES OF PROTEIN ENCODED BY GENE NO: 131
This gene is expressed only in infant early brain. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: development and diseases of the nervous system. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the brain and nervous system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treating diseases of the brain in children and in treating nervous system disorders such as Alzheimer's disease, schizophrenia, dementia, depression, etc.
FEATURES OF PROTEIN ENCODED BY GENE NO: 132
This gene is expressed primarily in brain and to a lesser extent in glioblastoma. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: Alzheimer's disease,
schizophrenia, depression, mania, and dementia. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the brain and nervous system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treating brain disorders such as Alzheimer's disease, schizophrenia, depression, mania, and dementia.
FEATURES OF PROTEIN ENCODED BY GENE NO: 133
The translation product of this gene shares sequence homology with ribitol dehydrogenase of bacteria which is thought to be important in metabolism of sugars. This gene is expressed primarily in macrophage and to a lesser extent in T-cell lymphoma and lung. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: tissue destruction in inflammation. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells and lung, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to ribitol dehydrogenase indicate that polynucleotides and polypeptides corresponding to this gene are useful for altering macrophage metabolism in diseases such as inflammation where macrophages are causing excess tissue destruction.
FEATURES OF PROTEIN ENCODED BY GENE NO: 134
This gene is expressed primarily in pancreatic tumor and to a lesser extent in synovial sarcoma.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions which include, but are not limited to,. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the endocrine and connective tissue systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., pancreas, and synovial tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treating and diagnosing various cancers.
FEATURES OF PROTEIN ENCODED BY GENE NO: 135
This gene is expressed primarily in T cell lines such as Raji and to a lesser extent in infant brain.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: immune system disorders and inflammation. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, and brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treating and diagnosing inflammatory diseases
such as rheumatoid arthritis, sepsis, inflammatory bowel disease, and psoriasis, as well as neutropenia.
FEATURES OF PROTEIN ENCODED BY GENE NO: 136 The translation product of this gene shares high sequence homology with SARI subfamily of GTP-binding proteins which is thought to be important in vesicular transport in mammalian cells.
This gene is expressed primarily in serum-stimulated smooth muscle cells and to a lesser extent in a T-cell lymphoma. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: diseases affecting vesicular transport. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the muscular system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, and smooth muscle, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to GTP-binding proteins indicate that polynucleotides and polypeptides corresponding to this gene are useful for gene therapy in treating the large number of diseases involved in defective vesicular transport within cells..
FEATURES OF PROTEIN ENCODED BY GENE NO: 137
The translation product of this gene shares sequence homology with a protein found in C. elegans cosmid F25B5.
This gene is expressed primarily in a fetal tissues and to a lesser extent in melanocytes.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: abnormal fetal development, especially of the pulmonary system. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes
for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the fetal pulmonary system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., fetal tissue, pulmonary tissue, and melanocytes, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment and diagnosis of diseases affecting the pulmonary system, such as emphysema.
FEATURES OF PROTEIN ENCODED BY GENE NO: 138
This gene is expressed primarily in gall bladder and to a lesser extent in smooth muscle.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: digestive system disease and gall bladder problems. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the digestive system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., gall bladder and tissue of the digestive system, and smooth muscle, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treating diseases of the digestive system.
FEATURES OF PROTEIN ENCODED BY GENE NO: 139
This gene is expressed primarily in placenta and to a lesser extent in brain.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: abnormal fetal development. Similarly, polypeptides and antibodies directed to these polypeptides are
useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of developing tissues, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., placenta, and brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treating and diagnosing abnormal fetal development.
FEATURES OF PROTEIN ENCODED BY GENE NO: 140 This gene is expressed primarily in smooth muscle and to a lesser extent in ovary, prostate cancer, and activated monocytes.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: hypertension and atherosclerosis. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the circulatory system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., smooth muscle, ovary and other reproductive tissue, prostate, and blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treating diseases of the circulatory system, such as hypertension, atherosclerosis, etc.
FEATURES OF PROTEIN ENCODED BY GENE NO: 141 This gene is expressed primarily in fetal spleen and to a lesser extent in placenta and bone marrow.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: anemia and other diseases affecting blood cells. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the circulatory and pulmonary systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., spleen, placenta, bone marrow, and blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for the generation of red and white blood cells and for the diagnosis of disease of these cells.
FEATURES OF PROTEIN ENCODED BY GENE NO: 142
The predicted translation product of this contig is a human homolog of the murine tetracycline/sugar transporter molecule recently reported by Matsuo and colleagues (Biochem. Biophys. Res. Commun. 238 (1), 126-129 (1997)).
This gene is expressed primarily in synovium and to a lesser extent in endothelial cells.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: rheumatoid arthritis and inflammation. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune and lymphatic systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., synovial tissue, and endothelial cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment and diagnosis of inflammatory diseases, such as rheumatoid arthritis, leukemia, neutropenia, inflammatory bowel disease, psoriasis, sepsis, and the like.
FEATURES OF PROTEIN ENCODED BY GENE NO: 143
This gene is expressed primarily in placenta and to a lesser extent in melanocyte, fetal liver and spleen, and bone marrow.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: abnormal early development. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, lower levels may be routinely detected in certain tissues and cell types (e.g., placenta, melanocytes, liver, spleen, and bone marrow, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for the treatment and diagnosis of abnormal early development phenomena and diseases.
FEATURES OF PROTEIN ENCODED BY GENE NO: 144
This gene is expressed primarily in fetal liver and spleen. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: anemia and neutropenia. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune and blood systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., liver and spleen, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the
expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful in hematopoeisis and bone marrow regeneration as it is most abundant in fetal tissues responsible for the generation of hematopoeitic cells.
FEATURES OF PROTEIN ENCODED BY GENE NO: 145
The translation product of this gene shares sequence homology with protein tyrosine phosphatase which is thought to be important in transducing signal to activate cells such as T cell, B cell and other cell types.
This gene is expressed primarily in T cells and tissues in early stages of development and to a lesser extent in cancers.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: immuno-related diseases and cancer. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., embryonic and fetal tissue, undifferentiated cells, and blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to the protein tyrosine phosphatase family indicate that polynucleotides and polypeptides corresponding to this gene are useful for modulating the immune system.
FEATURES OF PROTEIN ENCODED BY GENE NO: 146
This gene is expressed primarily in T cell and to a lesser extent in B cell, macrophages and tumor tissues.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: immuno-disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in
providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for regulating the immune system therefore can be used in treating diseases such as autoimmune diseases and cancers.
FEATURES OF PROTEIN ENCODED BY GENE NO: 147
This gene is expressed primarily in placenta and to a lesser extent in endothelial cells, testis tumor, ovarian cancer, uterine cancer.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions which include, but are not limited to cancer. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., placenta, endothelial cells, testis and ovary and other reproductive tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of cancers.
FEATURES OF PROTEIN ENCODED BY GENE NO: 148
This sequence has significant homology to mouse torsin A. Recently, another group cloned the human Torsin A gene. (See, Accession No. 2358279; see also Nature Genet. 17, 40-48 (1997).)
This gene is expressed primarily in osteoclastoma, T-cell, and placenta and to a lesser extent in fetal lung, fetal liver, fetal brain, adult brain and tumor tissues
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: disease conditions in hematopoiesis and cancers. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the hematopoiesis system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, bone, placenta, lung, liver, and brain and other tissues of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treating blood related diseases such as deficiencies in red blood cell, white blood cell, platelet and other hematopoiesis cells.
FEATURES OF PROTEIN ENCODED BY GENE NO: 149 This gene is expressed primarily in T cell, prostate and prostate cancer, endothelial cells and to a lesser extent in monocyte, dendritic cell, bone marrow, salivary gland, colon cancer, stomach cancer, pancreatic tumor, uterine cancer, fetal spleen and osteoclastoma.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: immuno-related diseases and cancers. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, prostate, endothelial cells, dendritic cells, bone marrow, salivary gland, colon, stomach, pancreas, uterus, spleen and bone, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment of cancers.
FEATURES OF PROTEIN ENCODED BY GENE NO: 150 This gene was recently cloned by another group, calling it eEF3-p66. (See
Accession No. 2351378.) This gene plays a role in RNA binding and macromolecular assembly, and therefore, any mutations in this gene would likely result in a diseased phenotype. Preferred polypeptide fragments comprise the amino acid sequence: MAKFMTPVIQDNPSGWGPCAVPEQFRDMPYQPFSKGDRLGKVADWTGATYQDKRYTNKYSS QFGGGSQYAYFHEEDESSFQLVDTARTQKTAYQRNRMRFAQRNLRRDKDRRNMLQFNLQILP KSAKQKERERIRLQKKFQKQFGVRQKWDQKSQKPRDSSVEVRSDWEVKEEMDFPQLMKMRY LEVSEPQDIECCGALEYYDKAI^RITTRSEKPLRXXKRIFHTVTTTDDPVIRKLAKTQGNVFATD AILATLMSCTRSVYSWDIVVQRVGSKLFFDKRDNSDFDLLTVSETANEPPQDEGNSFNSPRNL AMEATYINHNFSQQCLRMGKERYNFPNPNPFVEDDMDKNEIASVAYRYRSGKLGDDIDLΓVRC EHDGVMTGANGEVSFIMKTLNEWDSRHCNGVDVVTIQKLDSQRGAVIATELKNNS YKLARWTC CALLAGSEYLKLGYVSRYHVKDSSRHVILGTQQFKPNEFASQINLSVENAWGILRCVIDICMKL EEGKYLILKDPNKQVIRVYSLPDGTFSS (SEQ ID NO: 638), as well as N-terminal and C- terminal deletions of this polypeptide fragment.
This gene is expressed primarily in T cell, bone marrow, embryo and endothelial cells and to a lesser extent in testis tumor and endometrial tumor.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: immune diseases and tumors. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system and reproductive system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for immune disorders and cancers.
FEATURES OF PROTEIN ENCODED BY GENE NO: 151
This gene is expressed primarily in testis and to a lesser extent in T cell, spinal cord, placenta, neutrophil and monocyte.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: male reproductive and endocrine disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the reproductive, immune and endocrine systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., testis and other reproductive tissue, blood cells, tissue of the nervous system, and placenta, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for regulating immune and reproductive functions.
FEATURES OF PROTEIN ENCODED BY GENE NO: 152
The translation product of this gene shares sequence homology with tyrosyl- tRNA synthetase which is thought to be important in cell growth.
This gene is expressed primarily in brain, liver, keratinocytes, tonsils, and heart.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions which include, but are not limited to, cancer autoimmune diseases. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the brain, liver, keratinocytes, tonsils, heart expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissues of the nervous system, liver, keratinocytes, tonsils and heart, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard
gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to tyrosyl-tRNA synthetase indicate that polynucleotides and polypeptides corresponding to this gene are useful for modulating cell growth.
FEATURES OF PROTEIN ENCODED BY GENE NO: 153
This gene is homologous to the Drosophila transcriptional regulator dre4. (See Accession No. 2511745.) Dre4 is a gene required for steroidogenesis in Drosophila melanogaster and encodes a developmentally expressed homologue of the yeast transcriptional regulator CDC68. Preferred polypeptide fragments comprise the amino acid sequence: KKRHTDVQF TEVGEITTDLGKHQHMHDRDDLYAEQMEREMRHKLKTAFKN FIEKVEALTKEELEFEVPFRDLGFNGAPYRSTCLLQPTSSALVNATEWPPFVVTLDEVELIHFXR VQFHLKNFDMVIVYKDYSKKVTM AJPVASLDPIKEWLNSCDLKYTEGVQSLNWTKIMKTlVD DPEGFFEQGGWSFL (SEQ ID NO: 639), as well as N-terminal and C-terminal deletions of this fragments. Also preferred are polynucleotide fragments encoding this polypeptide fragment.
This gene is expressed primarily in fetal liver, spleen, placenta, lung, T cell, thyroid, testes. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: brain tumor, heart and liver diseases. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the fetal liver, spleen, placenta, lung, T cell, thyroid, testes expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., liver, spleen, placenta, lung, blood cells, thyroid, and testes and other reproductive tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
FEATURES OF PROTEIN ENCODED BY GENE NO: 154
This gene is expressed primarily in brain and to a lesser extent in fetal heart, testis, spleen, lung.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: heart, liver and spleen diseases, immunological diseases. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the brain, fetal heart, testis, spleen, lung expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, heart, testes and other reproductive tissue, spleen, and lung, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
FEATURES OF PROTEIN ENCODED BY GENE NO: 155
Activation of T cells through the T cell antigen receptor (TCR) results in the rapid tyrosine phosphorylation of a number of cellular proteins, one of the earliest being a 100 kDa protein. This gene is the human equivalent of murine valosin containing protein (VCP). VCP is a member of a family of ATP binding, homo-oligomeric proteins, and the mammalian homolog of Saccharomyces cerevisiae cdc48p, a protein essential to the completion of mitosis in yeast. Both endogenous and expressed murine VCP are tyrosine phosphorylated in response to T cell activation. Thus we have identified a novel component of the TCR mediated tyrosine kinase activation pathway that may provide a link between TCR activation and cell cycle control.
This gene is expressed primarily in brain, liver, spleen, placenta. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions which include, but are not limited to, cancer immunological disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the brain, liver, spleen, placenta expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, liver, spleen, and placenta, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from
an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to VCR indicate that polynucleotides and polypeptides corresponding to this gene are useful for treating cancer.
FEATURES OF PROTEIN ENCODED BY GENE NO: 156
The translation product of this gene shares sequence homology with rat growth response protein which is thought to be important in cell growth. A group recently cloned the human homolog of this gene, calling it insulin induced protein 1. (See Accession No. 2358269, see also, Genomics 43 (3), 278-284 (1997).) Preferred polypeptide fragments comprise the amino acid sequence: RSGLGLGITIAFLATLITQF LVYNGVYQYTSPDFLYIRSWLPCIFFSGGVTVGNIGRQLAMGVPEKPHSD (SEQ ID NO: 640), as well as N-terminal and C-terminal deletions of this polypeptide fragment. Also preferred are polynucleotide fragments encoding these polypeptide fragments. This gene is expressed primarily in brain, liver, placenta, heart, spleen, lymphoma.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions which include, but are not limited to, cancer immunological disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the brain, liver, placenta, heart, spleen. expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, liver, placenta, heart, spleen, and lymphoid tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to growth-response protein indicate that polynucleotides and polypeptides corresponding to this gene are useful for modulating cell growth.
FEATURES OF PROTEIN ENCODED BY GENE NO: 157
This gene is expressed primarily in Glioblastoma, endometrial tumor, lymphoma and pancreas tumor.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: Glioblastoma, Endometrial tumor, lymphoma and pancreas tumor. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., endometrium, lymphoid tissue, pancreas, and tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
FEATURES OF PROTEIN ENCODED BY GENE NO: 158 The translation product of this gene shares sequence homology with IGE receptor which is thought to be important in allergy and asthma. This gene is expressed primarily in T cell, and fetal liver. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: allergy and asthma and other immunological disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, and liver, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to IgE receptor indicate that polynucleotides and polypeptides corresponding to this gene are useful for allergy and asthma.
FEATURES OF PROTEIN ENCODED BY GENE NO: 159
The translation product of this gene shares sequence homology with immunoglobin heavy chain which is thought to be important in immune response to the antigen.
This gene is expressed primarily in activated neutrophil and to a lesser extent in activated T cell, monocyte and heart.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: infection , inflammation and cancer. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, and heart, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to immunoglobin heavy chain variable region indicate that polynucleotides and polypeptides corresponding to this gene are useful for making the ligand to block specific antigen which cause certain disease.
FEATURES OF PROTEIN ENCODED BY GENE NO: 160
The translation product of this gene shares sequence homology with mouse X inactive specific transcript protein which is thought to be important in X chromosome inactivation.
This gene is expressed primarily in HSA172 cell and to a lesser extent in normal ovary tissue, ovarian cancer, frontal cortex and brain.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: ovarian tumor, schizophrenia and other neurological disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for
differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune and neural system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., ovary and other reproductive tissue, and brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution and homology to X inactive specific transcript protein indicate that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of reproductive system tumors and CNS tumors.
FEATURES OF PROTEIN ENCODED BY GENE NO: 161 This gene is expressed primarily in adipose cell and to a lesser extent in liver and prostate.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: obesity and liver disorder. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the adipose cell, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., adipose cells, liver, and prostate, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for treatment of obesity and liver disorder.
FEATURES OF PROTEIN ENCODED BY GENE NO: 162
The translation product of this gene shares sequence homology with yeast ubiquitin activating enzyme homolog which is thought to be important in protein posttraslation processing.
This gene is expressed primarily in stromal cell and to a lesser extent in retina, H. Atrophic Endometrium, colon carcinoma and myeloid progenitor cell.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: defects of stromal cell development, neuronal growth disorders and tumors. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune and neural system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., retinal cells, endometrium, colon, and bone marrow, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to ubiquitin-activating enzyme homolog indicate that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis or treatment of some type of tumors , fucosidosis and neuronal growth disorders.
FEATURES OF PROTEIN ENCODED BY GENE NO: 163 This gene is expressed primarily in primary breast cancer and hemangiopericytoma and to a lesser extent in adult brain and cerebellum.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: breast cancer, leukemia and cerebellum disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system and neural system , expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., mammary tissue, brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis or treatment of various tumors and disease involved in neural system.
FEATURES OF PROTEIN ENCODED BY GENE NO: 164
The translation product of this gene shares sequence homology with proline rich proteins. Recently, another group has also cloned this gene, calling it CD84 leukocyte antigen, a new member of the Ig superfamily. (See Accession No. U82988, see also, Blood 90 (6), 2398-2405 (1997).) This gene is expressed primarily in Weizmann olfactory tissue and osteoclastoma and to a lesser extent in anergic T-cell.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: ostsis and immune disease. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., olfactory tissue, bone, and blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution and homology to the Ig superfamily indicate that the protein product of this clone is useful for treatment of osteoporosis, autoimmune disease, and other immune disorders.
FEATURES OF PROTEIN ENCODED BY GENE NO: 165 This gene is expressed primarily in atrophic endometrium and colon cancer and to a lesser extent in some fetal tissues.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: tumors. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system,
expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., endometrium, colon, and fetal tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of tumors, specifically endometrium and colon tumors.
FEATURES OF PROTEIN ENCODED BY GENE NO: 166 This gene is expressed primarily in human primary breast cancer and to a lesser extent in activated monocyte. Although the predicted signal sequence is identified in Table 1 , other upstream sequences are also relevant. Preferred polypeptide fragments comprise the amino acid sequence: VTQPKHLSASMGGSVEIPFSFYYPWELAXXPXVRISWRRGHFHG QSFYSTRPPSIHKDYVNRLFLNWTEGQESGFLRISNLRKEDQSVYFCRVELDTRRSG (SEQ ID NO: 641), as well as N-terminal and C-terminal deletions. Also preferred are polynucleotide fragments encoding these polypeptide fragments.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: breast cancer. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., mammary tissue, and blood cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis of breast cancer.
FEATURES OF PROTEIN ENCODED BY GENE NO: 167 This gene is expressed primarily in fetal tissues and to a lesser extent in adult lung. This gene has also been mapped to chromosomal location 9q34, and thus, can be used as a marker for linkage analysis for chromosome 9.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the embryo tissues, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., fetal tissues, and lung, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
FEATURES OF PROTEIN ENCODED BY GENE NO: 168 The translation product of this gene shares sequence homology with Ig Heavy
Chain which is thought to be important in immune response.
This gene is expressed primarily in prostate cancer tissue specifically Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: prostate cancer. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the prostate, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., prostate, tissue and cells of the immune system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
FEATURES OF PROTEIN ENCODED BY GENE NO: 169
The translation product of this gene shares sequence homology with cytosolic acyl coenzyme-A hydrolase, which is thought to be important in neuron-specific fatty acid metabolism. The gene represented by this contig has since been published by Hajra and colleagues (GenBank Accession No. U91316).
This gene is expressed primarily in human pituitary gland and to a lesser extent in colorectal cancer tissue. This gene has also been observed in the LNCAP cell line.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: hyperlipidemias of familial and/or idiopathic origins. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly blood, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., pituitary and colon, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to rat cytosolic acyl coenzyme-A hydrolase indicate that polynucleotides and polypeptides corresponding to this gene are useful for the detection or treatment of hyperlipidemia disease states by virtue of the ability of specific drugs to activate the enzyme.
FEATURES OF PROTEIN ENCODED BY GENE NO: 170
The translation product of this gene shares sequence homology with a Caenorhabditis elegans gene which is thought to be important in organism development. This gene is expressed primarily in human synovial sarcoma tissue, bone marrow, and to a lesser extent in human brain.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions which include, but are not limited to, of bone, specifically synovial sarcoma. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the bone, connective tissues and possibly immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., synovial tissue, bone marrow, brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another
tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to Caenorhabditis elegans indicate that polynucleotides and polypeptides corresponding to this gene are useful as a diagnostic and/or therapeutic modality directed at the detection and/or treatment of connective tissue sarcomas or other related bone diseases.
FEATURES OF PROTEIN ENCODED BY GENE NO: 171 The translation product of this gene shares sequence homology with betal-
6GlcNAc transferase which is thought to be important in the transfer and metabolism of beta 1-6, N-acetylglucosamine. This gene product has previously been shown to suppress melanoma lung metastasis in both syngeneic and nude mice, decreased invasiveness into the matrigel, and inhibition of cell attachment to collagen and laminin without affecting cell growth.
This gene is expressed primarily in human testes and prostate tissues, and to a lesser extent in kidney, medulla, and pancreas.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions which include, but are not limited to, cancer particularly melanoma. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., testes and other reproductive tissue, prostate, kidney, pancreas, brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to betal-6GlcNAc transferase indicate that the protein product of this clone is useful for the development of diagnostic and/or therapeutic modalities directed at the detection and/or treatment of cancer, the metastasis of malignant tissue or cells. Defects in this potentially secreted enzyme may play a role in metastasis.
FEATURES OF PROTEIN ENCODED BY GENE NO: 172
This gene is expressed primarily in fetal spleen and liver. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: immune disorders, Wilm's tumor disease, hepatic disorders, and hematopoietic disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the hematopoiesis and immune systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., spleen and liver, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that polynucleotides and polypeptides corresponding to this gene are useful for the treatment and identification of fetal defects along with correcting diseases that affect hematopoiesis and the immune system.
FEATURES OF PROTEIN ENCODED BY GENE NO: 173
The translation product of this gene shares sequence homology with ret II oncogene which is thought to be important in Hirschsprung disease and many types of cancers. This gene is expressed in multiple tissues including the lymphatic system, brain, and thyroid.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: Hirschsprung disease and multiple cancers. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune and central nervous system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., lymphoid tissue, thyroid, and brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to
the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to ret II oncogene indicate that polynucleotides and polypeptides corresponding to this gene are useful for diagnosis and treatment of various cancers. It would also be useful for the diagnosis and treatment of Hirschsprung disease. Preferred polypeptides of the invention comprise the amino acid sequence: MEAQQVNEAESAREQLQXLHDQIAGQKASKQELETELERLKQEFHYEEEDLY RTKNTLQSRIKDRDEEIQKLRNQLTNKTLSNSSQSELENRLHQLTETLIQKQTMLESLSTEKNSL VFQLERLEQQMNSASGSSSNGSSINMSGIDNGEGTRLRNVPVLFNDTETNLAGMYGKVRKAAS SIDQFSIRLGIFLRRYPIARVFVIIYM ALLHLWVMIVLLTYTPEM HHDQPYGK (SEQ ID NO: 642).
FEATURES OF PROTEIN ENCODED BY GENE NO: 174
The translation product of this gene shares sequence homology with testis enhanced gene transcript which is thought to be important in regulation of human development.
This gene is expressed primarily in infant brain and to a lesser extent in a variety of other tissues and cell types, including the prostate, testes, monocytes, macrophages, dendritic cells, keratinocytes, and adipocytes. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: neurological, developmental, immune and inflammation disorders. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the brain and immune systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, prostate, testes and other reproductive tissue, blood cells, keratinocytes, and adipocytes, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to testis enhanced gene transcript indicate that the protein product of this clone is useful for diagnosis and treatment of disorders involving the developing brain and the immune system.
FEATURES OF PROTEIN ENCODED BY GENE NO: 175
This gene is expressed primarily in prostate and to a lesser extent in various other tissues, including placenta. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions which include, but are not limited to, cancers, especially of the prostate. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the prostate, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., prostate and placenta, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution indicates that the protein product of this clone is useful for diagnosis and treatment of prostate disorders and cancer. It may also be useful for the diagnosis and treatment of endocrine disorders.
FEATURES OF PROTEIN ENCODED BY GENE NO: 176
The translation product of this gene shares sequence homology with Sacchromyces cerevisiae YNT20 gene which is thought to be important in mitochondrial function.
This gene is expressed at a particularly high level in muscle tissue. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases related to such tissues and cell types including: muscle wasting diseases. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the neuromuscular system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., muscle and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e.,
the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to the YNT20 gene indicate that this protein is useful for treatment and detection of neuromuscular diseases caused by loss of mitochondrial function. For example this gene or its protein product could be used in replacement therapy for such diseases.
FEATURES OF PROTEIN ENCODED BY GENE NO: 177
This gene is expressed primarily in the brain and to a lesser extent in kidney, placenta, smooth muscle, heart and lung.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: neuromuscular diseases, degenerative diseases of the central nervous system, and heart disease. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the neuromuscular system, central nervous system, and heart, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, kidney, placenta, muscle, heart and lung, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
This gene or its protein product could also be used for replacement therapy for the above mentioned diseases.
FEATURES OF PROTEIN ENCODED BY GENE NO: 178 The translation product of this gene shares sequence homology with caldesmon which is thought to be important in the cellular response to changes in glucose levels. This gene is expressed primarily in multiple tissues including brain and retina. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: central nervous system disorders and retinopathy. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for identification of the tissue(s) or cell
i l l
type(s). For a number of disorders of the above tissues or cells, particularly of the CNS disorders and retinopathy, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, and retinal tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution and homology to caldesmon indicate that polynucleotides and polypeptides corresponding to this gene are useful for treatment of retinopathies.
FEATURES OF PROTEIN ENCODED BY GENE NO: 179
The translation product of this gene shares sequence homology with mouse fibrosin protein which is thought to be important in regulation of fibrinogenesis in certain chronic inflammatory diseases.
This gene is expressed primarily in amniotic cells and breast tissue. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of breast cancer and abnormal embryo development. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the reproductive system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., amniotic cells, and mammary tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution and homology to fibrosin indicate that the protein product of this clone is useful for treatment of breast cancer. This gene or its protein product could be used in replacement therapy for breast cancer. In addition the protein product of this gene is useful in the treatment of chronic inflammatory diseases.
FEATURES OF PROTEIN ENCODED BY GENE NO: 180
This gene is expressed several infant tissues including brain and liver and various adult tissues including brain, lung, liver, testes, and prostate.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions which include, but are not limited to, brain cancer, lung cancer, liver cancer and cancers of the reproductive system. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the central nervous system, hepatic system, and reproductive system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, lung, liver, testes and other reproductive tissue, and prostate, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The tissue distribution of this gene product indicates that the protein product of this clone is involved in growth regulation and could be used as a growth factor or growth blocker in a variety of settings including treatment of cancers.
FEATURES OF PROTEIN ENCODED BY GENE NO: 181
This gene is expressed primarily in activated monocytes and to a lesser extent in melanocytes and dendritic cells.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of immune system diseases and cancer. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., blood cells, melanocytes, and dendritic cells, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution indicates that the protein product of this clone could be involved in growth regulation and could be used as a growth factor or growth blocker in a variety of settings including treatment of cancers.
FEATURES OF PROTEIN ENCODED BY GENE NO: 182
This gene is expressed primarily in placenta and several tumors of various tissue origin and to a lesser extent in normal tissues including liver, lung, brain, and skin, Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of cancers of all kinds. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the central nervous system, respiratory system and skin, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., liver, lung, brain and other tissues of the nervous system, and skin, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The high expression of this gene in multiple tumors indicates that the protein product of the clone may be involved in cell growth control and therefore would be useful for treatment of certain cancers. Likewise molecules developed to block the activity of the protein product of this clone could be used to block its potential role in tumor growth promotion.
FEATURES OF PROTEIN ENCODED BY GENE NO: 183 The translation product of this gene shares sequence homology with the mouse
Ndrl gene which is thought to be important in cancer progression.
This gene is expressed multiple cell types and tissues including brain, lung, kidney, bone marrow, liver, and spleen.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of all types of cancers. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the nervous, immune, and endocrine systems, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, lung, kidney, bone marrow, liver and spleen, and cancerous and wounded
tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The tissue distribution and homology to Ndrl gene, which is thought to be involved in cancer progression, indicate that polynucleotides and polypeptides corresponding to this gene are useful for treatment of certain cancers. Likewise molecules developed to block the activity of the protein product of this clone could be used to block its potential role in tumor growth promotion.
FEATURES OF PROTEIN ENCODED BY GENE NO: 184
This gene is expressed primarily in early stage human brain and liver and to a lesser extent in several other fetal tissues.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of diseases and conditions: brain and liver cancers. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the central nervous system and immune system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., brain and other tissue of the nervous system, liver, and fetal tissue, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The expression of this gene in embryonic tissues indicates that the protein could be involved in growth regulation and could be used as a growth factor or growth blocker in a variety of settings including treatment of cancers.
FEATURES OF PROTEIN ENCODED BY GENE NO: 185 This gene is expressed primarily in infant and embryonic brain. Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of degenerative nervous system disorders and brain cancer. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell
type(s). For a number of disorders of the above tissues or cells, particularly of the central nervous system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., embryonic tissue, brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder.
The expression of this gene in embryonic tissues indicates that the protein could be involved in growth regulation and could be used as a growth factor or growth blocker in a variety of settings including treatment of cancers.
FEATURES OF PROTEIN ENCODED BY GENE NO: 186
This gene is expressed primarily in multiple tissues including placenta, fetal lung, fetal liver, and brain.
Therefore, polynucleotides and polypeptides of the invention are useful as reagents for differential identification of the tissue(s) or cell type(s) present in a biological sample and for diagnosis of all types of cancers including liver, brain and lung. Similarly, polypeptides and antibodies directed to these polypeptides are useful in providing immunological probes for differential identification of the tissue(s) or cell type(s). For a number of disorders of the above tissues or cells, particularly of the central nervous system, pulmonary system, and hepatic system, expression of this gene at significantly higher or lower levels may be routinely detected in certain tissues and cell types (e.g., placenta, lung, liver, and brain and other tissue of the nervous system, and cancerous and wounded tissues) or bodily fluids (e.g., serum, plasma, urine, synovial fluid or spinal fluid) or another tissue or cell sample taken from an individual having such a disorder, relative to the standard gene expression level, i.e., the expression level in healthy tissue or bodily fluid from an individual not having the disorder. The expression of this gene in embryonic tissues indicates that the protein could be involved in growth regulation and could be used as a growth factor or growth blocker in a variety of settings including treatment of cancers.

Table 1 summarizes the information corresponding to each "Gene No." described above. The nucleotide sequence identified as "NT SEQ ID NO:X" was assembled from partially homologous ("overlapping") sequences obtained from the "cDNA clone ID" identified in Table 1 and, in some cases, from additional related DNA clones. The overlapping sequences were assembled into a single contiguous sequence of high redundancy (usually three to five overlapping sequences at each nucleotide position), resulting in a final sequence identified as SEQ ID NO:X.
The cDNA Clone ID was deposited on the date and given the corresponding deposit number listed in "ATCC Deposit No:Z and Date." Some of the deposits contain multiple different clones corresponding to the same gene. "Vector" refers to the type of vector contained in the cDNA Clone ID.
"Total NT Seq." refers to the total number of nucleotides in the contig identified by "Gene No." The deposited clone may contain all or most of these sequences, reflected by the nucleotide position indicated as "5' NT of Clone Seq." and the "3' NT of Clone Seq." of SEQ ID NO:X. The nucleotide position of SEQ ID NO:X of the putative start codon (methionine) is identified as "5' NT of Start Codon." Similarly , the nucleotide position of SEQ ID NO:X of the predicted signal sequence is identified as "5' NT of First AA of Signal Pep."
The translated amino acid sequence, beginning with the methionine, is identified as "AA SEQ ID NO:Y," although other reading frames can also be easily translated using known molecular biology techniques. The polypeptides produced by these alternative open reading frames are specifically contemplated by the present invention.
The first and last amino acid position of SEQ ID NO:Y of the predicted signal peptide is identified as "First AA of Sig Pep" and "Last AA of Sig Pep." The predicted first amino acid position of SEQ ID NO: Y of the secreted portion is identified as
"Predicted First AA of Secreted Portion." Finally, the amino acid position of SEQ ID NO: Y of the last amino acid in the open reading frame is identified as "Last AA of ORF."
SEQ ID NO:X and the translated SEQ ID NO:Y are sufficiently accurate and otherwise suitable for a variety of uses well known in the art and described further below. For instance, SEQ ID NO:X is useful for designing nucleic acid hybridization probes that will detect nucleic acid sequences contained in SEQ ID NO:X or the cDNA contained in the deposited clone. These probes will also hybridize to nucleic acid molecules in biological samples, thereby enabling a variety of forensic and diagnostic methods of the invention. Similarly, polypeptides identified from SEQ ID NO: Y may be used to generate antibodies which bind specifically to the secreted proteins encoded by the cDNA clones identified in Table 1.
Nevertheless, DNA sequences generated by sequencing reactions can contain sequencing errors. The errors exist as misidentified nucleotides, or as insertions or deletions of nucleotides in the generated DNA sequence. The erroneously inserted or deleted nucleotides cause frame shifts in the reading frames of the predicted amino acid sequence. In these cases, the predicted amino acid sequence diverges from the actual amino acid sequence, even though the generated DNA sequence may be greater than 99.9% identical to the actual DNA sequence (for example, one base insertion or deletion in an open reading frame of over 1000 bases).
Accordingly, for those applications requiring precision in the nucleotide sequence or the amino acid sequence, the present invention provides not only the generated nucleotide sequence identified as SEQ ID NO:X and the predicted translated amino acid sequence identified as SEQ ID NO: Y, but also a sample of plasmid DNA containing a human cDNA of the invention deposited with the ATCC, as set forth in Table 1. The nucleotide sequence of each deposited clone can readily be determined by sequencing the deposited clone in accordance with known methods. The predicted amino acid sequence can then be verified from such deposits. Moreover, the amino acid sequence of the protein encoded by a particular clone can also be directly determined by peptide sequencing or by expressing the protein in a suitable host cell containing the deposited human cDNA, collecting the protein, and determining its sequence.
The present invention also relates to the genes corresponding to SEQ ID NO:X, SEQ ID NO:Y, or the deposited clone. The corresponding gene can be isolated in accordance with known methods using the sequence information disclosed herein. Such methods include preparing probes or primers from the disclosed sequence and identifying or amplifying the corresponding gene from appropriate sources of genomic material.
Also provided in the present invention are species homologs. Species homologs may be isolated and identified by making suitable probes or primers from the sequences provided herein and screening a suitable nucleic acid source for the desired homologue.
The polypeptides of the invention can be prepared in any suitable manner. Such polypeptides include isolated naturally occurring polypeptides, recombinantly produced polypeptides, synthetically produced polypeptides, or polypeptides produced by a combination of these methods. Means for preparing such polypeptides are well understood in the art.
The polypeptides may be in the form of the secreted protein, including the mature form, or may be a part of a larger protein, such as a fusion protein (see below).
It is often advantageous to include an additional amino acid sequence which contains secretory or leader sequences, pro-sequences, sequences which aid in purification , such as multiple histidine residues, or an additional sequence for stability during recombinant production. The polypeptides of the present invention are preferably provided in an isolated form, and preferably are substantially purified. A recombinantly produced version of a polypeptide, including the secreted polypeptide, can be substantially purified by the one-step method described in Smith and Johnson, Gene 67:31-40 (1988). Polypeptides of the invention also can be purified from natural or recombinant sources using antibodies of the invention raised against the secreted protein in methods which are well known in the art.
Signal Sequences
Methods for predicting whether a protein has a signal sequence, as well as the cleavage point for that sequence, are available. For instance, the method of McGeoch, Virus Res. 3:271-286 (1985), uses the information from a short N-terminal charged region and a subsequent uncharged region of the complete (uncleaved) protein. The method of von Heinje, Nucleic Acids Res. 14:4683-4690 (1986) uses the information from the residues surrounding the cleavage site, typically residues -13 to +2, where +1 indicates the amino terminus of the secreted protein. The accuracy of predicting the cleavage points of known mammalian secretory proteins for each of these methods is in the range of 75-80%. (von Heinje, supra.) However, the two methods do not always produce the same predicted cleavage point(s) for a given protein.
In the present case, the deduced amino acid sequence of the secreted polypeptide was analyzed by a computer program called SignalP (Henrik Nielsen et al., Protein Engineering 10:1-6 (1997)), which predicts the cellular location of a protein based on the amino acid sequence. As part of this computational prediction of localization, the methods of McGeoch and von Heinje are incorporated. The analysis of the amino acid sequences of the secreted proteins described herein by this program provided the results shown in Table 1.
As one of ordinary skill would appreciate, however, cleavage sites sometimes vary from organism to organism and cannot be predicted with absolute certainty. Accordingly, the present invention provides secreted polypeptides having a sequence shown in SEQ ID NO:Y which have an N-terminus beginning within 5 residues (i.e., + or - 5 residues) of the predicted cleavage point. Similarly, it is also recognized that in some cases, cleavage of the signal sequence from a secreted protein is not entirely
uniform, resulting in more than one secreted species. These polypeptides, and the polynucleotides encoding such polypeptides, are contemplated by the present invention.
Moreover, the signal sequence identified by the above analysis may not necessarily predict the naturally occurring signal sequence. For example, the naturally occurring signal sequence may be further upstream from the predicted signal sequence. However, it is likely that the predicted signal sequence will be capable of directing the secreted protein to the ER. These polypeptides, and the polynucleotides encoding such polypeptides, are contemplated by the present invention.
Polynucleotide and Polypeptide Variants
"Variant" refers to a polynucleotide or polypeptide differing from the polynucleotide or polypeptide of the present invention, but retaining essential properties thereof. Generally, variants are overall closely similar, and, in many regions, identical to the polynucleotide or polypeptide of the present invention. "Identity" per se has an art-recognized meaning and can be calculated using published techniques. (See, e.g.: (COMPUTATIONAL MOLECULAR BIOLOGY, Lesk, A.M., ed., Oxford University Press, New York, (1988); BIOCOMPUTING: INFORMATICS AND GENOME PROJECTS, Smith, D.W., ed., Academic Press, New York, (1993); COMPUTER ANALYSIS OF SEQUENCE DATA, PART I, Griffin, A.M., and Griffin, H.G., eds., Humana Press, New Jersey, (1994);
SEQUENCE ANALYSIS IN MOLECULAR BIOLOGY, von Heinje, G., Academic Press, (1987); and SEQUENCE ANALYSIS PRIMER, Gribskov, M. and Devereux, J., eds., M Stockton Press, New York, (1991).) While there exists a number of methods to measure identity between two polynucleotide or polypeptide sequences, the term "identity" is well known to skilled artisans. (Carillo, H., and Lipton, D., SIAM J Applied Math 48: 1073 (1988).) Methods commonly employed to determine identity or similarity between two sequences include, but are not limited to, those disclosed in "Guide to Huge Computers," Martin J. Bishop, ed., Academic Press, San Diego, (1994), and Carillo, H., and Lipton, D., SIAM J Applied Math 48: 1073 (1988). Methods for aligning polynucleotides or polypeptides are codified in computer programs, including the GCG program package (Devereux, J., et al., Nucleic Acids Research (1984) 12(1):387 (1984)), BLASTP, BLASTN, FASTA (Atschul, S.F. et al., J. Molec. Biol. 215:403 (1990), Bestfit program (Wisconsin Sequence Analysis Package, Version 8 for Unix, Genetics Computer Group, University Research Park, 575 Science Drive, Madison, WI 53711 (using the local homology algorithm of Smith and Waterman, Advances in Applied Mathematics 2:482-489 (1981).)
When using any of the sequence alignment programs to determine whether a particular sequence is, for instance, 95% identical to a reference sequence, the parameters are set so that the percentage of identity is calculated over the full length of the reference polynucleotide and that gaps in identity of up to 5% of the total number of nucleotides in the reference polynucleotide are allowed.
A preferred method for determing the best overall match between a query sequence (a sequence of the present invention) and a subject sequence, also referred to as a global sequence alignment, can be determined using the FASTDB computer program based on the algorithm of Brutlag et al. (Comp. App. Biosci. 6:237-245 (1990).) The term "sequence" includes nucleotide and amino acid sequences. In a sequence alignment the query and subject sequences are either both nucleotide sequences or both amino acid sequences. The result of said global sequence alignment is in percent identity. Preferred parameters used in a FASTDB search of a DNA sequence to calculate percent identiy are: Matrix=Unitary, k-tuple=4, Mismatch Penalty= 1 , Joining Penalty=30, Randomization Group Length=0, and Cutoff Score= 1 , Gap Penalty=5, Gap Size Penalty 0.05, and Window Size=500 or query sequence length in nucleotide bases, whichever is shorter. Preferred parameters employed to calculate percent identity and similarity of an amino acid alignment are: Matrix=PAM 150, k-tuple=2, Mismatch Penalty=l, Joining Penalty=20, Randomization Group Length=0, Cutoff Score= 1 , Gap Penalty=5, Gap Size Penalty=0.05, and Window Size=500 or query sequence length in amino acid residues, whichever is shorter.
As an illustration, a polynucleotide having a nucleotide sequence of at least 95% "identity" to a sequence contained in SEQ ID NO:X or the cDNA contained in the deposited clone, means that the polynucleotide is identical to a sequence contained in SEQ ID NO:X or the cDNA except that the polynucleotide sequence may include up to five point mutations per each 100 nucleotides of the total length (not just within a given 100 nucleotide stretch). In other words, to obtain a polynucleotide having a nucleotide sequence at least 95% identical to SEQ ID NO:X or the deposited clone, up to 5% of the nucleotides in the sequence contained in SEQ ID NO:X or the cDNA can be deleted, inserted, or substituted with other nucleotides. These changes may occur anywhere throughout the polynucleotide.
Further embodiments of the present invention include polynucleotides having at least 85% identity, more preferably at least 90% identity, and most preferably at least 95%, 96%, 97%, 98% or 99% identity to a sequence contained in SEQ ID NO:X or the cDNA contained in the deposited clone. Of course, due to the degeneracy of the genetic code, one of ordinary skill in the art will immediately recognize that a large number of the polynucleotides having at least 85%, 90%, 95%, 96%, 97%, 98%, or 99% identity
will encode a polypeptide identical to an amino acid sequence contained in SEQ ID NON or the expressed protein produced by the deposited clone.
Similarly, by a polypeptide having an amino acid sequence having at least, for example, 95% "identity" to a reference polypeptide, is intended that the amino acid sequence of the polypeptide is identical to the reference polypeptide except that the polypeptide sequence may include up to five amino acid alterations per each 100 amino acids of the total length of the reference polypeptide. In other words, to obtain a polypeptide having an amino acid sequence at least 95% identical to a reference amino acid sequence, up to 5% of the amino acid residues in the reference sequence may be deleted or substituted with another amino acid, or a number of amino acids up to 5% of the total amino acid residues in the reference sequence may be inserted into the reference sequence. These alterations of the reference sequence may occur at the amino or carboxy terminal positions of the reference amino acid sequence or anywhere between those terminal positions, interspersed either individually among residues in the reference sequence or in one or more contiguous groups within the reference sequence. Further embodiments of the present invention include polypeptides having at least 80% identity, more preferably at least 85% identity, more preferably at least 90% identity, and most preferably at least 95%, 96%, 97%, 98% or 99% identity to an amino acid sequence contained in SEQ ID ΝO:Y or the expressed protein produced by the deposited clone. Preferably, the above polypeptides should exhibit at least one biological activity of the protein.
In a preferred embodiment, polypeptides of the present invention include polypeptides having at least 90% similarity, more preferably at least 95% similarity, and still more preferably at least 96%, 97%, 98%, or 99% similarity to an amino acid sequence contained in SEQ ID NO:Y or the expressed protein produced by the deposited clone.
The variants may contain alterations in the coding regions, non-coding regions, or both. Especially preferred are polynucleotide variants containing alterations which produce silent substitutions, additions, or deletions, but do not alter the properties or activities of the encoded polypeptide. Nucleotide variants produced by silent substitutions due to the degeneracy of the genetic code are preferred. Moreover, variants in which 5-10, 1-5, or 1-2 amino acids are substituted, deleted, or added in any combination are also preferred. Polynucleotide variants can be produced for a variety of reasons, e.g., to optimize codon expression for a particular host (change codons in the human mRNA to those preferred by a bacterial host such as E. coli).
Naturally occurring variants are called "allelic variants," and refer to one of several alternate forms of a gene occupying a given locus on a chromosome of an
organism. (Genes II, Lewin, B., ed., John Wiley & Sons, New York (1985).) These allelic variants can vary at either the polynucleotide and/or polypeptide level. Alternatively, non-naturally occurring variants may be produced by mutagenesis techniques or by direct synthesis. Using known methods of protein engineering and recombinant DNA technology, variants may be generated to improve or alter the characteristics of the polypeptides of the present invention. For instance, one or more amino acids can be deleted from the N-terminus or C-terminus of the secreted protein without substantial loss of biological function. The authors of Ron et al., J. Biol. Chem. 268: 2984-2988 (1993), reported variant KGF proteins having heparin binding activity even after deleting 3, 8, or 27 amino-terminal amino acid residues. Similarly, Interferon gamma exhibited up to ten times higher activity after deleting 8-10 amino acid residues from the carboxy terminus of this protein. (Dobeli et al., J. Biotechnology 7: 199-216 (1988).) Moreover, ample evidence demonstrates that variants often retain a biological activity similar to that of the naturally occurring protein. For example, Gayle and coworkers (J. Biol. Chem 268:22105-22111 (1993)) conducted extensive mutational analysis of human cytokine IL-la. They used random mutagenesis to generate over 3,500 individual IL-la mutants that averaged 2.5 amino acid changes per variant over the entire length of the molecule. Multiple mutations were examined at every possible amino acid position. The investigators found that "[m]ost of the molecule could be altered with little effect on either [binding or biological activity]." (See, Abstract.) In fact, only 23 unique amino acid sequences, out of more than 3,500 nucleotide sequences examined, produced a protein that significantly differed in activity from wild- type. Furthermore, even if deleting one or more amino acids from the N-terminus or
C-terminus of a polypeptide results in modification or loss of one or more biological functions, other biological activities may still be retained. For example, the ability of a deletion variant to induce and/or to bind antibodies which recognize the secreted form will likely be retained when less than the majority of the residues of the secreted form are removed from the N-terminus or C-terminus. Whether a particular polypeptide lacking N- or C-terminal residues of a protein retains such immunogenic activities can readily be determined by routine methods described herein and otherwise known in the art.
Thus, the invention further includes polypeptide variants which show substantial biological activity. Such variants include deletions, insertions, inversions, repeats, and substitutions selected according to general rules known in the art so as have little effect on activity. For example, guidance concerning how to make
phenotypically silent amino acid substitutions is provided in Bowie, J. U. et al., Science 247: 1306-1310 (1990), wherein the authors indicate that there are two main strategies for studying the tolerance of an amino acid sequence to change.
The first strategy exploits the tolerance of amino acid substitutions by natural selection during the process of evolution. By comparing amino acid sequences in different species, conserved amino acids can be identified. These conserved amino acids are likely important for protein function. In contrast, the amino acid positions where substitutions have been tolerated by natural selection indicates that these positions are not critical for protein function. Thus, positions tolerating amino acid substitution could be modified while still maintaining biological activity of the protein. The second strategy uses genetic engineering to introduce amino acid changes at specific positions of a cloned gene to identify regions critical for protein function. For example, site directed mutagenesis or alanine-scanning mutagenesis (introduction of single alanine mutations at every residue in the molecule) can be used. (Cunningham and Wells, Science 244:1081-1085 (1989).) The resulting mutant molecules can then be tested for biological activity.
As the authors state, these two strategies have revealed that proteins are surprisingly tolerant of amino acid substitutions. The authors further indicate which amino acid changes are likely to be permissive at certain amino acid positions in the protein. For example, most buried (within the tertiary structure of the protein) amino acid residues require nonpolar side chains, whereas few features of surface side chains are generally conserved. Moreover, tolerated conservative amino acid substitutions involve replacement of the aliphatic or hydrophobic amino acids Ala, Val, Leu and lie; replacement of the hydroxyl residues Ser and Thr; replacement of the acidic residues Asp and Glu; replacement of the amide residues Asn and Gin, replacement of the basic residues Lys, Arg, and His; replacement of the aromatic residues Phe, Tyr, and Trp, and replacement of the small-sized amino acids Ala, Ser, Thr, Met, and Gly.
Besides conservative amino acid substitution, variants of the present invention include (i) substitutions with one or more of the non-conserved amino acid residues, where the substituted amino acid residues may or may not be one encoded by the genetic code, or (ii) substitution with one or more of amino acid residues having a substituent group, or (iii) fusion of the mature polypeptide with another compound, such as a compound to increase the stability and/or solubility of the polypeptide (for example, polyethylene glycol), or (iv) fusion of the polypeptide with additional amino acids, such as an IgG Fc fusion region peptide, or leader or secretory sequence, or a sequence facilitating purification. Such variant polypeptides are deemed to be within the scope of those skilled in the art from the teachings herein.
For example, polypeptide variants containing amino acid substitutions of charged amino acids with other charged or neutral amino acids may produce proteins with improved characteristics, such as less aggregation. Aggregation of pharmaceutical formulations both reduces activity and increases clearance due to the aggregate's immunogenic activity. (Pinckard et al., Clin. Exp. Immunol. 2:331-340 (1967); Robbins et al., Diabetes 36: 838-845 (1987); Cleland et al., Crit. Rev. Therapeutic Drug Carrier Systems 10:307-377 (1993).)
Polynucleotide and Polypeptide Fragments In the present invention, a "polynucleotide fragment" refers to a short polynucleotide having a nucleic acid sequence contained in the deposited clone or shown in SEQ ID NO:X. The short nucleotide fragments are preferably at least about 15 nt, and more preferably at least about 20 nt, still more preferably at least about 30 nt, and even more preferably, at least about 40 nt in length. A fragment "at least 20 nt in length," for example, is intended to include 20 or more contiguous bases from the cDNA sequence contained in the deposited clone or the nucleotide sequence shown in SEQ ID NO:X. These nucleotide fragments are useful as diagnostic probes and primers as discussed herein. Of course, larger fragments (e.g., 50, 150, 500, 600, 2000 nucleotides) are preferred. Moreover, representative examples of polynucleotide fragments of the invention, include, for example, fragments having a sequence from about nucleotide number 1-50, 51-100, 101-150, 151-200, 201-250, 251-300, 301-350, 351-400, 401- 450, 451-500, 501-550, 551-600, 651-700, and 701 to the end of SEQ ID NO:X or the cDNA contained in the deposited clone. In this context "about" includes the particularly recited ranges, larger or smaller by several (5, 4, 3, 2, or 1) nucleotides, at either terminus or at both termini. Preferably, these fragments encode a polypeptide which has biological activity.
In the present invention, a "polypeptide fragment" refers to a short amino acid sequence contained in SEQ ID NO:Y or encoded by the cDNA contained in the deposited clone. Protein fragments may be "free-standing," or comprised within a larger polypeptide of which the fragment forms a part or region, most preferably as a single continuous region. Representative examples of polypeptide fragments of the invention, include, for example, fragments from about amino acid number 1-20, 21-40, 41-60, 61-80, 81-100, 102-120, 121-140, 141-160, and 161 to the end of the coding region. Moreover, polypeptide fragments can be about 20, 30, 40, 50, 60, 70, 80, 90, 100, 110, 120, 130, 140, or 150 amino acids in length. In this context "about"
includes the particularly recited ranges, larger or smaller by several (5, 4, 3, 2, or 1) amino acids, at either extreme or at both extremes.
Preferred polypeptide fragments include the secreted protein as well as the mature form. Further preferred polypeptide fragments include the secreted protein or the mature form having a continuous series of deleted residues from the amino or the carboxy terminus, or both. For example, any number of amino acids, ranging from 1- 60, can be deleted from the amino terminus of either the secreted polypeptide or the mature form. Similarly, any number of amino acids, ranging from 1-30, can be deleted from the carboxy terminus of the secreted protein or mature form. Furthermore, any combination of the above amino and carboxy terminus deletions are preferred.
Similarly, polynucleotide fragments encoding these polypeptide fragments are also preferred.
Also preferred are polypeptide and polynucleotide fragments characterized by structural or functional domains, such as fragments that comprise alpha-helix and alpha- helix forming regions, beta-sheet and beta-sheet-forming regions, turn and turn- forming regions, coil and coil-forming regions, hydrophilic regions, hydrophobic regions, alpha amphipathic regions, beta amphipathic regions, flexible regions, surface- forming regions, substrate binding region, and high antigenic index regions. Polypeptide fragments of SEQ ID NON falling within conserved domains are specifically contemplated by the present invention. Moreover, polynucleotide fragments encoding these domains are also contemplated.
Other preferred fragments are biologically active fragments. Biologically active fragments are those exhibiting activity similar, but not necessarily identical, to an activity of the polypeptide of the present invention. The biological activity of the fragments may include an improved desired activity, or a decreased undesirable activity.
Epitopes & Antibodies
In the present invention, "epitopes" refer to polypeptide fragments having antigenic or immunogenic activity in an animal, especially in a human. A preferred embodiment of the present invention relates to a polypeptide fragment comprising an epitope, as well as the polynucleotide encoding this fragment. A region of a protein molecule to which an antibody can bind is defined as an "antigenic epitope." In contrast, an "immunogenic epitope" is defined as a part of a protein that elicits an antibody response. (See, for instance, Geysen et al., Proc. Νatl. Acad. Sci. USA 81:3998- 4002 (1983).)
Fragments which function as epitopes may be produced by any conventional means. (See, e.g., Houghten, R. A., Proc. Natl. Acad. Sci. USA 82:5131-5135 (1985) further described in U.S. Patent No. 4,631,211.)
In the present invention, antigenic epitopes preferably contain a sequence of at least seven, more preferably at least nine, and most preferably between about 15 to about 30 amino acids. Antigenic epitopes are useful to raise antibodies, including monoclonal antibodies, that specifically bind the epitope. (See, for instance, Wilson et al., Cell 37:767-778 (1984); Sutcliffe, J. G. et al., Science 219:660-666 (1983).)
Similarly, immunogenic epitopes can be used to induce antibodies according to methods well known in the art. (See, for instance, Sutcliffe et al., supra; Wilson et al., supra; Chow, M. et al., Proc. Natl. Acad. Sci. USA 82:910-914; and Bittle, F. J. et al., J. Gen. Virol. 66:2347-2354 (1985).) A preferred immunogenic epitope includes the secreted protein. The immunogenic epitopes may be presented together with a carrier protein, such as an albumin, to an animal system (such as rabbit or mouse) or, if it is long enough (at least about 25 amino acids), without a carrier. However, immunogenic epitopes comprising as few as 8 to 10 amino acids have been shown to be sufficient to raise antibodies capable of binding to, at the very least, linear epitopes in a denatured polypeptide (e.g., in Western blotting.)
As used herein, the term "antibody" (Ab) or "monoclonal antibody" (Mab) is meant to include intact molecules as well as antibody fragments (such as, for example, Fab and F(ab')2 fragments) which are capable of specifically binding to protein. Fab and F(ab')2 fragments lack the Fc fragment of intact antibody, clear more rapidly from the circulation, and may have less non-specific tissue binding than an intact antibody. (Wahl et al., J. Nucl. Med. 24:316-325 (1983).) Thus, these fragments are preferred, as well as the products of a FAB or other immunoglobulin expression library. Moreover, antibodies of the present invention include chimeric, single chain, and humanized antibodies.
Fusion Proteins Any polypeptide of the present invention can be used to generate fusion proteins. For example, the polypeptide of the present invention, when fused to a second protein, can be used as an antigenic tag. Antibodies raised against the polypeptide of the present invention can be used to indirectly detect the second protein by binding to the polypeptide. Moreover, because secreted proteins target cellular locations based on trafficking signals, the polypeptides of the present invention can be used as targeting molecules once fused to other proteins.
Examples of domains that can be fused to polypeptides of the present invention include not only heterologous signal sequences, but also other heterologous functional regions. The fusion does not necessarily need to be direct, but may occur through linker sequences. Moreover, fusion proteins may also be engineered to improve characteristics of the polypeptide of the present invention. For instance, a region of additional amino acids, particularly charged amino acids, may be added to the N-terminus of the polypeptide to improve stability and persistence during purification from the host cell or subsequent handling and storage. Also, peptide moieties may be added to the polypeptide to facilitate purification. Such regions may be removed prior to final preparation of the polypeptide. The addition of peptide moieties to facilitate handling of polypeptides are familiar and routine techniques in the art.
Moreover, polypeptides of the present invention, including fragments, and specifically epitopes, can be combined with parts of the constant domain of immunoglobulins (IgG), resulting in chimeric polypeptides. These fusion proteins facilitate purification and show an increased half-life in vivo. One reported example describes chimeric proteins consisting of the first two domains of the human CD4- polypeptide and various domains of the constant regions of the heavy or light chains of mammalian immunoglobulins. (EP A 394,827; Traunecker et al., Nature 331 :84-86 (1988).) Fusion proteins having disulfide-linked dimeric structures (due to the IgG) can also be more efficient in binding and neutralizing other molecules, than the monomeric secreted protein or protein fragment alone. (Fountoulakis et al., J. Biochem. 270:3958-3964 (1995).)
Similarly, EP-A-O 464 533 (Canadian counterpart 2045869) discloses fusion proteins comprising various portions of constant region of immunoglobulin molecules together with another human protein or part thereof. In many cases, the Fc part in a fusion protein is beneficial in therapy and diagnosis, and thus can result in, for example, improved pharmacokinetic properties. (EP-A 0232 262.) Alternatively, deleting the Fc part after the fusion protein has been expressed, detected, and purified, would be desired. For example, the Fc portion may hinder therapy and diagnosis if the fusion protein is used as an antigen for immunizations. In drug discovery, for example, human proteins, such as hIL-5, have been fused with Fc portions for the purpose of high-throughput screening assays to identify antagonists of hIL-5. (See, D. Bennett et al., J. Molecular Recognition 8:52-58 (1995); K. Johanson et al., J. Biol. Chem. 270:9459-9471 (1995).)
Moreover, the polypeptides of the present invention can be fused to marker sequences, such as a peptide which facilitates purification of the fused polypeptide. In
preferred embodiments, the marker amino acid sequence is a hexa-histidine peptide, such as the tag provided in a pQE vector (QIAGEN, Inc., 9259 Eton Avenue, Chatsworth, CA, 91311), among others, many of which are commercially available. As described in Gentz et al., Proc. Natl. Acad. Sci. USA 86:821-824 (1989), for instance, hexa-histidine provides for convenient purification of the fusion protein. Another peptide tag useful for purification, the "HA" tag, corresponds to an epitope derived from the influenza hemagglutinin protein. (Wilson et al., Cell 37:767 (1984).)
Thus, any of these above fusions can be engineered using the polynucleotides or the polypeptides of the claimed invention.
Vectors. Host Cells, and Protein Production
The present invention also relates to vectors containing the polynucleotide of the present invention, host cells, and the production of polypeptides by recombinant techniques. The vector may be, for example, a phage, plasmid, viral, or retroviral vector. Retroviral vectors may be replication competent or replication defective. In the latter case, viral propagation generally will occur only in complementing host cells.
The polynucleotides may be joined to a vector containing a selectable marker for propagation in a host. Generally, a plasmid vector is introduced in a precipitate, such as a calcium phosphate precipitate, or in a complex with a charged lipid. If the vector is a virus, it may be packaged in vitro using an appropriate packaging cell line and then transduced into host cells.
The polynucleotide insert should be operatively linked to an appropriate promoter, such as the phage lambda PL promoter, the E. coli lac, trp, phoA and tac promoters, the SV40 early and late promoters and promoters of retroviral LTRs, to name a few. Other suitable promoters will be known to the skilled artisan. The expression constructs will further contain sites for transcription initiation, termination, and, in the transcribed region, a ribosome binding site for translation. The coding portion of the transcripts expressed by the constructs will preferably include a translation initiating codon at the beginning and a termination codon (UAA, UGA or UAG) appropriately positioned at the end of the polypeptide to be translated. As indicated, the expression vectors will preferably include at least one selectable marker. Such markers include dihydrofolate reductase, G418 or neomycin resistance for eukaryotic cell culture and tetracycline, kanamycin or ampicillin resistance genes for culturing in E. coli and other bacteria. Representative examples of appropriate hosts include, but are not limited to, bacterial cells, such as E. coli,
Streptomyces and Salmonella typhimurium cells; fungal cells, such as yeast cells; insect cells such as Drosophila S2 and Spodoptera Sf9 cells; animal cells such as CHO, COS,
293, and Bowes melanoma cells; and plant cells. Appropriate culture mediums and conditions for the above-described host cells are known in the art.
Among vectors preferred for use in bacteria include pQE70, pQE60 and pQE-9, available from QIAGEN, Inc.; pBluescript vectors, Phagescript vectors, pNH8A, pNHlόa, pNH18A, pNH46A, available from Stratagene Cloning Systems, Inc.; and ptrc99a, pKK223-3, pKK233-3, pDR540, pRIT5 available from Pharmacia Biotech, Inc. Among preferred eukaryotic vectors are pWLNEO, pSV2CAT, pOG44, pXTl and pSG available from Stratagene; and pSVK3, pBPV, pMSG and pSVL available from Pharmacia. Other suitable vectors will be readily apparent to the skilled artisan. Introduction of the construct into the host cell can be effected by calcium phosphate transfection, DEAE-dextran mediated transfection, cationic lipid-mediated transfection, electroporation, transduction, infection, or other methods. Such methods are described in many standard laboratory manuals, such as Davis et al., Basic Methods In Molecular Biology (1986). It is specifically contemplated that the polypeptides of the present invention may in fact be expressed by a host cell lacking a recombinant vector. A polypeptide of this invention can be recovered and purified from recombinant cell cultures by well-known methods including ammonium sulfate or ethanol precipitation, acid extraction, anion or cation exchange chromatography, phosphocellulose chromatography, hydrophobic interaction chromatography, affinity chromatography, hydroxylapatite chromatography and lectin chromatography. Most preferably, high performance liquid chromatography ("HPLC") is employed for purification.
Polypeptides of the present invention, and preferably the secreted form, can also be recovered from: products purified from natural sources, including bodily fluids, tissues and cells, whether directly isolated or cultured; products of chemical synthetic procedures; and products produced by recombinant techniques from a prokaryotic or eukaryotic host, including, for example, bacterial, yeast, higher plant, insect, and mammalian cells. Depending upon the host employed in a recombinant production procedure, the polypeptides of the present invention may be glycosylated or may be non-glycosylated. In addition, polypeptides of the invention may also include an initial modified methionine residue, in some cases as a result of host-mediated processes. Thus, it is well known in the art that the N-terminal methionine encoded by the translation initiation codon generally is removed with high efficiency from any protein after translation in all eukaryotic cells. While the N-terminal methionine on most proteins also is efficiently removed in most prokaryotes, for some proteins, this prokaryotic removal process is inefficient, depending on the nature of the amino acid to which the N-terminal methionine is covalently linked.
Uses of the Polynucleotides
Each of the polynucleotides identified herein can be used in numerous ways as reagents. The following description should be considered exemplary and utilizes known techniques.
The polynucleotides of the present invention are useful for chromosome identification. There exists an ongoing need to identify new chromosome markers, since few chromosome marking reagents, based on actual sequence data (repeat polymorphisms), are presently available. Each polynucleotide of the present invention can be used as a chromosome marker.
Briefly, sequences can be mapped to chromosomes by preparing PCR primers (preferably 15-25 bp) from the sequences shown in SEQ ID NO:X. Primers can be selected using computer analysis so that primers do not span more than one predicted exon in the genomic DNA. These primers are then used for PCR screening of somatic cell hybrids containing individual human chromosomes. Only those hybrids containing the human gene corresponding to the SEQ ID NO:X will yield an amplified fragment.
Similarly, somatic hybrids provide a rapid method of PCR mapping the polynucleotides to particular chromosomes. Three or more clones can be assigned per day using a single thermal cycler. Moreover, sublocalization of the polynucleotides can be achieved with panels of specific chromosome fragments. Other gene mapping strategies that can be used include in situ hybridization, prescreening with labeled flow- sorted chromosomes, and preselection by hybridization to construct chromosome specific-cDNA libraries.
Precise chromosomal location of the polynucleotides can also be achieved using fluorescence in situ hybridization (FISH) of a metaphase chromosomal spread. This technique uses polynucleotides as short as 500 or 600 bases; however, polynucleotides 2,000-4,000 bp are preferred. For a review of this technique, see Verma et al., "Human Chromosomes: a Manual of Basic Techniques," Pergamon Press, New York (1988). For chromosome mapping, the polynucleotides can be used individually (to mark a single chromosome or a single site on that chromosome) or in panels (for marking multiple sites and/or multiple chromosomes). Preferred polynucleotides correspond to the noncoding regions of the cDNAs because the coding sequences are more likely conserved within gene families, thus increasing the chance of cross hybridization during chromosomal mapping.
Once a polynucleotide has been mapped to a precise chromosomal location, the physical position of the polynucleotide can be used in linkage analysis. Linkage
analysis establishes coinheritance between a chromosomal location and presentation of a particular disease. (Disease mapping data are found, for example, in V. McKusick, Mendelian Inheritance in Man (available on line through Johns Hopkins University Welch Medical Library) .) Assuming 1 megabase mapping resolution and one gene per 20 kb, a cDNA precisely localized to a chromosomal region associated with the disease could be one of 50-500 potential causative genes.
Thus, once coinheritance is established, differences in the polynucleotide and the corresponding gene between affected and unaffected individuals can be examined. First, visible structural alterations in the chromosomes, such as deletions or translocations, are examined in chromosome spreads or by PCR. If no structural alterations exist, the presence of point mutations are ascertained. Mutations observed in some or all affected individuals, but not in normal individuals, indicates that the mutation may cause the disease. However, complete sequencing of the polypeptide and the corresponding gene from several normal individuals is required to distinguish the mutation from a polymorphism. If a new polymorphism is identified, this polymoφhic polypeptide can be used for further linkage analysis.
Furthermore, increased or decreased expression of the gene in affected individuals as compared to unaffected individuals can be assessed using polynucleotides of the present invention. Any of these alterations (altered expression, chromosomal rearrangement, or mutation) can be used as a diagnostic or prognostic marker.
In addition to the foregoing, a polynucleotide can be used to control gene expression through triple helix formation or antisense DNA or RNA. Both methods rely on binding of the polynucleotide to DNA or RNA. For these techniques, preferred polynucleotides are usually 20 to 40 bases in length and complementary to either the region of the gene involved in transcription (triple helix - see Lee et al., Nucl. Acids Res. 6:3073 (1979); Cooney et al., Science 241:456 (1988); and Dervan et al., Science 251: 1360 (1991) ) or to the mRNA itself (antisense - Okano, J. Neurochem. 56:560 (1991); Oligodeoxy-nucleotides as Antisense Inhibitors of Gene Expression, CRC Press, Boca Raton, FL (1988).) Triple helix formation optimally results in a shut-off of RNA transcription from DNA, while antisense RNA hybridization blocks translation of an mRNA molecule into polypeptide. Both techniques are effective in model systems, and the information disclosed herein can be used to design antisense or triple helix polynucleotides in an effort to treat disease. Polynucleotides of the present invention are also useful in gene therapy. One goal of gene therapy is to insert a normal gene into an organism having a defective gene, in an effort to correct the genetic defect. The polynucleotides disclosed in the
present invention offer a means of targeting such genetic defects in a highly accurate manner. Another goal is to insert a new gene that was not present in the host genome, thereby producing a new trait in the host cell.
The polynucleotides are also useful for identifying individuals from minute biological samples. The United States military, for example, is considering the use of restriction fragment length polymoφhism (RFLP) for identification of its personnel. In this technique, an individual's genomic DNA is digested with one or more restriction enzymes, and probed on a Southern blot to yield unique bands for identifying personnel. This method does not suffer from the current limitations of "Dog Tags" which can be lost, switched, or stolen, making positive identification difficult. The polynucleotides of the present invention can be used as additional DNA markers for RFLP.
The polynucleotides of the present invention can also be used as an alternative to RFLP, by determining the actual base-by-base DNA sequence of selected portions of an individual's genome. These sequences can be used to prepare PCR primers for amplifying and isolating such selected DNA, which can then be sequenced. Using this technique, individuals can be identified because each individual will have a unique set of DNA sequences. Once an unique ID database is established for an individual, positive identification of that individual, living or dead, can be made from extremely small tissue samples.
Forensic biology also benefits from using DNA-based identification techniques as disclosed herein. DNA sequences taken from very small biological samples such as tissues, e.g., hair or skin, or body fluids, e.g., blood, saliva, semen, etc., can be amplified using PCR. In one prior art technique, gene sequences amplified from polymoφhic loci, such as DQa class II HLA gene, are used in forensic biology to identify individuals. (Erlich, H., PCR Technology, Freeman and Co. (1992).) Once these specific polymoφhic loci are amplified, they are digested with one or more restriction enzymes, yielding an identifying set of bands on a Southern blot probed with DNA corresponding to the DQa class II HLA gene. Similarly, polynucleotides of the present invention can be used as polymoφhic markers for forensic puφoses.
There is also a need for reagents capable of identifying the source of a particular tissue. Such need arises, for example, in forensics when presented with tissue of unknown origin. Appropriate reagents can comprise, for example, DNA probes or primers specific to particular tissue prepared from the sequences of the present invention. Panels of such reagents can identify tissue by species and/or by organ type. In a similar fashion, these reagents can be used to screen tissue cultures for contamination.
In the very least, the polynucleotides of the present invention can be used as molecular weight markers on Southern gels, as diagnostic probes for the presence of a specific mRNA in a particular cell type, as a probe to "subtract-out" known sequences in the process of discovering novel polynucleotides, for selecting and making oligomers for attachment to a "gene chip" or other support, to raise anti-DNA antibodies using DNA immunization techniques, and as an antigen to elicit an immune response.
Uses of the Polypeptides
Each of the polypeptides identified herein can be used in numerous ways. The following description should be considered exemplary and utilizes known techniques. A polypeptide of the present invention can be used to assay protein levels in a biological sample using antibody-based techniques. For example, protein expression in tissues can be studied with classical immunohistological methods. (Jalkanen, M., et al., J. Cell. Biol. 101:976-985 (1985); Jalkanen, M., et al., J. Cell . Biol. 105:3087- 3096 (1987).) Other antibody-based methods useful for detecting protein gene expression include immunoassays, such as the enzyme linked immunosorbent assay (ELISA) and the radioimmunoassay (RJA). Suitable antibody assay labels are known in the art and include enzyme labels, such as, glucose oxidase, and radioisotopes, such as iodine (1251, 1211), carbon (14C), sulfur (35S), tritium (3H), indium (112In), and technetium (99mTc), and fluorescent labels, such as fluorescein and rhodamine, and biotin.
In addition to assaying secreted protein levels in a biological sample, proteins can also be detected in vivo by imaging. Antibody labels or markers for in vivo imaging of protein include those detectable by X-radiography, NMR or ESR. For X- radiography, suitable labels include radioisotopes such as barium or cesium, which emit detectable radiation but are not overtly harmful to the subject. Suitable markers for NMR and ESR include those with a detectable characteristic spin, such as deuterium, which may be incoφorated into the antibody by labeling of nutrients for the relevant hybridoma. A protein-specific antibody or antibody fragment which has been labeled with an appropriate detectable imaging moiety, such as a radioisotope (for example, 1311, 112In, 99mTc), a radio-opaque substance, or a material detectable by nuclear magnetic resonance, is introduced (for example, parenterally, subcutaneously, or intraperitoneally) into the mammal. It will be understood in the art that the size of the subject and the imaging system used will determine the quantity of imaging moiety needed to produce diagnostic images. In the case of a radioisotope moiety, for a human subject, the quantity of radioactivity injected will normally range from about 5 to 20
millicuries of 99mTc. The labeled antibody or antibody fragment will then preferentially accumulate at the location of cells which contain the specific protein. In vivo tumor imaging is described in S.W. Burchiel et al., "Immunopharmacokinetics of Radiolabeled Antibodies and Their Fragments." (Chapter 13 in Tumor Imaging: The Radiochemical Detection of Cancer, S.W. Burchiel and B. A. Rhodes, eds., Masson Publishing Inc. (1982).)
Thus, the invention provides a diagnostic method of a disorder, which involves (a) assaying the expression of a polypeptide of the present invention in cells or body fluid of an individual; (b) comparing the level of gene expression with a standard gene expression level, whereby an increase or decrease in the assayed polypeptide gene expression level compared to the standard expression level is indicative of a disorder. Moreover, polypeptides of the present invention can be used to treat disease. For example, patients can be administered a polypeptide of the present invention in an effort to replace absent or decreased levels of the polypeptide (e.g., insulin), to supplement absent or decreased levels of a different polypeptide (e.g., hemoglobin S for hemoglobin B), to inhibit the activity of a polypeptide (e.g., an oncogene), to activate the activity of a polypeptide (e.g., by binding to a receptor), to reduce the activity of a membrane bound receptor by competing with it for free ligand (e.g., soluble TNF receptors used in reducing inflammation), or to bring about a desired response (e.g., blood vessel growth).
Similarly, antibodies directed to a polypeptide of the present invention can also be used to treat disease. For example, administration of an antibody directed to a polypeptide of the present invention can bind and reduce oveφroduction of the polypeptide. Similarly, administration of an antibody can activate the polypeptide, such as by binding to a polypeptide bound to a membrane (receptor).
At the very least, the polypeptides of the present invention could be used as molecular weight markers on SDS-PAGE gels or on molecular sieve gel filtration columns using methods well known to those of skill in the art. Polypeptides can also be used to raise antibodies, which in turn are used to measure protein expression from a recombinant cell, as a way of assessing transformation of the host cell. Moreover, the polypeptides of the present invention can be used to test the following biological activities.
Biological Activities The polynucleotides and polypeptides of the present invention can be used in assays to test for one or more biological activities. If these polynucleotides and polypeptides do exhibit activity in a particular assay, it is likely that these molecules
may be involved in the diseases associated with the biological activity. Thus, the polynucleotides and polypeptides could be used to treat the associated disease.
Immune Activity A polypeptide or polynucleotide of the present invention may be useful in treating deficiencies or disorders of the immune system, by activating or inhibiting the proliferation, differentiation, or mobilization (chemotaxis) of immune cells. Immune cells develop through a process called hematopoiesis, producing myeloid (platelets, red blood cells, neutrophils, and macrophages) and lymphoid (B and T lymphocytes) cells from pluripotent stem cells. The etiology of these immune deficiencies or disorders may be genetic, somatic, such as cancer or some autoimmune disorders, acquired (e.g., by chemotherapy or toxins), or infectious. Moreover, a polynucleotide or polypeptide of the present invention can be used as a marker or detector of a particular immune system disease or disorder. A polynucleotide or polypeptide of the present invention may be useful in treating or detecting deficiencies or disorders of hematopoietic cells. A polypeptide or polynucleotide of the present invention could be used to increase differentiation and proliferation of hematopoietic cells, including the pluripotent stem cells, in an effort to treat those disorders associated with a decrease in certain (or many) types hematopoietic cells. Examples of immunologic deficiency syndromes include, but are not limited to: blood protein disorders (e.g. agammaglobulinemia, dysgammaglobulinemia), ataxia telangiectasia, common variable immunodeficiency, Digeorge Syndrome, HIV infection, HTLV-BLV infection, leukocyte adhesion deficiency syndrome, lymphopenia, phagocyte bactericidal dysfunction, severe combined immunodeficiency (SCIDs), Wiskott-Aldrich Disorder, anemia, thrombocytopenia, or hemoglobinuria. Moreover, a polypeptide or polynucleotide of the present invention could also be used to modulate hemostatic (the stopping of bleeding) or thrombolytic activity (clot formation). For example, by increasing hemostatic or thrombolytic activity, a polynucleotide or polypeptide of the present invention could be used to treat blood coagulation disorders (e.g., afibrinogenemia, factor deficiencies), blood platelet disorders (e.g. thrombocytopenia), or wounds resulting from trauma, surgery, or other causes. Alternatively, a polynucleotide or polypeptide of the present invention that can decrease hemostatic or thrombolytic activity could be used to inhibit or dissolve clotting. These molecules could be important in the treatment of heart attacks (infarction), strokes, or scarring.
A polynucleotide or polypeptide of the present invention may also be useful in treating or detecting autoimmune disorders. Many autoimmune disorders result from
inappropriate recognition of self as foreign material by immune cells. This inappropriate recognition results in an immune response leading to the destruction of the host tissue. Therefore, the administration of a polypeptide or polynucleotide of the present invention that inhibits an immune response, particularly the proliferation, differentiation, or chemotaxis of T-cells, may be an effective therapy in preventing autoimmune disorders.
Examples of autoimmune disorders that can be treated or detected by the present invention include, but are not limited to: Addison's Disease, hemolytic anemia, antiphospholipid syndrome, rheumatoid arthritis, dermatitis, allergic encephalomyelitis, glomerulonephritis, Goodpasture' s Syndrome, Graves' Disease, Multiple Sclerosis, Myasthenia Gravis, Neuritis, Ophthalmia, Bullous Pemphigoid, Pemphigus, Polyendocrinopathies, Puφura, Reiter's Disease, Stiff-Man Syndrome, Autoimmune Thyroiditis, Systemic Lupus Erythematosus, Autoimmune Pulmonary Inflammation, Guillain-Barre Syndrome, insulin dependent diabetes mellitis, and autoimmune inflammatory eye disease.
Similarly, allergic reactions and conditions, such as asthma (particularly allergic asthma) or other respiratory problems, may also be treated by a polypeptide or polynucleotide of the present invention. Moreover, these molecules can be used to treat anaphylaxis, hypersensitivity to an antigenic molecule, or blood group incompatibility. A polynucleotide or polypeptide of the present invention may also be used to treat and/or prevent organ rejection or graft- versus-host disease (GVHD). Organ rejection occurs by host immune cell destruction of the transplanted tissue through an immune response. Similarly, an immune response is also involved in GVHD, but, in this case, the foreign transplanted immune cells destroy the host tissues. The administration of a polypeptide or polynucleotide of the present invention that inhibits an immune response, particularly the proliferation, differentiation, or chemotaxis of T- cells, may be an effective therapy in preventing organ rejection or GVHD.
Similarly, a polypeptide or polynucleotide of the present invention may also be used to modulate inflammation. For example, the polypeptide or polynucleotide may inhibit the proliferation and differentiation of cells involved in an inflammatory response. These molecules can be used to treat inflammatory conditions, both chronic and acute conditions, including inflammation associated with infection (e.g., septic shock, sepsis, or systemic inflammatory response syndrome (SIRS)), ischemia- reperfusion injury, endotoxin lethality, arthritis, complement-mediated hyperacute rejection, nephritis, cytokine or chemokine induced lung injury, inflammatory bowel disease, Crohn's disease, or resulting from over production of cytokines (e.g., TNF or IL-1.)
Hvperproliferative Disorders
A polypeptide or polynucleotide can be used to treat or detect hypeφroliferative disorders, including neoplasms. A polypeptide or polynucleotide of the present invention may inhibit the proliferation of the disorder through direct or indirect interactions. Alternatively, a polypeptide or polynucleotide of the present invention may proliferate other cells which can inhibit the hypeφroliferative disorder.
For example, by increasing an immune response, particularly increasing antigenic qualities of the hypeφroliferative disorder or by proliferating, differentiating, or mobilizing T-cells, hypeφroliferative disorders can be treated. This immune response may be increased by either enhancing an existing immune response, or by initiating a new immune response. Alternatively, decreasing an immune response may also be a method of treating hypeφroliferative disorders, such as a chemotherapeutic agent. Examples of hypeφroliferative disorders that can be treated or detected by a polynucleotide or polypeptide of the present invention include, but are not limited to neoplasms located in the: abdomen, bone, breast, digestive system, liver, pancreas, peritoneum, endocrine glands (adrenal, parathyroid, pituitary, testicles, ovary, thymus, thyroid), eye, head and neck, nervous (central and peripheral), lymphatic system, pelvic, skin, soft tissue, spleen, thoracic, and urogenital.
Similarly, other hypeφroliferative disorders can also be treated or detected by a polynucleotide or polypeptide of the present invention. Examples of such hypeφroliferative disorders include, but are not limited to: hypergammaglobulinemia, lymphoproliferative disorders, paraproteinemias, puφura, sarcoidosis, Sezary Syndrome, Waldenstron's Macroglobulinemia, Gaucher's Disease, histiocytosis, and any other hypeφroliferative disease, besides neoplasia, located in an organ system listed above.
Infectious Disease A polypeptide or polynucleotide of the present invention can be used to treat or detect infectious agents. For example, by increasing the immune response, particularly increasing the proliferation and differentiation of B and/or T cells, infectious diseases may be treated. The immune response may be increased by either enhancing an existing immune response, or by initiating a new immune response. Alternatively, the polypeptide or polynucleotide of the present invention may also directly inhibit the infectious agent, without necessarily eliciting an immune response.
Viruses are one example of an infectious agent that can cause disease or symptoms that can be treated or detected by a polynucleotide or polypeptide of the present invention. Examples of viruses, include, but are not limited to the following DNA and RNA viral families: Arbovirus, Adenoviridae, Arenaviridae, Arterivirus, Birnaviridae, Bunyaviridae, Caliciviridae, Circoviridae, Coronaviridae, Flaviviridae, Hepadnaviridae (Hepatitis), Herpesviridae (such as, Cytomegalovirus, Herpes Simplex, Heφes Zoster), Mononegavirus (e.g., Paramyxoviridae, Morbillivirus, Rhabdoviridae), Orthomyxoviridae (e.g., Influenza), Papovaviridae, Parvoviridae, Picornaviridae, Poxviridae (such as Smallpox or Vaccinia), Reoviridae (e.g., Rotavirus), Retroviridae (HTLV-I, HTLV-II, Lentivirus), and Togaviridae (e.g., Rubivirus). Viruses falling within these families can cause a variety of diseases or symptoms, including, but not limited to: arthritis, bronchiollitis, encephalitis, eye infections (e.g., conjunctivitis, keratitis), chronic fatigue syndrome, hepatitis (A, B, C, E, Chronic Active, Delta), meningitis, opportunistic infections (e.g., AIDS), pneumonia, Burkitt's Lymphoma, chickenpox , hemorrhagic fever, Measles, Mumps, Parainfluenza, Rabies, the common cold, Polio, leukemia, Rubella, sexually transmitted diseases, skin diseases (e.g., Kaposi's, warts), and viremia. A polypeptide or polynucleotide of the present invention can be used to treat or detect any of these symptoms or diseases. Similarly, bacterial or fungal agents that can cause disease or symptoms and that can be treated or detected by a polynucleotide or polypeptide of the present invention include, but not limited to, the following Gram-Negative and Gram-positive bacterial families and fungi: Actinomycetales (e.g., Corynebacterium, Mycobacterium, Norcardia), Aspergillosis, Bacillaceae (e.g., Anthrax, Clostridium), Bacteroidaceae, Blastomycosis, Bordetella, Borrelia, Brucellosis, Candidiasis, Campylobacter,
Coccidioidomycosis, Cryptococcosis, Dermatocycoses, Enterobacteriaceae (Klebsiella, Salmonella, Serratia, Yersinia), Erysipelothrix, Helicobacter, Legionellosis, Leptospirosis, Listeria, Mycoplasmatales, Neisseriaceae (e.g., Acinetobacter, Gonorrhea, Menigococcal), Pasteurellacea Infections (e.g., Actinobacillus, Heamophilus, Pasteurella), Pseudomonas, Rickettsiaceae, Chlamydiaceae, Syphilis, and Staphylococcal. These bacterial or fungal families can cause the following diseases or symptoms, including, but not limited to: bacteremia, endocarditis, eye infections (conjunctivitis, tuberculosis, uveitis), gingivitis, opportunistic infections (e.g., AIDS related infections), paronychia, prosthesis-related infections, Reiter's Disease, respiratory tract infections, such as Whooping Cough or Empyema, sepsis, Lyme Disease, Cat-Scratch Disease, Dysentery, Paratyphoid Fever, food poisoning, Typhoid, pneumonia, Gonorrhea, meningitis, Chlamydia, Syphilis, Diphtheria,
Leprosy, Paratuberculosis, Tuberculosis, Lupus, Botulism, gangrene, tetanus, impetigo, Rheumatic Fever, Scarlet Fever, sexually transmitted diseases, skin diseases (e.g., cellulitis, dermatocycoses), toxemia, urinary tract infections, wound infections. A polypeptide or polynucleotide of the present invention can be used to treat or detect any of these symptoms or diseases.
Moreover, parasitic agents causing disease or symptoms that can be treated or detected by a polynucleotide or polypeptide of the present invention include, but not limited to, the following families: Amebiasis, Babesiosis, Coccidiosis, Cryptosporidiosis, Dientamoebiasis, Dourine, Ectoparasitic, Giardiasis, Helminthiasis, Leishmaniasis, Theileriasis, Toxoplasmosis, Trypanosomiasis, and Trichomonas.
These parasites can cause a variety of diseases or symptoms, including, but not limited to: Scabies, Trombiculiasis, eye infections, intestinal disease (e.g., dysentery, giardiasis), liver disease, lung disease, opportunistic infections (e.g., AIDS related), Malaria, pregnancy complications, and toxoplasmosis. A polypeptide or polynucleotide of the present invention can be used to treat or detect any of these symptoms or diseases.
Preferably, treatment using a polypeptide or polynucleotide of the present invention could either be by administering an effective amount of a polypeptide to the patient, or by removing cells from the patient, supplying the cells with a polynucleotide of the present invention, and returning the engineered cells to the patient (ex vivo therapy). Moreover, the polypeptide or polynucleotide of the present invention can be used as an antigen in a vaccine to raise an immune response against infectious disease.
Regeneration A polynucleotide or polypeptide of the present invention can be used to differentiate, proliferate, and attract cells, leading to the regeneration of tissues. (See, Science 276:59-87 (1997).) The regeneration of tissues could be used to repair, replace, or protect tissue damaged by congenital defects, trauma (wounds, burns, incisions, or ulcers), age, disease (e.g. osteoporosis, osteocarthritis, periodontal disease, liver failure), surgery, including cosmetic plastic surgery, fibrosis, reperfusion injury, or systemic cytokine damage.
Tissues that could be regenerated using the present invention include organs (e.g., pancreas, liver, intestine, kidney, skin, endothelium), muscle (smooth, skeletal or cardiac), vascular (including vascular endothelium), nervous, hematopoietic, and skeletal (bone, cartilage, tendon, and ligament) tissue. Preferably, regeneration occurs without or decreased scarring. Regeneration also may include angiogenesis.
Moreover, a polynucleotide or polypeptide of the present invention may increase regeneration of tissues difficult to heal. For example, increased tendon/ligament regeneration would quicken recovery time after damage. A polynucleotide or polypeptide of the present invention could also be used prophylactically in an effort to avoid damage. Specific diseases that could be treated include of tendinitis, caφal tunnel syndrome, and other tendon or ligament defects. A further example of tissue regeneration of non-healing wounds includes pressure ulcers, ulcers associated with vascular insufficiency, surgical, and traumatic wounds.
Similarly, nerve and brain tissue could also be regenerated by using a polynucleotide or polypeptide of the present invention to proliferate and differentiate nerve cells. Diseases that could be treated using this method include central and peripheral nervous system diseases, neuropathies, or mechanical and traumatic disorders (e.g., spinal cord disorders, head trauma, cerebrovascular disease, and stoke). Specifically, diseases associated with peripheral nerve injuries, peripheral neuropathy (e.g., resulting from chemotherapy or other medical therapies), localized neuropathies, and central nervous system diseases (e.g., Alzheimer's disease, Parkinson's disease, Huntington's disease, amyotrophic lateral sclerosis, and Shy- Drager syndrome), could all be treated using the polynucleotide or polypeptide of the present invention.
Chemotaxis
A polynucleotide or polypeptide of the present invention may have chemotaxis activity. A chemotaxic molecule attracts or mobilizes cells (e.g., monocytes, fibroblasts, neutrophils, T-cells, mast cells, eosinophils, epithelial and/or endothelial cells) to a particular site in the body, such as inflammation, infection, or site of hypeφrøliferation. The mobilized cells can then fight off and/or heal the particular trauma or abnormality.
A polynucleotide or polypeptide of the present invention may increase chemotaxic activity of particular cells. These chemotactic molecules can then be used to treat inflammation, infection, hypeφroliferative disorders, or any immune system disorder by increasing the number of cells targeted to a particular location in the body. For example, chemotaxic molecules can be used to treat wounds and other trauma to tissues by attracting immune cells to the injured location. Chemotactic molecules of the present invention can also attract fibroblasts, which can be used to treat wounds. It is also contemplated that a polynucleotide or polypeptide of the present invention may inhibit chemotactic activity. These molecules could also be used to treat
disorders. Thus, a polynucleotide or polypeptide of the present invention could be used as an inhibitor of chemotaxis.
Binding Activity A polypeptide of the present invention may be used to screen for molecules that bind to the polypeptide or for molecules to which the polypeptide binds. The binding of the polypeptide and the molecule may activate (agonist), increase, inhibit (antagonist), or decrease activity of the polypeptide or the molecule bound. Examples of such molecules include antibodies, oligonucleotides, proteins (e.g., receptors),or small molecules.
Preferably, the molecule is closely related to the natural ligand of the polypeptide, e.g., a fragment of the ligand, or a natural substrate, a ligand, a structural or functional mimetic. (See, Coligan et al., Current Protocols in Immunology l(2):Chapter 5 (1991).) Similarly, the molecule can be closely related to the natural receptor to which the polypeptide binds, or at least, a fragment of the receptor capable of being bound by the polypeptide (e.g., active site). In either case, the molecule can be rationally designed using known techniques.
Preferably, the screening for these molecules involves producing appropriate cells which express the polypeptide, either as a secreted protein or on the cell membrane. Preferred cells include cells from mammals, yeast, Drosophila, or E. coli. Cells expressing the polypeptide (or cell membrane containing the expressed polypeptide) are then preferably contacted with a test compound potentially containing the molecule to observe binding, stimulation, or inhibition of activity of either the polypeptide or the molecule. The assay may simply test binding of a candidate compound to the polypeptide, wherein binding is detected by a label, or in an assay involving competition with a labeled competitor. Further, the assay may test whether the candidate compound results in a signal generated by binding to the polypeptide.
Alternatively, the assay can be carried out using cell-free preparations, polypeptide/molecule affixed to a solid support, chemical libraries, or natural product mixtures. The assay may also simply comprise the steps of mixing a candidate compound with a solution containing a polypeptide, measuring polypeptide/molecule activity or binding, and comparing the polypeptide/molecule activity or binding to a standard. Preferably, an ELISA assay can measure polypeptide level or activity in a sample (e.g., biological sample) using a monoclonal or polyclonal antibody. The
antibody can measure polypeptide level or activity by either binding, directly or indirectly, to the polypeptide or by competing with the polypeptide for a substrate.
All of these above assays can be used as diagnostic or prognostic markers. The molecules discovered using these assays can be used to treat disease or to bring about a particular result in a patient (e.g., blood vessel growth) by activating or inhibiting the polypeptide/molecule. Moreover, the assays can discover agents which may inhibit or enhance the production of the polypeptide from suitably manipulated cells or tissues. Therefore, the invention includes a method of identifying compounds which bind to a polypeptide of the invention comprising the steps of: (a) incubating a candidate binding compound with a polypeptide of the invention; and (b) determining if binding has occurred. Moreover, the invention includes a method of identifying agonists/antagonists comprising the steps of: (a) incubating a candidate compound with a polypeptide of the invention, (b) assaying a biological activity , and (b) determining if a biological activity of the polypeptide has been altered.
Other Activities
A polypeptide or polynucleotide of the present invention may also increase or decrease the differentiation or proliferation of embryonic stem cells, besides, as discussed above, hematopoietic lineage. A polypeptide or polynucleotide of the present invention may also be used to modulate mammalian characteristics, such as body height, weight, hair color, eye color, skin, percentage of adipose tissue, pigmentation, size, and shape (e.g., cosmetic surgery). Similarly, a polypeptide or polynucleotide of the present invention may be used to modulate mammalian metabolism affecting catabolism, anabolism, processing, utilization, and storage of energy.
A polypeptide or polynucleotide of the present invention may be used to change a mammal's mental state or physical state by influencing biorhythms, caricadic rhythms, depression (including depressive disorders), tendency for violence, tolerance for pain, reproductive capabilities (preferably by Activin or Inhibin-like activity), hormonal or endocrine levels, appetite, libido, memory, stress, or other cognitive qualities.
A polypeptide or polynucleotide of the present invention may also be used as a food additive or preservative, such as to increase or decrease storage capabilities, fat content, lipid, protein, carbohydrate, vitamins, minerals, cofactors or other nutritional components.
Other Preferred Embodiments
Other preferred embodiments of the claimed invention include an isolated nucleic acid molecule comprising a nucleotide sequence which is at least 95% identical to a sequence of at least about 50 contiguous nucleotides in the nucleotide sequence of SEQ ID NO:X wherein X is any integer as defined in Table 1.
Also preferred is a nucleic acid molecule wherein said sequence of contiguous nucleotides is included in the nucleotide sequence of SEQ ID NO:X in the range of positions beginning with the nucleotide at about the position of the 5' Nucleotide of the Clone Sequence and ending with the nucleotide at about the position of the 3' Nucleotide of the Clone Sequence as defined for SEQ ID NO:X in Table 1.
Also preferred is a nucleic acid molecule wherein said sequence of contiguous nucleotides is included in the nucleotide sequence of SEQ ID NO:X in the range of positions beginning with the nucleotide at about the position of the 5' Nucleotide of the Start Codon and ending with the nucleotide at about the position of the 3' Nucleotide of the Clone Sequence as defined for SEQ ID NO:X in Table 1.
Similarly preferred is a nucleic acid molecule wherein said sequence of contiguous nucleotides is included in the nucleotide sequence of SEQ ID NO:X in the range of positions beginning with the nucleotide at about the position of the 5' Nucleotide of the First Amino Acid of the Signal Peptide and ending with the nucleotide at about the position of the 3' Nucleotide of the Clone Sequence as defined for SEQ ID NO:X in Table 1.
Also preferred is an isolated nucleic acid molecule comprising a nucleotide sequence which is at least 95% identical to a sequence of at least about 150 contiguous nucleotides in the nucleotide sequence of SEQ ID NO:X. Further preferred is an isolated nucleic acid molecule comprising a nucleotide sequence which is at least 95% identical to a sequence of at least about 500 contiguous nucleotides in the nucleotide sequence of SEQ ID NO:X.
A further preferred embodiment is a nucleic acid molecule comprising a nucleotide sequence which is at least 95% identical to the nucleotide sequence of SEQ ID NO:X beginning with the nucleotide at about the position of the 5' Nucleotide of the First Amino Acid of the Signal Peptide and ending with the nucleotide at about the position of the 3' Nucleotide of the Clone Sequence as defined for SEQ ID NO:X in Table 1.
A further preferred embodiment is an isolated nucleic acid molecule comprising a nucleotide sequence which is at least 95% identical to the complete nucleotide sequence of SEQ ID NO:X.
Also preferred is an isolated nucleic acid molecule which hybridizes under stringent hybridization conditions to a nucleic acid molecule, wherein said nucleic acid molecule which hybridizes does not hybridize under stringent hybridization conditions to a nucleic acid molecule having a nucleotide sequence consisting of only A residues or of only T residues.
Also preferred is a composition of matter comprising a DNA molecule which comprises a human cDNA clone identified by a cDNA Clone Identifier in Table 1, which DNA molecule is contained in the material deposited with the American Type Culture Collection and given the ATCC Deposit Number shown in Table 1 for said cDNA Clone Identifier.
Also preferred is an isolated nucleic acid molecule comprising a nucleotide sequence which is at least 95% identical to a sequence of at least 50 contiguous nucleotides in the nucleotide sequence of a human cDNA clone identified by a cDNA Clone Identifier in Table 1 , which DNA molecule is contained in the deposit given the ATCC Deposit Number shown in Table 1.
Also preferred is an isolated nucleic acid molecule, wherein said sequence of at least 50 contiguous nucleotides is included in the nucleotide sequence of the complete open reading frame sequence encoded by said human cDNA clone.
Also preferred is an isolated nucleic acid molecule comprising a nucleotide sequence which is at least 95% identical to sequence of at least 150 contiguous nucleotides in the nucleotide sequence encoded by said human cDNA clone.
A further preferred embodiment is an isolated nucleic acid molecule comprising a nucleotide sequence which is at least 95% identical to sequence of at least 500 contiguous nucleotides in the nucleotide sequence encoded by said human cDNA clone. A further preferred embodiment is an isolated nucleic acid molecule comprising a nucleotide sequence which is at least 95% identical to the complete nucleotide sequence encoded by said human cDNA clone.
A further preferred embodiment is a method for detecting in a biological sample a nucleic acid molecule comprising a nucleotide sequence which is at least 95% identical to a sequence of at least 50 contiguous nucleotides in a sequence selected from the group consisting of: a nucleotide sequence of SEQ ID NO:X wherein X is any integer as defined in Table 1 ; and a nucleotide sequence encoded by a human cDNA clone identified by a cDNA Clone Identifier in Table 1 and contained in the deposit with the ATCC Deposit Number shown for said cDNA clone in Table 1 ; which method comprises a step of comparing a nucleotide sequence of at least one nucleic acid molecule in said sample with a sequence selected from said group and determining
whether the sequence of said nucleic acid molecule in said sample is at least 95% identical to said selected sequence.
Also preferred is the above method wherein said step of comparing sequences comprises determining the extent of nucleic acid hybridization between nucleic acid molecules in said sample and a nucleic acid molecule comprising said sequence selected from said group. Similarly, also preferred is the above method wherein said step of comparing sequences is performed by comparing the nucleotide sequence determined from a nucleic acid molecule in said sample with said sequence selected from said group. The nucleic acid molecules can comprise DNA molecules or RNA molecules. A further preferred embodiment is a method for identifying the species, tissue or cell type of a biological sample which method comprises a step of detecting nucleic acid molecules in said sample, if any, comprising a nucleotide sequence that is at least 95% identical to a sequence of at least 50 contiguous nucleotides in a sequence selected from the group consisting of: a nucleotide sequence of SEQ ID NO:X wherein X is any integer as defined in Table 1; and a nucleotide sequence encoded by a human cDNA clone identified by a cDNA Clone Identifier in Table 1 and contained in the deposit with the ATCC Deposit Number shown for said cDNA clone in Table 1.
The method for identifying the species, tissue or cell type of a biological sample can comprise a step of detecting nucleic acid molecules comprising a nucleotide sequence in a panel of at least two nucleotide sequences, wherein at least one sequence in said panel is at least 95% identical to a sequence of at least 50 contiguous nucleotides in a sequence selected from said group.
Also preferred is a method for diagnosing in a subject a pathological condition associated with abnormal structure or expression of a gene encoding a secreted protein identified in Table 1 , which method comprises a step of detecting in a biological sample obtained from said subject nucleic acid molecules, if any, comprising a nucleotide sequence that is at least 95% identical to a sequence of at least 50 contiguous nucleotides in a sequence selected from the group consisting of: a nucleotide sequence of SEQ ID NO:X wherein X is any integer as defined in Table 1 ; and a nucleotide sequence encoded by a human cDNA clone identified by a cDNA Clone Identifier in Table 1 and contained in the deposit with the ATCC Deposit Number shown for said cDNA clone in Table 1.
The method for diagnosing a pathological condition can comprise a step of detecting nucleic acid molecules comprising a nucleotide sequence in a panel of at least two nucleotide sequences, wherein at least one sequence in said panel is at least 95% identical to a sequence of at least 50 contiguous nucleotides in a sequence selected from said group.
Also preferred is a composition of matter comprising isolated nucleic acid molecules wherein the nucleotide sequences of said nucleic acid molecules comprise a panel of at least two nucleotide sequences, wherein at least one sequence in said panel is at least 95% identical to a sequence of at least 50 contiguous nucleotides in a sequence selected from the group consisting of: a nucleotide sequence of SEQ ID NO:X wherein X is any integer as defined in Table 1 ; and a nucleotide sequence encoded by a human cDNA clone identified by a cDNA Clone Identifier in Table 1 and contained in the deposit with the ATCC Deposit Number shown for said cDNA clone in Table 1. The nucleic acid molecules can comprise DNA molecules or RNA molecules. Also preferred is an isolated polypeptide comprising an amino acid sequence at least 90% identical to a sequence of at least about 10 contiguous amino acids in the amino acid sequence of SEQ ID NO:Y wherein Y is any integer as defined in Table 1.
Also preferred is a polypeptide, wherein said sequence of contiguous amino acids is included in the amino acid sequence of SEQ ID NO:Y in the range of positions beginning with the residue at about the position of the First Amino Acid of the Secreted Portion and ending with the residue at about the Last Amino Acid of the Open Reading Frame as set forth for SEQ ID NO:Y in Table 1.
Also preferred is an isolated polypeptide comprising an amino acid sequence at least 95% identical to a sequence of at least about 30 contiguous amino acids in the amino acid sequence of SEQ ID NO: Y.
Further preferred is an isolated polypeptide comprising an amino acid sequence at least 95% identical to a sequence of at least about 100 contiguous amino acids in the amino acid sequence of SEQ ID NO: Y.
Further preferred is an isolated polypeptide comprising an amino acid sequence at least 95% identical to the complete amino acid sequence of SEQ ID NO:Y.
Further preferred is an isolated polypeptide comprising an amino acid sequence at least 90% identical to a sequence of at least about 10 contiguous amino acids in the complete amino acid sequence of a secreted protein encoded by a human cDNA clone identified by a cDNA Clone Identifier in Table 1 and contained in the deposit with the ATCC Deposit Number shown for said cDNA clone in Table 1.
Also preferred is a polypeptide wherein said sequence of contiguous amino acids is included in the amino acid sequence of a secreted portion of the secreted protein encoded by a human cDNA clone identified by a cDNA Clone Identifier in Table 1 and contained in the deposit with the ATCC Deposit Number shown for said cDNA clone in Table 1.
Also preferred is an isolated polypeptide comprising an amino acid sequence at least 95% identical to a sequence of at least about 30 contiguous amino acids in the
amino acid sequence of the secreted portion of the protein encoded by a human cDNA clone identified by a cDNA Clone Identifier in Table 1 and contained in the deposit with the ATCC Deposit Number shown for said cDNA clone in Table 1.
Also preferred is an isolated polypeptide comprising an amino acid sequence at least 95% identical to a sequence of at least about 100 contiguous amino acids in the amino acid sequence of the secreted portion of the protein encoded by a human cDNA clone identified by a cDNA Clone Identifier in Table 1 and contained in the deposit with the ATCC Deposit Number shown for said cDNA clone in Table 1.
Also preferred is an isolated polypeptide comprising an amino acid sequence at least 95% identical to the amino acid sequence of the secreted portion of the protein encoded by a human cDNA clone identified by a cDNA Clone Identifier in Table 1 and contained in the deposit with the ATCC Deposit Number shown for said cDNA clone in Table 1.
Further preferred is an isolated antibody which binds specifically to a polypeptide comprising an amino acid sequence that is at least 90% identical to a sequence of at least 10 contiguous amino acids in a sequence selected from the group consisting of: an amino acid sequence of SEQ ID NO: Y wherein Y is any integer as defined in Table 1 ; and a complete amino acid sequence of a protein encoded by a human cDNA clone identified by a cDNA Clone Identifier in Table 1 and contained in the deposit with the ATCC Deposit Number shown for said cDNA clone in Table 1.
Further preferred is a method for detecting in a biological sample a polypeptide comprising an amino acid sequence which is at least 90% identical to a sequence of at least 10 contiguous amino acids in a sequence selected from the group consisting of: an amino acid sequence of SEQ ID NO:Y wherein Y is any integer as defined in Table 1; and a complete amino acid sequence of a protein encoded by a human cDNA clone identified by a cDNA Clone Identifier in Table 1 and contained in the deposit with the ATCC Deposit Number shown for said cDNA clone in Table 1 ; which method comprises a step of comparing an amino acid sequence of at least one polypeptide molecule in said sample with a sequence selected from said group and determining whether the sequence of said polypeptide molecule in said sample is at least 90% identical to said sequence of at least 10 contiguous amino acids.
Also preferred is the above method wherein said step of comparing an amino acid sequence of at least one polypeptide molecule in said sample with a sequence selected from said group comprises determining the extent of specific binding of polypeptides in said sample to an antibody which binds specifically to a polypeptide comprising an amino acid sequence that is at least 90% identical to a sequence of at least 10 contiguous amino acids in a sequence selected from the group consisting of: an
amino acid sequence of SEQ ID NON wherein Y is any integer as defined in Table 1 ; and a complete amino acid sequence of a protein encoded by a human cDΝA clone identified by a cDΝA Clone Identifier in Table 1 and contained in the deposit with the ATCC Deposit Number shown for said cDNA clone in Table 1. Also preferred is the above method wherein said step of comparing sequences is performed by comparing the amino acid sequence determined from a polypeptide molecule in said sample with said sequence selected from said group.
Also preferred is a method for identifying the species, tissue or cell type of a biological sample which method comprises a step of detecting polypeptide molecules in said sample, if any, comprising an amino acid sequence that is at least 90% identical to a sequence of at least 10 contiguous amino acids in a sequence selected from the group consisting of: an amino acid sequence of SEQ ID NO: Y wherein Y is any integer as defined in Table 1 ; and a complete amino acid sequence of a secreted protein encoded by a human cDNA clone identified by a cDNA Clone Identifier in Table 1 and contained in the deposit with the ATCC Deposit Number shown for said cDNA clone in Table 1. Also preferred is the above method for identifying the species, tissue or cell type of a biological sample, which method comprises a step of detecting polypeptide molecules comprising an amino acid sequence in a panel of at least two amino acid sequences, wherein at least one sequence in said panel is at least 90% identical to a sequence of at least 10 contiguous amino acids in a sequence selected from the above group.
Also preferred is a method for diagnosing in a subject a pathological condition associated with abnormal structure or expression of a gene encoding a secreted protein identified in Table 1 , which method comprises a step of detecting in a biological sample obtained from said subject polypeptide molecules comprising an amino acid sequence in a panel of at least two amino acid sequences, wherein at least one sequence in said panel is at least 90% identical to a sequence of at least 10 contiguous amino acids in a sequence selected from the group consisting of: an amino acid sequence of SEQ ID NO:Y wherein Y is any integer as defined in Table 1; and a complete amino acid sequence of a secreted protein encoded by a human cDNA clone identified by a cDNA Clone Identifier in Table 1 and contained in the deposit with the ATCC Deposit Number shown for said cDNA clone in Table 1.
In any of these methods, the step of detecting said polypeptide molecules includes using an antibody. Also preferred is an isolated nucleic acid molecule comprising a nucleotide sequence which is at least 95% identical to a nucleotide sequence encoding a polypeptide wherein said polypeptide comprises an amino acid sequence that is at least
90% identical to a sequence of at least 10 contiguous amino acids in a sequence selected from the group consisting of: an amino acid sequence of SEQ ID NON wherein Y is any integer as defined in Table 1 ; and a complete amino acid sequence of a secreted protein encoded by a human cDΝA clone identified by a cDΝA Clone Identifier in Table 1 and contained in the deposit with the ATCC Deposit Number shown for said cDNA clone in Table 1.
Also preferred is an isolated nucleic acid molecule, wherein said nucleotide sequence encoding a polypeptide has been optimized for expression of said polypeptide in a prokaryotic host. Also preferred is an isolated nucleic acid molecule, wherein said polypeptide comprises an amino acid sequence selected from the group consisting of: an amino acid sequence of SEQ ID NO:Y wherein Y is any integer as defined in Table 1; and a complete amino acid sequence of a secreted protein encoded by a human cDNA clone identified by a cDNA Clone Identifier in Table 1 and contained in the deposit with the ATCC Deposit Number shown for said cDNA clone in Table 1.
Further preferred is a method of making a recombinant vector comprising inserting any of the above isolated nucleic acid molecule into a vector. Also preferred is the recombinant vector produced by this method. Also preferred is a method of making a recombinant host cell comprising introducing the vector into a host cell, as well as the recombinant host cell produced by this method.
Also preferred is a method of making an isolated polypeptide comprising culturing this recombinant host cell under conditions such that said polypeptide is expressed and recovering said polypeptide. Also preferred is this method of making an isolated polypeptide, wherein said recombinant host cell is a eukaryotic cell and said polypeptide is a secreted portion of a human secreted protein comprising an amino acid sequence selected from the group consisting of: an amino acid sequence of SEQ ID NO: Y beginning with the residue at the position of the First Amino Acid of the Secreted Portion of SEQ ID NO: Y wherein Y is an integer set forth in Table 1 and said position of the First Amino Acid of the Secreted Portion of SEQ ID NO: Y is defined in Table 1 ; and an amino acid sequence of a secreted portion of a protein encoded by a human cDNA clone identified by a cDNA Clone Identifier in Table 1 and contained in the deposit with the ATCC Deposit Number shown for said cDNA clone in Table 1. The isolated polypeptide produced by this method is also preferred.
Also preferred is a method of treatment of an individual in need of an increased level of a secreted protein activity, which method comprises administering to such an individual a pharmaceutical composition comprising an amount of an isolated
polypeptide, polynucleotide, or antibody of the claimed invention effective to increase the level of said protein activity in said individual.
Having generally described the invention, the same will be more readily understood by reference to the following examples, which are provided by way of illustration and are not intended as limiting.
Examples Example 1; Isolation of a Selected cDNA Clone From the Deposited Sample Each cDNA clone in a cited ATCC deposit is contained in a plasmid vector.
Table 1 identifies the vectors used to construct the cDNA library from which each clone was isolated. In many cases, the vector used to construct the library is a phage vector from which a plasmid has been excised. The table immediately below correlates the related plasmid for each phage vector used in constructing the cDNA library. For example, where a particular clone is identified in Table 1 as being isolated in the vector "Lambda Zap," the corresponding deposited clone is in "pBluescript."
Vector Used to Construct Library Corresponding Deposited Plasmid
Lambda Zap pBluescript (pBS)
Uni-Zap XR pBluescript (pBS) Zap Express pBK lafmid BA plafmid BA pSportl pSportl pCMVSport 2.0 pCMVSport 2.0 pCMVSport 3.0 pCMVSport 3.0 pCR®2.1 pCR®2.1
Vectors Lambda Zap (U.S. Patent Nos. 5,128,256 and 5,286,636), Uni-Zap XR (U.S. Patent Nos. 5,128, 256 and 5,286,636), Zap Express (U.S. Patent Nos. 5,128,256 and 5,286,636), pBluescript (pBS) (Short, J. M. et al., Nucleic Acids Res. 16:7583-7600 (1988); Alting-Mees, M. A. and Short, J. M., Nucleic Acids Res. 17:9494 (1989)) and pBK (Alting-Mees, M. A. et al., Strategies 5:58-61 (1992)) are commercially available from Stratagene Cloning Systems, Inc., 11011 N. Torrey Pines Road, La Jolla, CA, 92037. pBS contains an ampicillin resistance gene and pBK contains a neomycin resistance gene. Both can be transformed into E. coli strain XL- 1 Blue, also available from Stratagene. pBS comes in 4 forms SK+, SK-, KS+ and KS. The S and K refers to the orientation of the polylinker to the T7 and T3 primer sequences which flank the polylinker region ("S" is for Sad and "K" is for Kpnl which are the first sites on each respective end of the linker). "+" or "-" refer to the orientation
of the fl origin of replication ("ori"), such that in one orientation, single stranded rescue initiated from the f 1 ori generates sense strand DNA and in the other, antisense.
Vectors pSportl, pCMVSport 2.0 and pCMVSport 3.0, were obtained from Life Technologies, Inc., P. O. Box 6009, Gaithersburg, MD 20897. All Sport vectors contain an ampicillin resistance gene and may be transformed into E. coli strain
DH10B, also available from Life Technologies. (See, for instance, Gruber, C. E., et al., Focus 15:59 (1993).) Vector lafmid BA (Bento Soares, Columbia University, NY) contains an ampicillin resistance gene and can be transformed into E. coli strain XL-1 Blue. Vector pCR®2.1, which is available from Invitrogen, 1600 Faraday Avenue, Carlsbad, CA 92008, contains an ampicillin resistance gene and may be transformed into E. coli strain DH10B, available from Life Technologies. (See, for instance, Clark, J. M., Nuc. Acids Res. 16:9677-9686 (1988) and Mead, D. et al., Bio/Technology 9: (1991).) Preferably, a polynucleotide of the present invention does not comprise the phage vector sequences identified for the particular clone in Table 1, as well as the corresponding plasmid vector sequences designated above.
The deposited material in the sample assigned the ATCC Deposit Number cited in Table 1 for any given cDNA clone also may contain one or more additional plasmids, each comprising a cDNA clone different from that given clone. Thus, deposits sharing the same ATCC Deposit Number contain at least a plasmid for each cDNA clone identified in Table 1. Typically, each ATCC deposit sample cited in Table 1 comprises a mixture of approximately equal amounts (by weight) of about 50 plasmid DNAs, each containing a different cDNA clone; but such a deposit sample may include plasmids for more or less than 50 cDNA clones, up to about 500 cDNA clones.
Two approaches can be used to isolate a particular clone from the deposited sample of plasmid DNAs cited for that clone in Table 1. First, a plasmid is directly isolated by screening the clones using a polynucleotide probe corresponding to SEQ ID NO:X.
Particularly, a specific polynucleotide with 30-40 nucleotides is synthesized using an Applied Biosystems DNA synthesizer according to the sequence reported. The oligonucleotide is labeled, for instance, with 32P-γ-ATP using T4 polynucleotide kinase and purified according to routine methods. (E.g., Maniatis et al., Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Press, Cold Spring, NY (1982).) The plasmid mixture is transformed into a suitable host, as indicated above (such as XL-1 Blue (Stratagene)) using techniques known to those of skill in the art, such as those provided by the vector supplier or in related publications or patents cited above. The transformants are plated on 1.5% agar plates (containing the appropriate selection
agent, e.g., ampicillin) to a density of about 150 transformants (colonies) per plate. These plates are screened using Nylon membranes according to routine methods for bacterial colony screening (e.g., Sambrook et al., Molecular Cloning: A Laboratory Manual, 2nd Edit., (1989), Cold Spring Harbor Laboratory Press, pages 1.93 to 1.104), or other techniques known to those of skill in the art.
Alternatively, two primers of 17-20 nucleotides derived from both ends of the SEQ ID NO:X (i.e., within the region of SEQ ID NO:X bounded by the 5' NT and the 3' NT of the clone defined in Table 1) are synthesized and used to amplify the desired cDNA using the deposited cDNA plasmid as a template. The polymerase chain reaction is carried out under routine conditions, for instance, in 25 μl of reaction mixture with 0.5 ug of the above cDNA template. A convenient reaction mixture is 1.5-5 mM MgCl2, 0.01% (w/v) gelatin, 20 μM each of dATP, dCTP, dGTP, dTTP, 25 pmol of each primer and 0.25 Unit of Taq polymerase. Thirty five cycles of PCR (denaturation at 94°C for 1 min; annealing at 55°C for 1 min; elongation at 72°C for 1 min) are performed with a Perkin-Elmer Cetus automated thermal cycler. The amplified product is analyzed by agarose gel electrophoresis and the DNA band with expected molecular weight is excised and purified. The PCR product is verified to be the selected sequence by subcloning and sequencing the DNA product.
Several methods are available for the identification of the 5' or 3' non-coding portions of a gene which may not be present in the deposited clone. These methods include but are not limited to, filter probing, clone enrichment using specific probes, and protocols similar or identical to 5' and 3' "RACE" protocols which are well known in the art. For instance, a method similar to 5' RACE is available for generating the missing 5' end of a desired full-length transcript. (Fromont-Racine et al., Nucleic Acids Res. 21(7): 1683-1684 (1993).)
Briefly, a specific RNA oligonucleotide is ligated to the 5' ends of a population of RNA presumably containing full-length gene RNA transcripts. A primer set containing a primer specific to the ligated RNA oligonucleotide and a primer specific to a known sequence of the gene of interest is used to PCR amplify the 5' portion of the desired full-length gene. This amplified product may then be sequenced and used to generate the full length gene.
This above method starts with total RNA isolated from the desired source, although poly-A+ RNA can be used. The RNA preparation can then be treated with phosphatase if necessary to eliminate 5' phosphate groups on degraded or damaged RNA which may interfere with the later RNA ligase step. The phosphatase should then be inactivated and the RNA treated with tobacco acid pyrophosphatase in order to
remove the cap structure present at the 5' ends of messenger RNAs. This reaction leaves a 5' phosphate group at the 5' end of the cap cleaved RNA which can then be ligated to an RNA oligonucleotide using T4 RNA ligase.
This modified RNA preparation is used as a template for first strand cDNA synthesis using a gene specific oligonucleotide. The first strand synthesis reaction is used as a template for PCR amplification of the desired 5' end using a primer specific to the ligated RNA oligonucleotide and a primer specific to the known sequence of the gene of interest. The resultant product is then sequenced and analyzed to confirm that the 5' end sequence belongs to the desired gene.
Example 2: Isolation of Genomic Clones Corresponding to a
Polynucleotide
A human genomic PI library (Genomic Systems, Inc.) is screened by PCR using primers selected for the cDNA sequence corresponding to SEQ ID NO:X., according to the method described in Example 1. (See also, Sambrook.)
Example 3: Tissue Distribution of Polypeptide
Tissue distribution of mRNA expression of polynucleotides of the present invention is determined using protocols for Northern blot analysis, described by, among others, Sambrook et al. For example, a cDNA probe produced by the method described in Example 1 is labeled with P32 using the rediprime™ DNA labeling system (Amersham Life Science), according to manufacturer's instructions. After labeling, the probe is purified using CHROMA SPIN- 100™ column (Clontech Laboratories, Inc.), according to manufacturer's protocol number PT 1200-1. The purified labeled probe is then used to examine various human tissues for mRNA expression.
Multiple Tissue Northern (MTN) blots containing various human tissues (H) or human immune system tissues (IM) (Clontech) are examined with the labeled probe using ExpressHyb™ hybridization solution (Clontech) according to manufacturer's protocol number PT1190-1. Following hybridization and washing, the blots are mounted and exposed to film at -70°C overnight, and the films developed according to standard procedures.
Example 4: Chromosomal Mapping of the Polynucleotides
An oligonucleotide primer set is designed according to the sequence at the 5' end of SEQ ID NO:X. This primer preferably spans about 100 nucleotides. This primer set is then used in a polymerase chain reaction under the following set of
conditions : 30 seconds, 95°C; 1 minute, 56°C; 1 minute, 70°C. This cycle is repeated
32 times followed by one 5 minute cycle at 70°C. Human, mouse, and hamster DNA is used as template in addition to a somatic cell hybrid panel containing individual chromosomes or chromosome fragments (Bios, Inc). The reactions is analyzed on either 8% polyacrylamide gels or 3.5 % agarose gels. Chromosome mapping is determined by the presence of an approximately 100 bp PCR fragment in the particular somatic cell hybrid.
Example 5: Bacterial Expression of a Polypeptide A polynucleotide encoding a polypeptide of the present invention is amplified using PCR oligonucleotide primers corresponding to the 5' and 3' ends of the DNA sequence, as outlined in Example 1, to synthesize insertion fragments. The primers used to amplify the cDNA insert should preferably contain restriction sites, such as BamHl and Xbal, at the 5' end of the primers in order to clone the amplified product into the expression vector. For example, BamHl and Xbal correspond to the restriction enzyme sites on the bacterial expression vector pQE-9. (Qiagen, Inc., Chatsworth,
CA). This plasmid vector encodes antibiotic resistance (Amp1*), a bacterial origin of replication (ori), an IPTG-regulatable promoter/operator (P/O), a ribosome binding site (RBS), a 6-histidine tag (6-His), and restriction enzyme cloning sites. The pQE-9 vector is digested with BamHl and Xbal and the amplified fragment is ligated into the pQE-9 vector maintaining the reading frame initiated at the bacterial RBS. The ligation mixture is then used to transform the E. coli strain M15/rep4 (Qiagen, Inc.) which contains multiple copies of the plasmid pREP4, which expresses the lad repressor and also confers kanamycin resistance (Kan1)- Transformants are identified by their ability to grow on LB plates and ampicillin/kanamycin resistant colonies are selected. Plasmid DNA is isolated and confirmed by restriction analysis. Clones containing the desired constructs are grown overnight (O/N) in liquid culture in LB media supplemented with both Amp (100 ug/ml) and Kan (25 ug/ml).
The O/N culture is used to inoculate a large culture at a ratio of 1:100 to 1:250. The cells are grown to an optical density 600 (O.D.600) of between 0.4 and 0.6. IPTG
(Isopropyl-B-D-thiogalacto pyranoside) is then added to a final concentration of 1 mM.
IPTG induces by inactivating the lad repressor, clearing the P/O leading to increased gene expression.
Cells are grown for an extra 3 to 4 hours. Cells are then harvested by centrifugation (20 mins at 6000Xg). The cell pellet is solubilized in the chaotropic
agent 6 Molar Guanidine HC1 by stirring for 3-4 hours at 4°C. The cell debris is removed by centrifugation, and the supernatant containing the polypeptide is loaded onto a nickel-nitrilo-tri-acetic acid ("Ni-NTA") affinity resin column (available from QIAGEN, Inc., supra). Proteins with a 6 x His tag bind to the Ni-NTA resin with high affinity and can be purified in a simple one-step procedure (for details see: The QIAexpressionist (1995) QIAGEN, Inc., supra).
Briefly, the supernatant is loaded onto the column in 6 M guanidine-HCl, pH 8, the column is first washed with 10 volumes of 6 M guanidine-HCl, pH 8, then washed with 10 volumes of 6 M guanidine-HCl pH 6, and finally the polypeptide is eluted with 6 M guanidine-HCl, pH 5.
The purified protein is then renatured by dialyzing it against phosphate-buffered saline (PBS) or 50 mM Na-acetate, pH 6 buffer plus 200 mM NaCl. Alternatively, the protein can be successfully refolded while immobilized on the Ni-NTA column. The recommended conditions are as follows: renature using a linear 6M-1M urea gradient in 500 mM NaCl, 20% glycerol, 20 mM Tris/HCl pH 7.4, containing protease inhibitors. The renaturation should be performed over a period of 1.5 hours or more. After renaturation the proteins are eluted by the addition of 250 mM immidazole. Immidazole is removed by a final dialyzing step against PBS or 50 mM sodium acetate pH 6 buffer plus 200 mM NaCl. The purified protein is stored at 4°C or frozen at -80° C. In addition to the above expression vector, the present invention further includes an expression vector comprising phage operator and promoter elements operatively linked to a polynucleotide of the present invention, called pHE4a. (ATCC Accession Number XXXXXX.) This vector contains: 1) a neomycinphosphotransferase gene as a selection marker, 2) an E. coli origin of replication, 3) a T5 phage promoter sequence, 4) two lac operator sequences, 5) a Shine-Delgarno sequence, and 6) the lactose operon repressor gene (laclq). The origin of replication (oriC) is derived from pUC19 (LTI, Gaithersburg, MD). The promoter sequence and operator sequences are made synthetically.
DNA can be inserted into the pHEa by restricting the vector with Ndel and Xbal, BamHl, Xhol, or Asp718, running the restricted product on a gel, and isolating the larger fragment (the stuffer fragment should be about 310 base pairs). The DNA insert is generated according to the PCR protocol described in Example 1, using PCR primers having restriction sites for Ndel (5' primer) and Xbal, BamHl, Xhol, or Asp718 (3' primer). The PCR insert is gel purified and restricted with compatible enzymes. The insert and vector are ligated according to standard protocols.
The engineered vector could easily be substituted in the above protocol to express protein in a bacterial system.
Example 6; Purification of a Polypeptide from an Inclusion Body The following alternative method can be used to purify a polypeptide expressed in E coli when it is present in the form of inclusion bodies. Unless otherwise specified, all of the following steps are conducted at 4-10°C.
Upon completion of the production phase of the E. coli fermentation, the cell culture is cooled to 4-10°C and the cells harvested by continuous centrifugation at 15,000 φm (Heraeus Sepatech). On the basis of the expected yield of protein per unit weight of cell paste and the amount of purified protein required, an appropriate amount of cell paste, by weight, is suspended in a buffer solution containing 100 mM Tris, 50 mM EDTA, pH 7.4. The cells are dispersed to a homogeneous suspension using a high shear mixer. The cells are then lysed by passing the solution through a microfluidizer
(Microfuidics, Coφ. or APV Gaulin, Inc.) twice at 4000-6000 psi. The homogenate is then mixed with NaCl solution to a final concentration of 0.5 M NaCl, followed by centrifugation at 7000 xg for 15 min. The resultant pellet is washed again using 0.5M NaCl, 100 mM Tris, 50 mM EDTA, pH 7.4. The resulting washed inclusion bodies are solubilized with 1.5 M guanidine hydrochloride (GuHCl) for 2-4 hours. After 7000 xg centrifugation for 15 min., the pellet is discarded and the polypeptide containing supernatant is incubated at 4°C overnight to allow further GuHCl extraction.
Following high speed centrifugation (30,000 xg) to remove insoluble particles, the GuHCl solubilized protein is refolded by quickly mixing the GuHCl extract with 20 volumes of buffer containing 50 mM sodium, pH 4.5, 150 mM NaCl, 2 mM EDTA by vigorous stirring. The refolded diluted protein solution is kept at 4°C without mixing for 12 hours prior to further purification steps.
To clarify the refolded polypeptide solution, a previously prepared tangential filtration unit equipped with 0.16 μm membrane filter with appropriate surface area
(e.g., Filtron), equilibrated with 40 mM sodium acetate, pH 6.0 is employed. The filtered sample is loaded onto a cation exchange resin (e.g., Poros HS-50, Perseptive Biosystems). The column is washed with 40 mM sodium acetate, pH 6.0 and eluted with 250 mM, 500 mM, 1000 mM, and 1500 mM NaCl in the same buffer, in a
stepwise manner. The absorbance at 280 nm of the effluent is continuously monitored. Fractions are collected and further analyzed by SDS-PAGE.
Fractions containing the polypeptide are then pooled and mixed with 4 volumes of water. The diluted sample is then loaded onto a previously prepared set of tandem columns of strong anion (Poros HQ-50, Perseptive Biosystems) and weak anion
(Poros CM-20, Perseptive Biosystems) exchange resins. The columns are equilibrated with 40 mM sodium acetate, pH 6.0. Both columns are washed with 40 mM sodium acetate, pH 6.0, 200 mM NaCl. The CM-20 column is then eluted using a 10 column volume linear gradient ranging from 0.2 M NaCl, 50 mM sodium acetate, pH 6.0 to 1.0 M NaCl, 50 mM sodium acetate, pH 6.5. Fractions are collected under constant A280 monitoring of the effluent. Fractions containing the polypeptide (determined, for instance, by 16% SDS-PAGE) are then pooled.
The resultant polypeptide should exhibit greater than 95% purity after the above refolding and purification steps. No major contaminant bands should be observed from Commassie blue stained 16% SDS-PAGE gel when 5 μg of purified protein is loaded.
The purified protein can also be tested for endotoxin LPS contamination, and typically the LPS content is less than 0.1 ng/ml according to LAL assays.
Example 7: Cloning and Expression of a Polypeptide in a Baculovirus Expression System
In this example, the plasmid shuttle vector pA2 is used to insert a polynucleotide into a baculovirus to express a polypeptide. This expression vector contains the strong polyhedrin promoter of the Autographa californica nuclear polyhedrosis virus (AcMNPV) followed by convenient restriction sites such as BamHl, Xba I and Asp718. The polyadenylation site of the simian virus 40 ("S V40") is used for efficient polyadenylation. For easy selection of recombinant virus, the plasmid contains the beta-galactosidase gene from E. coli under control of a weak Drosophila promoter in the same orientation, followed by the polyadenylation signal of the polyhedrin gene. The inserted genes are flanked on both sides by viral sequences for cell-mediated homologous recombination with wild-type viral DNA to generate a viable virus that express the cloned polynucleotide.
Many other baculovirus vectors can be used in place of the vector above, such as pAc373, pVL941, and pAcIMl, as one skilled in the art would readily appreciate, as long as the construct provides appropriately located signals for transcription, translation, secretion and the like, including a signal peptide and an in- frame AUG as
required. Such vectors are described, for instance, in Luckow et al., Virology 170:31- 39 (1989).
Specifically, the cDNA sequence contained in the deposited clone, including the AUG initiation codon and the naturally associated leader sequence identified in Table 1, is amplified using the PCR protocol described in Example 1. If the naturally occurring signal sequence is used to produce the secreted protein, the pA2 vector does not need a second signal peptide. Alternatively, the vector can be modified (pA2 GP) to include a baculovirus leader sequence, using the standard methods described in Summers et al., "A Manual of Methods for Baculovirus Vectors and Insect Cell Culture Procedures," Texas Agricultural Experimental Station Bulletin No. 1555 (1987).
The amplified fragment is isolated from a 1 % agarose gel using a commercially available kit ("Geneclean," BIO 101 Inc., La Jolla, Ca.). The fragment then is digested with appropriate restriction enzymes and again purified on a 1% agarose gel.
The plasmid is digested with the corresponding restriction enzymes and optionally, can be dephosphorylated using calf intestinal phosphatase, using routine procedures known in the art. The DNA is then isolated from a 1 % agarose gel using a commercially available kit ("Geneclean" BIO 101 Inc., La Jolla, Ca.).
The fragment and the dephosphorylated plasmid are ligated together with T4 DNA ligase. E. coli HB101 or other suitable E. coli hosts such as XL-1 Blue (Stratagene Cloning Systems, La Jolla, CA) cells are transformed with the ligation mixture and spread on culture plates. Bacteria containing the plasmid are identified by digesting DNA from individual colonies and analyzing the digestion product by gel electrophoresis. The sequence of the cloned fragment is confirmed by DNA sequencing. Five μg of a plasmid containing the polynucleotide is co-transfected with 1.0 μg of a commercially available linearized baculovirus DNA ("BaculoGold™ baculovirus DNA", Pharmingen, San Diego, CA), using the lipofection method described by Feigner et al., Proc. Natl. Acad. Sci. USA 84:7413-7417 (1987). One μg of BaculoGold™ virus DNA and 5 μg of the plasmid are mixed in a sterile well of a microtiter plate containing 50 μl of serum-free Grace's medium (Life Technologies
Inc., Gaithersburg, MD). Afterwards, 10 μl Lipofectin plus 90 μl Grace's medium are added, mixed and incubated for 15 minutes at room temperature. Then the transfection mixture is added drop- wise to Sf9 insect cells (ATCC CRL 1711) seeded in a 35 mm tissue culture plate with 1 ml Grace's medium without serum. The plate is then incubated for 5 hours at 27° C. The transfection solution is then removed from the plate and 1 ml of Grace's insect medium supplemented with 10% fetal calf serum is added. Cultivation is then continued at 27° C for four days.
After four days the supernatant is collected and a plaque assay is performed, as described by Summers and Smith, supra. An agarose gel with "Blue Gal" (Life Technologies Inc., Gaithersburg) is used to allow easy identification and isolation of gal-expressing clones, which produce blue-stained plaques. (A detailed description of a "plaque assay" of this type can also be found in the user's guide for insect cell culture and baculovirology distributed by Life Technologies Inc., Gaithersburg, page 9-10.) After appropriate incubation, blue stained plaques are picked with the tip of a micropipettor (e.g., Eppendorf). The agar containing the recombinant viruses is then resuspended in a microcentrifuge tube containing 200 μl of Grace's medium and the suspension containing the recombinant baculovirus is used to infect Sf9 cells seeded in 35 mm dishes. Four days later the supernatants of these culture dishes are harvested and then they are stored at 4° C.
To verify the expression of the polypeptide, Sf9 cells are grown in Grace's medium supplemented with 10% heat-inactivated FBS. The cells are infected with the recombinant baculovirus containing the polynucleotide at a multiplicity of infection ("MOI") of about 2. If radiolabeled proteins are desired, 6 hours later the medium is removed and is replaced with SF900 II medium minus methionine and cysteine (available from Life Technologies Inc., Rockville, MD). After 42 hours, 5 μCi of 35S- methionine and 5 μCi 35S-cysteine (available from Amersham) are added. The cells are further incubated for 16 hours and then are harvested by centrifugation. The proteins in the supernatant as well as the intracellular proteins are analyzed by SDS-PAGE followed by autoradiography (if radiolabeled).
Microsequencing of the amino acid sequence of the amino terminus of purified protein may be used to determine the amino terminal sequence of the produced protein.
Example 8: Expression of a Polypeptide in Mammalian Cells
The polypeptide of the present invention can be expressed in a mammalian cell. A typical mammalian expression vector contains a promoter element, which mediates the initiation of transcription of mRNA, a protein coding sequence, and signals required for the termination of transcription and polyadenylation of the transcript. Additional elements include enhancers, Kozak sequences and intervening sequences flanked by donor and acceptor sites for RNA splicing. Highly efficient transcription is achieved with the early and late promoters from SV40, the long terminal repeats (LTRs) from Retroviruses, e.g., RSV, HTLVI, HIVI and the early promoter of the cytomegalovirus (CMV). However, cellular elements can also be used (e.g., the human actin promoter). Suitable expression vectors for use in practicing the present invention include, for example, vectors such as pSVL and pMSG (Pharmacia, Uppsala, Sweden),
pRSVcat (ATCC 37152), pSV2dhfr (ATCC 37146), pBC12MI (ATCC 67109), pCMVSport 2.0, and pCMVSport 3.0. Mammalian host cells that could be used include, human Hela, 293, H9 and Jurkat cells, mouse NIH3T3 and C127 cells, Cos 1, Cos 7 and CV1, quail QC1-3 cells, mouse L cells and Chinese hamster ovary (CHO) cells.
Alternatively, the polypeptide can be expressed in stable cell lines containing the polynucleotide integrated into a chromosome. The co- transfection with a selectable marker such as dhfr, gpt, neomycin, hygromycin allows the identification and isolation of the transfected cells. The transfected gene can also be amplified to express large amounts of the encoded protein. The DHFR (dihydrofolate reductase) marker is useful in developing cell lines that carry several hundred or even several thousand copies of the gene of interest. (See, e.g., Alt, F. W., et al., J. Biol. Chem. 253: 1357-1370 (1978); Hamlin, J. L. and Ma, C, Biochem. et Biophys. Acta, 1097:107-143 (1990); Page, M. J. and Sydenham, M. A., Biotechnology 9:64-68 (1991).) Another useful selection marker is the enzyme glutamine synthase (GS) (Muφhy et al., Biochem J. 227:277-279 (1991); Bebbington et al., Bio/Technology 10: 169-175 (1992). Using these markers, the mammalian cells are grown in selective medium and the cells with the highest resistance are selected. These cell lines contain the amplified gene(s) integrated into a chromosome. Chinese hamster ovary (CHO) and NSO cells are often used for the production of proteins.
Derivatives of the plasmid ρSV2-dhfr (ATCC Accession No. 37146), the expression vectors pC4 (ATCC Accession No. 209646) and pC6 (ATCC Accession No. 209647) contain the strong promoter (LTR) of the Rous Sarcoma Virus (Cullen et al., Molecular and Cellular Biology, 438-447 (March, 1985)) plus a fragment of the CMV-enhancer (Boshart et al., Cell 41:521-530 (1985).) Multiple cloning sites, e.g., with the restriction enzyme cleavage sites BamHl, Xbal and Asp718, facilitate the cloning of the gene of interest. The vectors also contain the 3' intron, the polyadenylation and termination signal of the rat preproinsulin gene, and the mouse DHFR gene under control of the S V40 early promoter.
Specifically, the plasmid pC6, for example, is digested with appropriate restriction enzymes and then dephosphorylated using calf intestinal phosphates by procedures known in the art. The vector is then isolated from a 1% agarose gel.
A polynucleotide of the present invention is amplified according to the protocol outlined in Example 1. If the naturally occurring signal sequence is used to produce the secreted protein, the vector does not need a second signal peptide. Alternatively, if the
naturally occurring signal sequence is not used, the vector can be modified to include a heterologous signal sequence. (See, e.g., WO 96/34891.)
The amplified fragment is isolated from a 1 % agarose gel using a commercially available kit ("Geneclean," BIO 101 Inc., La Jolla, Ca.). The fragment then is digested with appropriate restriction enzymes and again purified on a 1% agarose gel.
The amplified fragment is then digested with the same restriction enzyme and purified on a 1% agarose gel. The isolated fragment and the dephosphorylated vector are then ligated with T4 DNA ligase. E. coli HB101 or XL-1 Blue cells are then transformed and bacteria are identified that contain the fragment inserted into plasmid pC6 using, for instance, restriction enzyme analysis.
Chinese hamster ovary cells lacking an active DHFR gene is used for transfection. Five μg of the expression plasmid pC6 is cotransfected with 0.5 μg of the plasmid pSVneo using lipofectin (Feigner et al., supra). The plasmid pSV2-neo contains a dominant selectable marker, the neo gene from Tn5 encoding an enzyme that confers resistance to a group of antibiotics including G418. The cells are seeded in alpha minus MEM supplemented with 1 mg/ml G418. After 2 days, the cells are trypsinized and seeded in hybridoma cloning plates (Greiner, Germany) in alpha minus MEM supplemented with 10, 25, or 50 ng/ml of metothrexate plus 1 mg/ml G418. After about 10-14 days single clones are trypsinized and then seeded in 6-well petri dishes or 10 ml flasks using different concentrations of methotrexate (50 nM, 100 nM, 200 nM, 400 nM, 800 nM). Clones growing at the highest concentrations of methotrexate are then transferred to new 6-well plates containing even higher concentrations of methotrexate (1 μM, 2 μM, 5 μM, 10 mM, 20 mM). The same procedure is repeated until clones are obtained which grow at a concentration of 100 - 200 μM. Expression of the desired gene product is analyzed, for instance, by SDS- PAGE and Western blot or by reversed phase HPLC analysis.
Example 9: Protein Fusions
The polypeptides of the present invention are preferably fused to other proteins. These fusion proteins can be used for a variety of applications. For example, fusion of the present polypeptides to His-tag, HA-tag, protein A, IgG domains, and maltose binding protein facilitates purification. (See Example 5; see also EP A 394,827; Traunecker, et al., Nature 331:84-86 (1988).) Similarly, fusion to IgG-1, IgG-3, and albumin increases the halflife time in vivo. Nuclear localization signals fused to the polypeptides of the present invention can target the protein to a specific subcellular localization, while covalent heterodimer or homodimers can increase or decrease the activity of a fusion protein. Fusion proteins can also create chimeric molecules having
more than one function. Finally, fusion proteins can increase solubility and/or stability of the fused protein compared to the non-fused protein. All of the types of fusion proteins described above can be made by modifying the following protocol, which outlines the fusion of a polypeptide to an IgG molecule, or the protocol described in Example 5.
Briefly, the human Fc portion of the IgG molecule can be PCR amplified, using primers that span the 5' and 3' ends of the sequence described below. These primers also should have convenient restriction enzyme sites that will facilitate cloning into an expression vector, preferably a mammalian expression vector. For example, if pC4 (Accession No.209646) is used, the human Fc portion can be ligated into the BamHl cloning site. Note that the 3' BamHl site should be destroyed. Next, the vector containing the human Fc portion is re-restricted with BamHl, linearizing the vector, and a polynucleotide of the present invention, isolated by the PCR protocol described in Example 1, is ligated into this BamHl site. Note that the polynucleotide is cloned without a stop codon, otherwise a fusion protein will not be produced.
If the naturally occurring signal sequence is used to produce the secreted protein, pC4 does not need a second signal peptide. Alternatively, if the naturally occurring signal sequence is not used, the vector can be modified to include a heterologous signal sequence. (See, e.g., WO 96/34891.)
Human IgG Fc region:
GGGATCCGGAGCCCAAATCTTCTGACAAAACTCACACATGCCCACCGTGCC
CAGCACCTGAATTCGAGGGTGCACCGTCAGTCTTCCTCTTCCCCCCAAAACC CAAGGACACCCTCATGATCTCCCGGACTCCTGAGGTCACATGCGTGGTGGT GGACGTAAGCCACGAAGACCCTGAGGTCAAGTTCAACTGGTACGTGGACG GCGTGGAGGTGCATAATGCCAAGACAAAGCCGCGGGAGGAGCAGTACAAC AGCACGTACCGTGTGGTCAGCGTCCTCACCGTCCTGCACCAGGACTGGCTG AATGGCAAGGAGTACAAGTGCAAGGTCTCCAACAAAGCCCTCCCAACCCCC ATCGAGAAAACCATCTCCAAAGCCAAAGGGCAGCCCCGAGAACCACAGGT GTACACCCTGCCCCCATCCCGGGATGAGCTGACCAAGAACCAGGTCAGCCT GACCTGCCTGGTCAAAGGCTTCTATCCAAGCGACATCGCCGTGGAGTGGGA GAGCAATGGGCAGCCGGAGAACAACTACAAGACCACGCCTCCCGTGCTGG ACTCCGACGGCTCCTTCTTCCTCTACAGCAAGCTCACCGTGGACAAGAGCA GGTGGCAGCAGGGGAACGTCTTCTCATGCTCCGTGATGCATGAGGCTCTGC ACAACCACTACACGCAGAAGAGCCTCTCCCTGTCTCCGGGTAAATGAGTGC GACGGCCGCGACTCTAGAGGAT (SEQ ID NO: 1 )
Example 10: Production of an Antibody from a Polypeptide
The antibodies of the present invention can be prepared by a variety of methods. (See, Current Protocols, Chapter 2.) For example, cells expressing a polypeptide of the present invention is administered to an animal to induce the production of sera containing polyclonal antibodies. In a preferred method, a preparation of the secreted protein is prepared and purified to render it substantially free of natural contaminants. Such a preparation is then introduced into an animal in order to produce polyclonal antisera of greater specific activity. In the most preferred method, the antibodies of the present invention are monoclonal antibodies (or protein binding fragments thereof). Such monoclonal antibodies can be prepared using hybridoma technology. (Kohler et al., Nature 256:495 (1975); Kohler et al., Eur. J. Immunol. 6:511 (1976); Kohler et al., Eur. J. Immunol. 6:292 (1976); Hammerling et al., in: Monoclonal Antibodies and T-Cell Hybridomas, Elsevier, N.Y., pp. 563-681 (1981).) In general, such procedures involve immunizing an animal (preferably a mouse) with polypeptide or, more preferably, with a secreted polypeptide-expressing cell. Such cells may be cultured in any suitable tissue culture medium; however, it is preferable to culture cells in Earle's modified Eagle's medium supplemented with 10% fetal bovine serum (inactivated at about 56°C), and supplemented with about 10 g/1 of nonessential amino acids, about
1,000 U/ml of penicillin, and about 100 μg/ml of streptomycin.
The splenocytes of such mice are extracted and fused with a suitable myeloma cell line. Any suitable myeloma cell line may be employed in accordance with the present invention; however, it is preferable to employ the parent myeloma cell line (SP2O), available from the ATCC. After fusion, the resulting hybridoma cells are selectively maintained in HAT medium, and then cloned by limiting dilution as described by Wands et al. (Gastroenterology 80:225-232 (1981).) The hybridoma cells obtained through such a selection are then assayed to identify clones which secrete antibodies capable of binding the polypeptide. Alternatively, additional antibodies capable of binding to the polypeptide can be produced in a two-step procedure using anti-idiotypic antibodies. Such a method makes use of the fact that antibodies are themselves antigens, and therefore, it is possible to obtain an antibody which binds to a second antibody. In accordance with this method, protein specific antibodies are used to immunize an animal, preferably a mouse. The splenocytes of such an animal are then used to produce hybridoma cells, and the hybridoma cells are screened to identify clones which produce an antibody
whose ability to bind to the protein-specific antibody can be blocked by the polypeptide. Such antibodies comprise anti-idiotypic antibodies to the protein-specific antibody and can be used to immunize an animal to induce formation of further protein-specific antibodies. It will be appreciated that Fab and F(ab')2 and other fragments of the antibodies of the present invention may be used according to the methods disclosed herein. Such fragments are typically produced by proteolytic cleavage, using enzymes such as papain (to produce Fab fragments) or pepsin (to produce F(ab')2 fragments). Alternatively, secreted protein-binding fragments can be produced through the application of recombinant DNA technology or through synthetic chemistry.
For in vivo use of antibodies in humans, it may be preferable to use "humanized" chimeric monoclonal antibodies. Such antibodies can be produced using genetic constructs derived from hybridoma cells producing the monoclonal antibodies described above. Methods for producing chimeric antibodies are known in the art. (See, for review, Morrison, Science 229: 1202 (1985); Oi et al., BioTechniques 4:214 (1986); Cabilly et al., U.S. Patent No. 4,816,567; Taniguchi et al., EP 171496; Morrison et al., EP 173494; Neuberger et al., WO 8601533; Robinson et al., WO 8702671; Boulianne et al., Nature 312:643 (1984); Neuberger et al., Nature 314:268 (1985).)
Example 11: Production Of Secreted Protein For High-Throughput Screening Assays
The following protocol produces a supernatant containing a polypeptide to be tested. This supernatant can then be used in the Screening Assays described in Examples 13-20.
First, dilute Poly-D-Lysine (644 587 Boehringer-Mannheim) stock solution (lmg/ml in PBS) 1:20 in PBS (w/o calcium or magnesium 17-516F Biowhittaker) for a working solution of 50ug/ml. Add 200 ul of this solution to each well (24 well plates) and incubate at RT for 20 minutes. Be sure to distribute the solution over each well (note: a 12-channel pipetter may be used with tips on every other channel). Aspirate off the Poly-D-Lysine solution and rinse with 1ml PBS (Phosphate Buffered Saline). The PBS should remain in the well until just prior to plating the cells and plates may be poly-lysine coated in advance for up to two weeks.
Plate 293T cells (do not carry cells past P+20) at 2 x 105 cells/well in .5ml DMEM(Dulbecco's Modified Eagle Medium)(with 4.5 G/L glucose and L-glutamine (12-604F Biowhittaker) )/10% heat inactivated FBS(14-503F Biowhittaker)/ lx Penstrep(17-602E Biowhittaker). Let the cells grow overnight.
The next day, mix together in a sterile solution basin: 300 ul Lipofectamine (18324-012 Gibco/BRL) and 5ml Optimem I (31985070 Gibco/BRL)/96-well plate. With a small volume multi-channel pipetter, aliquot approximately 2ug of an expression vector containing a polynucleotide insert, produced by the methods described in Examples 8 or 9, into an appropriately labeled 96-well round bottom plate. With a multi-channel pipetter, add 50ul of the Lipofectamine/Optimem I mixture to each well. Pipette up and down gently to mix. Incubate at RT 15-45 minutes. After about 20 minutes, use a multi-channel pipetter to add 150ul Optimem I to each well. As a control, one plate of vector DNA lacking an insert should be transfected with each set of transfections.
Preferably, the transfection should be performed by tag-teaming the following tasks. By tag-teaming, hands on time is cut in half, and the cells do not spend too much time on PBS. First, person A aspirates off the media from four 24-well plates of cells, and then person B rinses each well with .5- lml PBS. Person A then aspirates off PBS rinse, and person B, using al2-channel pipetter with tips on every other channel, adds the 200ul of DNA/Lipofectamine/Optimem I complex to the odd wells first, then to the even wells, to each row on the 24-well plates. Incubate at 37°C for 6 hours.
While cells are incubating, prepare appropriate media, either 1 %BS A in DMEM with lx penstrep, or CHO-5 media (see below) with 2mm glutamine and lx penstrep. (BSA (81-068-3 Bayer) lOOgm dissolved in IL DMEM for a 10% BSA stock solution). Filter the media and collect 50 ul for endotoxin assay in 15ml polystyrene conical.
The transfection reaction is terminated, preferably by tag-teaming, at the end of the incubation period. Person A aspirates off the transfection media, while person B adds 1.5ml appropriate media to each well. Incubate at 37°C for 45 or 72 hours depending on the media used: 1 %BSA for 45 hours or CHO-5 for 72 hours.
On day four, using a 300ul multichannel pipetter, aliquot 600ul in one lml deep well plate and the remaining supernatant into a 2ml deep well. The supernatants from each well can then be used in the assays described in Examples 13-20. It is specifically understood that when activity is obtained in any of the assays described below using a supernatant, the activity originates from either the polypeptide directly (e.g., as a secreted protein) or by the polypeptide inducing expression of other proteins, which are then secreted into the supernatant. Thus, the invention further provides a method of identifying the protein in the supernatant characterized by an activity in a particular assay.
HGS-CHO-5 medium formulation: Inorganic Salts
Other Components
Adjust osmolarity to 327 mOsm
Example 12: Construction of GAS Reporter Construct
One signal transduction pathway involved in the differentiation and proliferation of cells is called the Jaks-STATs pathway. Activated proteins in the Jaks-STATs pathway bind to gamma activation site "GAS" elements or interferon-sensitive responsive element ("ISRE"), located in the promoter of many genes. The binding of a protein to these elements alter the expression of the associated gene.
GAS and ISRE elements are recognized by a class of transcription factors called Signal Transducers and Activators of Transcription, or "STATs." There are six members of the STATs family. Statl and Stat3 are present in many cell types, as is Stat2 (as response to EFN-alpha is widespread). Stat4 is more restricted and is not in many cell types though it has been found in T helper class I, cells after treatment with IL-12. Stat5 was originally called mammary growth factor, but has been found at higher concentrations in other cells including myeloid cells. It can be activated in tissue culture cells by many cytokines. The STATs are activated to translocate from the cytoplasm to the nucleus upon tyrosine phosphorylation by a set of kinases known as the Janus Kinase ("Jaks") family. Jaks represent a distinct family of soluble tyrosine kinases and include Tyk2, Jakl, Jak2, and Jak3. These kinases display significant sequence similarity and are generally catalytically inactive in resting cells. The Jaks are activated by a wide range of receptors summarized in the Table below. (Adapted from review by Schidler and Darnell, Ann. Rev. Biochem. 64:621-51 (1995).) A cytokine receptor family, capable of activating Jaks, is divided into two groups: (a) Class 1 includes receptors for IL-2, IL-3, EL-4, IL-6, IL-7, IL-9, IL-11, IL- 12, IL-15, Epo, PRL, GH, G-CSF, GM-CSF, LIF, CNTF, and thrombopoietin; and (b) Class 2 includes IFN-a, IFN-g, and IL-10. The Class 1 receptors share a conserved cysteine motif (a set of four conserved cysteines and one tryptophan) and a WSXWS motif (a membrane proxial region encoding Tφ-Ser-Xxx-Tφ-Ser (SEQ ID NO:2)).
Thus, on binding of a ligand to a receptor, Jaks are activated, which in turn activate STATs, which then translocate and bind to GAS elements. This entire process is encompassed in the Jaks-STATs signal transduction pathway.
Therefore, activation of the Jaks-STATs pathway, reflected by the binding of the GAS or the ISRE element, can be used to indicate proteins involved in the proliferation and differentiation of cells. For example, growth factors and cytokines are known to activate the Jaks-STATs pathway. (See Table below.) Thus, by using GAS elements linked to reporter molecules, activators of the Jaks-STATs pathway can be identified.
JAKs STATS GAS (elements) or
ISRE
Ligand tyk2 Jakl Jak2 Jak3
IFN familv
IFN-a/B + + - - 1,2,3 ISRE
IFN-g + + - 1 GAS
(IRFl>Lys6>IFP)
11-10 + ? ? - 1 ,3 gp 130 familv
IL-6 (Pleiotrohic) + + + ? 1 ,3 GAS
(IRFl>Lys6>IFP)
11- 11 (Pleiotrohic) + ? ? 1 ,3
OnM(Pleiotrohic) ? + + 7 1 ,3
LIF(Pleiotrohic) ? + + ? 1 ,3
CNTF(Pleiotrohic) -/+ + + ? 1 ,3
G-CSF(Pleiotrohic) ? + ? ? 1 ,3
IL-12(Pleiotrohic) + - + + 1 ,3 g-C familv
IL-2 (lymphocytes) - + - + 1,3,5 GAS
IL-4 (lymph/myeloid) - + - + 6 GAS (IRF1 = IFP
»Ly6)(IgH)
IL-7 (lymphocytes) - + - + 5 GAS
IL-9 (lymphocytes) - + - + 5 GAS
IL-13 (lymphocyte) - + 7 ? 6 GAS
IL-15 ? + ? + 5 GAS gp 140 familv
IL-3 (myeloid) - - + - 5 GAS
(IRFl>IFP»Ly6)
IL-5 (myeloid) - - + - 5 GAS
GM-CSF (myeloid) - - + - 5 GAS
Growth hormone familv
GH ? - + - 5
PRL ? +/- + - 1 ,3,5
EPO ? - + - 5 GAS(B-
CAS>IRFl=IFP»Ly6)
Receptor Tvrosine Kinases
EGF ? + + - 1 ,3 GAS (IRF1)
PDGF 7 + + _ 1 ,3
CSF-1 7 + + - 1 ,3 GAS (not ΙRFl)
To construct a synthetic GAS containing promoter element, which is used in the Biological Assays described in Examples 13-14, a PCR based strategy is employed to generate a GAS-SV40 promoter sequence. The 5' primer contains four tandem copies of the GAS binding site found in the IRF1 promoter and previously demonstrated to bind STATs upon induction with a range of cytokines (Rothman et al., Immunity
1 :457-468 (1994).), although other GAS or ISRE elements can be used instead. The 5' primer also contains 18bp of sequence complementary to the SV40 early promoter sequence and is flanked with an Xhol site. The sequence of the 5' primer is: 5 ' :GCGCCTCGAGATTTCCCCGAAATCTAGATTTCCCCGAAATGATTTCCCCG AAATGATTTCCCCGAAATATCTGCCATCTCAATTAG:3' (SEQ ID NO:3)
The downstream primer is complementary to the SV40 promoter and is flanked with a Hind m site: 5':GCGGCAAGCTTTTTGCAAAGCCTAGGC:3' (SEQ ID NO:4)
PCR amplification is performed using the SV40 promoter template present in the B-gal:promoter plasmid obtained from Clontech. The resulting PCR fragment is digested with Xhol/Hind III and subcloned into BLSK2-. (Stratagene.) Sequencing with forward and reverse primers confirms that the insert contains the following sequence: 5 ' :CTCGAGATTTCCCCGAAATCTAGATTTCCCCGAAATGATTTCCCCGAAATG ATTTCCCCGAAATATCTGCCATCTCAATTAGTCAGCAACCATAGTCCCGCCC CTAACTCCGCCCATCCCGCCCCTAACTCCGCCCAGTTCCGCCCATTCTCCGC CCCATGGCTGACTAATTTTTTTTATTTATGCAGAGGCCGAGGCCGCCTCGGC CTCTGAGCTATTCCAGAAGTAGTGAGGAGGCITTTTTGGAGGCCTAGGCTTT TGCAAAAAGCTT:3' (SEQ ID NO:5)
With this GAS promoter element linked to the SV40 promoter, a GAS:SEAP2 reporter construct is next engineered. Here, the reporter molecule is a secreted alkaline phosphatase, or "SEAP." Clearly, however, any reporter molecule can be instead of SEAP, in this or in any of the other Examples. Well known reporter molecules that can be used instead of SEAP include chloramphenicol acetyltransferase (CAT), luciferase, alkaline phosphatase, B-galactosidase, green fluorescent protein (GFP), or any protein detectable by an antibody.
The above sequence confirmed synthetic GAS-SV40 promoter element is subcloned into the pSEAP-Promoter vector obtained from Clontech using Hindlll and Xhol, effectively replacing the SV40 promoter with the amplified GAS:SV40 promoter element, to create the GAS-SEAP vector. However, this vector does not contain a
neomycin resistance gene, and therefore, is not preferred for mammalian expression systems.
Thus, in order to generate mammalian stable cell lines expressing the GAS- SEAP reporter, the GAS-SEAP cassette is removed from the GAS-SEAP vector using Sail and Notl, and inserted into a backbone vector containing the neomycin resistance gene, such as pGFP-1 (Clontech), using these restriction sites in the multiple cloning site, to create the GAS-SEAP/Neo vector. Once this vector is transfected into mammalian cells, this vector can then be used as a reporter molecule for GAS binding as described in Examples 13-14. Other constructs can be made using the above description and replacing GAS with a different promoter sequence. For example, construction of reporter molecules containing NFK-B and EGR promoter sequences are described in Examples 15 and 16. However, many other promoters can be substituted using the protocols described in these Examples. For instance, SRE, IL-2, NFAT, or Osteocalcin promoters can be substituted, alone or in combination (e.g., GAS/NF-KB/EGR, GAS/NF-KB, II - 2/NFAT, or NF-KB/GAS). Similarly, other cell lines can be used to test reporter construct activity, such as HELA (epithelial), HUVEC (endothelial), Reh (B-cell), Saos-2 (osteoblast), HUVAC (aortic), or Cardiomyocyte.
Example 13: High-Throughput Screening Assay for T-cell Activity.
The following protocol is used to assess T-cell activity by identifying factors, such as growth factors and cytokines, that may proliferate or differentiate T-cells. T- cell activity is assessed using the GAS/SEAP/Neo construct produced in Example 12. Thus, factors that increase SEAP activity indicate the ability to activate the Jaks-STATS signal transduction pathway. The T-cell used in this assay is Jurkat T-cells (ATCC
Accession No. TIB- 152), although Molt-3 cells (ATCC Accession No. CRL-1552) and Molt-4 cells (ATCC Accession No. CRL-1582) cells can also be used.
Jurkat T-cells are lymphoblastic CD4+ Thl helper cells. In order to generate stable cell lines, approximately 2 million Jurkat cells are transfected with the GAS- SEAP/neo vector using DMRIE-C (Life Technologies)(transfection procedure described below). The transfected cells are seeded to a density of approximately 20,000 cells per well and transfectants resistant to 1 mg/ml genticin selected. Resistant colonies are expanded and then tested for their response to increasing concentrations of interferon gamma. The dose response of a selected clone is demonstrated. Specifically, the following protocol will yield sufficient cells for 75 wells containing 200 ul of cells. Thus, it is either scaled up, or performed in multiple to generate sufficient cells for multiple 96 well plates. Jurkat cells are maintained in RPMI
+ 10% serum with l%Pen-Strep. Combine 2.5 mis of OPTI-MEM (Life Technologies) with 10 ug of plasmid DNA in a T25 flask. Add 2.5 ml OPTI-MEM containing 50 ul of DMRIE-C and incubate at room temperature for 15-45 mins.
During the incubation period, count cell concentration, spin down the required number of cells ( 107 per transfection), and resuspend in OPTI-MEM to a final concentration of 107 cells/ml. Then add lml of 1 x 107 cells in OPTI-MEM to T25 flask and incubate at 37°C for 6 hrs. After the incubation, add 10 ml of RPMI + 15% serum.
The Jurkat:GAS-SEAP stable reporter lines are maintained in RPMI + 10% serum, 1 mg/ml Genticin, and 1% Pen-Strep. These cells are treated with supematants containing a polypeptide as produced by the protocol described in Example 11.
On the day of treatment with the supernatant, the cells should be washed and resuspended in fresh RPMI + 10% serum to a density of 500,000 cells per ml. The exact number of cells required will depend on the number of supematants being screened. For one 96 well plate, approximately 10 million cells (for 10 plates, 100 million cells) are required.
Transfer the cells to a triangular reservoir boat, in order to dispense the cells into a 96 well dish, using a 12 channel pipette. Using a 12 channel pipette, transfer 200 ul of cells into each well (therefore adding 100, 000 cells per well).
After all the plates have been seeded, 50 ul of the supematants are transferred directly from the 96 well plate containing the supematants into each well using a 12 channel pipette. In addition, a dose of exogenous interferon gamma (0.1, 1.0, 10 ng) is added to wells H9, H10, and HI 1 to serve as additional positive controls for the assay.
The 96 well dishes containing Jurkat cells treated with supematants are placed in an incubator for 48 hrs (note: this time is variable between 48-72 hrs). 35 ul samples from each well are then transferred to an opaque 96 well plate using a 12 channel pipette. The opaque plates should be covered (using sellophene covers) and stored at -
20°C until SEAP assays are performed according to Example 17. The plates containing the remaining treated cells are placed at 4°C and serve as a source of material for repeating the assay on a specific well if desired.
As a positive control, 100 Unit/ml interferon gamma can be used which is known to activate Jurkat T cells. Over 30 fold induction is typically observed in the positive control wells.
Example 14: High-Throughput Screening Assay Identifying Mveloid Activity
The following protocol is used to assess myeloid activity by identifying factors, such as growth factors and cytokines, that may proliferate or differentiate myeloid cells. Myeloid cell activity is assessed using the GAS/SEAP/Neo construct produced in
Example 12. Thus, factors that increase SEAP activity indicate the ability to activate the Jaks-STATS signal transduction pathway. The myeloid cell used in this assay is U937, a pre-monocyte cell line, although TF-1, HL60, or KG1 can be used.
To transiently transfect U937 cells with the GAS/SEAP/Neo construct produced in Example 12, a DEAE-Dextran method (Kharbanda et. al., 1994, Cell Growth &
Differentiation, 5:259-265) is used. First, harvest 2xl0e^ U937 cells and wash with PBS. The U937 cells are usually grown in RPMI 1640 medium containing 10% heat- inactivated fetal bovine serum (FBS) supplemented with 100 units/ml penicillin and 100 mg/ml streptomycin. Next, suspend the cells in 1 ml of 20 mM Tris-HCl (pH 7.4) buffer containing
0.5 mg/ml DEAE-Dextran, 8 ug GAS-SEAP2 plasmid DNA, 140 mM NaCl, 5 mM
KC1, 375 uM Na2HPO4.7H O, 1 mM MgCl2, and 675 uM CaCl2. Incubate at 37°C for 45 min.
Wash the cells with RPMI 1640 medium containing 10% FBS and then resuspend in 10 ml complete medium and incubate at 37°C for 36 hr.
The GAS-SEAP/U937 stable cells are obtained by growing the cells in 400 ug/ml G418. The G418-free medium is used for routine growth but every one to two months, the cells should be re-grown in 400 ug/ml G418 for couple of passages.
These cells are tested by harvesting 1x10 cells (this is enough for ten 96- well plates assay) and wash with PBS. Suspend the cells in 200 ml above described growth medium, with a final density of 5xl05 cells/ml. Plate 200 ul cells per well in the 96- well plate (or 1x10s cells/well).
Add 50 ul of the supernatant prepared by the protocol described in Example 11.
Incubate at 37°C for 48 to 72 hr. As a positive control, 100 Unit/ml interferon gamma can be used which is known to activate U937 cells. Over 30 fold induction is typically observed in the positive control wells. SEAP assay the supernatant according to the protocol described in Example 17.
Example 15: High-Throughput Screening Assay Identifying Neuronal Activity.
When cells undergo differentiation and proliferation, a group of genes are activated through many different signal transduction pathways. One of these genes, EGRl (early growth response gene 1), is induced in various tissues and cell types upon activation. The promoter of EGRl is responsible for such induction. Using the EGRl promoter linked to reporter molecules, activation of cells can be assessed.
Particularly, the following protocol is used to assess neuronal activity in PC 12 cell lines. PC 12 cells (rat phenochromocytoma cells) are known to proliferate and/or differentiate by activation with a number of mitogens, such as TPA (tetradecanoyl phorbol acetate), NGF (nerve growth factor), and EGF (epidermal growth factor). The EGRl gene expression is activated during this treatment. Thus, by stably transfecting PC 12 cells with a construct containing an EGR promoter linked to SEAP reporter, activation of PC 12 cells can be assessed. The EGR/SEAP reporter construct can be assembled by the following protocol.
The EGR-1 promoter sequence (-633 to +l)(Sakamoto K et al., Oncogene 6:867-871 (1991)) can be PCR amplified from human genomic DNA using the following primers:
5' GCGCTCGAGGGATGACAGCGATAGAACCCCGG -3' (SEQ ID NO:6) 5' GCGAAGCTTCGCGACTCCCCGGATCCGCCTC-3' (SEQ ID NO:7)
Using the GAS:SEAP/Neo vector produced in Example 12, EGRl amplified product can then be inserted into this vector. Linearize the GAS:SEAP/Neo vector using restriction enzymes Xhol Hindlll, removing the GAS/SV40 stuffer. Restrict the EGRl amplified product with these same enzymes. Ligate the vector and the EGRl promoter.
To prepare 96 well-plates for cell culture, two mis of a coating solution (1:30 dilution of collagen type I (Upstate Biotech Inc. Cat#08-115) in 30% ethanol (filter sterilized)) is added per one 10 cm plate or 50 ml per well of the 96-well plate, and allowed to air dry for 2 hr.
PC 12 cells are routinely grown in RPMI- 1640 medium (Bio Whittaker) containing 10% horse se m (JRH BIOSCIENCES, Cat. # 12449-78P), 5% heat- inactivated fetal bovine semm (FBS) supplemented with 100 units/ml penicillin and 100 ug/ml streptomycin on a precoated 10 cm tissue culture dish. One to four split is done every three to four days. Cells are removed from the plates by scraping and resuspended with pipetting up and down for more than 15 times.
Transfect the EGR/SEAP/Neo constmct into PC 12 using the Lipofectamine protocol described in Example 11. EGR-SEAP/PC12 stable cells are obtained by growing the cells in 300 ug/ml G418. The G418-free medium is used for routine growth but every one to two months, the cells should be re-grown in 300 ug/ml G418 for couple of passages.
To assay for neuronal activity, a 10 cm plate with cells around 70 to 80% confluent is screened by removing the old medium. Wash the cells once with PBS (Phosphate buffered saline). Then starve the cells in low semm medium (RPMI- 1640 containing 1% horse semm and 0.5% FBS with antibiotics) overnight. The next morning, remove the medium and wash the cells with PBS. Scrape off the cells from the plate, suspend the cells well in 2 ml low semm medium. Count the cell number and add more low semm medium to reach final cell density as 5x10^ cells/ml.
Add 200 ul of the cell suspension to each well of 96-well plate (equivalent to lxlO5 cells/well). Add 50 ul supernatant produced by Example 11, 37°C for 48 to 72 hr. As a positive control, a growth factor known to activate PC 12 cells through EGR can be used, such as 50 ng/ul of Neuronal Growth Factor (NGF). Over fifty-fold induction of SEAP is typically seen in the positive control wells. SEAP assay the supernatant according to Example 17.
Example 16: High-Throughput Screening Assay for T-cell Activity
NF-κB (Nuclear Factor KB) is a transcription factor activated by a wide variety of agents including the inflammatory cytokines IL- 1 and TNF, CD30 and CD40, lymphotoxin-alpha and lymphotoxin-beta, by exposure to LPS or thrombin, and by expression of certain viral gene products. As a transcription factor, NF-κB regulates the expression of genes involved in immune cell activation, control of apoptosis (NF-
KB appears to shield cells from apoptosis), B and T-cell development, anti-viral and antimicrobial responses, and multiple stress responses.
In non-stimulated conditions, NF- KB is retained in the cytoplasm with I-κB (Inhibitor KB). However, upon stimulation, I- KB is phosphorylated and degraded, causing NF- KB to shuttle to the nucleus, thereby activating transcription of target genes. Target genes activated by NF- KB include IL-2, IL-6, GM-CSF, ICAM-1 and class 1 MHC.
Due to its central role and ability to respond to a range of stimuli, reporter constmcts utilizing the NF-κB promoter element are used to screen the supematants produced in Example 11. Activators or inhibitors of NF-kB would be useful in treating diseases. For example, inhibitors of NF-κB could be used to treat those diseases related to the acute or chronic activation of NF-kB, such as rheumatoid arthritis.
To construct a vector containing the NF-κB promoter element, a PCR based strategy is employed. The upstream primer contains four tandem copies of the NF-κB binding site (GGGGACTTTCCC) (SEQ ID NO: 8), 18 bp of sequence complementary to the 5' end of the SV40 early promoter sequence, and is flanked with an Xhol site: 5' :GCGGCCTCGAGGGGACTTTCCCGGGGACTTTCCGGGGACTTTCCGGGAC TTTCCATCCTGCCATCTCAATTAG:3' (SEQ ID NO:9)
The downstream primer is complementary to the 3' end of the SV40 promoter and is flanked with a Hind III site: 5':GCGGCAAGC1TTTTGCAAAGCCTAGGC:3' (SEQ ID NO:4) PCR amplification is performed using the SV40 promoter template present in the pB-gal:promoter plasmid obtained from Clontech. The resulting PCR fragment is digested with Xhol and Hind HI and subcloned into BLSK2-. (Stratagene) Sequencing with the T7 and T3 primers confirms the insert contains the following sequence: 5 ' :CTCGAGGGGACTTTCCCGGGGACTTTCCGGGGACTTTCCGGGACTTTCC ATCTGCCATCTCAATTAGTCAGCAACCATAGTCCCGCCCCTAACTCCGCCCA TCCCGCCCCTAACTCCGCCCAGTTCCGCCCATTCTCCGCCCCATGGCTGACT AATTTTTTTTATTTATGCAGAGGCCGAGGCCGCCTCGGCCTCTGAGCTATTC CAGAAGTAGTGAGGAGGCTTTTTTGGAGGCCTAGGCTTTTGCAAAAAGCTT: 3' (SEQ ID NO: 10)
Next, replace the SV40 minimal promoter element present in the pSEAP2- promoter plasmid (Clontech) with this NF-KB/SV40 fragment using Xhol and Hindlll.
However, this vector does not contain a neomycin resistance gene, and therefore, is not preferred for mammalian expression systems.
In order to generate stable mammalian cell lines, the NF-KB/SV40/SEAP cassette is removed from the above NF-κB/SEAP vector using restriction enzymes Sail and Notl, and inserted into a vector containing neomycin resistance. Particularly, the
NF-KB/SV40/SEAP cassette was inserted into pGFP-1 (Clontech), replacing the GFP gene, after restricting pGFP-1 with Sail and Notl.
Once NF-κB/SV40/SEAP/Neo vector is created, stable Jurkat T-cells are created and maintained according to the protocol described in Example 13. Similarly, the method for assaying supematants with these stable Jurkat T-cells is also described in Example 13. As a positive control, exogenous TNF alpha (0.1,1, 10 ng) is added to wells H9, H10, and HI 1, with a 5-10 fold activation typically observed.
Example 17: Assay for SEAP Activity As a reporter molecule for the assays described in Examples 13-16, SEAP activity is assayed using the Tropix Phospho-light Kit (Cat. BP-400) according to the following general procedure. The Tropix Phospho-light Kit supplies the Dilution, Assay, and Reaction Buffers used below.
Prime a dispenser with the 2.5x Dilution Buffer and dispense 15 μl of 2.5x dilution buffer into Optiplates containing 35 μl of a supernatant. Seal the plates with a plastic sealer and incubate at 65°C for 30 min. Separate the Optiplates to avoid uneven heating.
Cool the samples to room temperature for 15 minutes. Empty the dispenser and prime with the Assay Buffer. Add 50 μl Assay Buffer and incubate at room temperature 5 min. Empty the dispenser and prime with the Reaction Buffer (see the table below). Add 50 μl Reaction Buffer and incubate at room temperature for 20 minutes. Since the intensity of the chemiluminescent signal is time dependent, and it takes about 10 minutes to read 5 plates on luminometer, one should treat 5 plates at each time and start the second set 10 minutes later. Read the relative light unit in the luminometer. Set H 12 as blank, and print the results. An increase in chemiluminescence indicates reporter activity.
Reaction Buffer Formulation:
# of plates Rxn buffer diluent (ml) CSPD (ml)
10 60 3
1 1 65 3.25
12 70 3.5
13 75 3.75
14 80 4
15 85 4.25 16 90 4.5
17 95 4.75 18 100 5 19 105 5.25 20 110 5.5 21 115 5.75 22 120 6 23 125 6.25 24 130 6.5 25 135 6.75 26 140 7 27 145 7.25 28 150 7.5 29 155 7.75 30 160 31 165 8.25 32 170 8.5 33 175 8.75 34 180 9 35 185 9.25 36 190 9.5 37 195 9.75 38 200 10 39 205 10.25 40 210 10.5 41 215 10.75 42 220 11 43 225 11.25 44 230 11.5 45 235 11.75 46 240 12 47 245 12.25 48 250 12.5 49 255 12.75 50 260 13
Example 18: High-Throughput Screening Assay Identifying Changes in Small Molecule Concentration and Membrane Permeability
Binding of a ligand to a receptor is known to alter intracellular levels of small molecules, such as calcium, potassium, sodium, and pH, as well as alter membrane potential. These alterations can be measured in an assay to identify supematants which bind to receptors of a particular cell. Although the following protocol describes an assay for calcium, this protocol can easily be modified to detect changes in potassium, sodium, pH, membrane potential, or any other small molecule which is detectable by a fluorescent probe.
The following assay uses Fluorometric Imaging Plate Reader ("FLIPR") to measure changes in fluorescent molecules (Molecular Probes) that bind small molecules. Clearly, any fluorescent molecule detecting a small molecule can be used instead of the calcium fluorescent molecule, fluo-3, used here. For adherent cells, seed the cells at 10,000 -20,000 cells/well in a Co-star black
96-well plate with clear bottom. The plate is incubated in a CO2 incubator for 20 hours. The adherent cells are washed two times in Biotek washer with 200 ul of HBSS (Hank's Balanced Salt Solution) leaving 100 ul of buffer after the final wash.
A stock solution of 1 mg/ml fluo-3 is made in 10% pluronic acid DMSO. To load the cells with fluo-3, 50 ul of 12 ug/ml fluo-3 is added to each well. The plate is incubated at 37°C in a CO2 incubator for 60 min. The plate is washed four times in the Biotek washer with HBSS leaving 100 ul of buffer.
For non-adherent cells, the cells are spun down from culture media. Cells are re-suspended to 2-5xl06 cells/ml with HBSS in a 50-ml conical tube. 4 ul of 1 mg/ml fluo-3 solution in 10% pluronic acid DMSO is added to each ml of cell suspension.
The tube is then placed in a 37°C water bath for 30-60 min. The cells are washed twice with HBSS, resuspended to lxlO6 cells/ml, and dispensed into a microplate, 100 ul/well. The plate is centrifuged at 1000 φm for 5 min. The plate is then washed once in Denley CellWash with 200 ul, followed by an aspiration step to 100 ul final volume. For a non-cell based assay, each well contains a fluorescent molecule, such as fluo-3. The supernatant is added to the well, and a change in fluorescence is detected.
To measure the fluorescence of intracellular calcium, the FLIPR is set for the following parameters: (1) System gain is 300-800 mW; (2) Exposure time is 0.4 second; (3) Camera F/stop is F/2; (4) Excitation is 488 nm; (5) Emission is 530 nm; and (6) Sample addition is 50 ul. Increased emission at 530 nm indicates an extracellular
signaling even which has resulted in an increase in the intracellular Ca*4" concentration.
Example 19: High-Throughput Screening Assay Identifying Tyrosine Kinase Activity
The Protein Tyrosine Kinases (PTK) represent a diverse group of transmembrane and cytoplasmic kinases. Within the Receptor Protein Tyrosine Kinase RPTK) group are receptors for a range of mitogenic and metabolic growth factors including the PDGF, FGF, EGF, NGF, HGF and Insulin receptor subfamilies. In addition there are a large family of RPTKs for which the corresponding ligand is unknown. Ligands for RPTKs include mainly secreted small proteins, but also membrane-bound and extracellular matrix proteins.
Activation of RPTK by ligands involves ligand-mediated receptor dimerization, resulting in transphosphorylation of the receptor subunits and activation of the cytoplasmic tyrosine kinases. The cytoplasmic tyrosine kinases include receptor associated tyrosine kinases of the src-family (e.g., src, yes, lck, lyn, fyn) and non- receptor linked and cytosolic protein tyrosine kinases, such as the Jak family, members of which mediate signal transduction triggered by the cytokine superfamily of receptors (e.g., the Interleukins, Interferons, GM-CSF, and Leptin). Because of the wide range of known factors capable of stimulating tyrosine kinase activity, the identification of novel human secreted proteins capable of activating tyrosine kinase signal transduction pathways are of interest. Therefore, the following protocol is designed to identify those novel human secreted proteins capable of activating the tyrosine kinase signal transduction pathways. Seed target cells (e.g., primary keratinocytes) at a density of approximately
25,000 cells per well in a 96 well Loprodyne Silent Screen Plates purchased from Nalge Nunc (Naperville, IL). The plates are sterilized with two 30 minute rinses with 100% ethanol, rinsed with water and dried overnight. Some plates are coated for 2 hr with 100 ml of cell culture grade type I collagen (50 mg/ml), gelatin (2%) or polylysine (50 mg/ml), all of which can be purchased from Sigma Chemicals (St. Louis, MO) or 10% Matrigel purchased from Becton Dickinson (Bedford,MA), or calf semm, rinsed with PBS and stored at 4°C. Cell growth on these plates is assayed by seeding 5,000 cells/well in growth medium and indirect quantitation of cell number through use of alamarBlue as described by the manufacturer Alamar Biosciences, Inc. (Sacramento, CA) after 48 hr. Falcon plate covers #3071 from Becton Dickinson (Bedford,MA) are
used to cover the Loprodyne Silent Screen Plates. Falcon Microtest III cell culture plates can also be used in some proliferation experiments.
To prepare extracts, A431 cells are seeded onto the nylon membranes of Loprodyne plates (20,000/200ml/well) and cultured overnight in complete medium. Cells are quiesced by incubation in serum-free basal medium for 24 hr. After 5-20 minutes treatment with EGF (60ng/ml) or 50 ul of the supernatant produced in Example 11, the medium was removed and 100 ml of extraction buffer ((20 mM HEPES pH 7.5, 0.15 M NaCl, 1% Triton X-100, 0.1% SDS, 2 mM Na3VO4, 2 mM Na4P2O7 and a cocktail of protease inhibitors (# 1836170) obtained from Boeheringer Mannheim (Indianapolis, IN) is added to each well and the plate is shaken on a rotating shaker for
5 minutes at 4°C. The plate is then placed in a vacuum transfer manifold and the extract filtered through the 0.45 mm membrane bottoms of each well using house vacuum. Extracts are collected in a 96-well catch/assay plate in the bottom of the vacuum manifold and immediately placed on ice. To obtain extracts clarified by centrifugation, the content of each well, after detergent solubihzation for 5 minutes, is removed and centrifuged for 15 minutes at 4°C at 16,000 x g.
Test the filtered extracts for levels of tyrosine kinase activity. Although many methods of detecting tyrosine kinase activity are known, one method is described here. Generally, the tyrosine kinase activity of a supernatant is evaluated by determining its ability to phosphorylate a tyrosine residue on a specific substrate (a biotinylated peptide). Biotinylated peptides that can be used for this puφose include PSK1 (corresponding to amino acids 6-20 of the cell division kinase cdc2-p34) and PSK2 (corresponding to amino acids 1-17 of gastrin). Both peptides are substrates for a range of tyrosine kinases and are available from Boehringer Mannheim. The tyrosine kinase reaction is set up by adding the following components in order. First, add lOul of 5uM Biotinylated Peptide, then lOul ATP/Mg2+ (5mM
ATP/50mM MgCl2), then lOul of 5x Assay Buffer (40mM imidazole hydrochloride, pH7.3, 40 mM beta-glycerophosphate, ImM EGTA, lOOmM MgCl2, 5 mM MnCl2?
0.5 mg/ml BSA), then 5ul of Sodium Vanadate(lmM), and then 5ul of water. Mix the components gently and preincubate the reaction mix at 30°C for 2 min. Initial the reaction by adding lOul of the control enzyme or the filtered supernatant.
The tyrosine kinase assay reaction is then terminated by adding 10 ul of 120mm EDTA and place the reactions on ice.
Tyrosine kinase activity is determined by transferring 50 ul aliquot of reaction mixture to a microtiter plate (MTP) module and incubating at 37°C for 20 min. This
allows the streptavadin coated 96 well plate to associate with the biotinylated peptide. Wash the MTP module with 300ul/well of PBS four times. Next add 75 ul of anti- phospotyrosine antibody conjugated to horse radish peroxidase(anti-P-Tyr-
POD(0.5u/ml)) to each well and incubate at 37°C for one hour. Wash the well as above.
Next add lOOul of peroxidase substrate solution (Boehringer Mannheim) and incubate at room temperature for at least 5 mins (up to 30 min). Measure the absorbance of the sample at 405 nm by using ELISA reader. The level of bound peroxidase activity is quantitated using an ELISA reader and reflects the level of tyrosine kinase activity.
Example 20: High-Throughput Screening Assay Identifying Phosphorylation Activity
As a potential alternative and/or compliment to the assay of protein tyrosine kinase activity described in Example 19, an assay which detects activation
(phosphorylation) of major intracellular signal transduction intermediates can also be used. For example, as described below one particular assay can detect tyrosine phosphorylation of the Erk-1 and Erk-2 kinases. However, phosphorylation of other molecules, such as Raf, JNK, p38 MAP, Map kinase kinase (MEK), MEK kinase, Src, Muscle specific kinase (MuSK), IRAK, Tec, and Janus, as well as any other phosphoserine, phosphotyrosine, or phosphothreonine molecule, can be detected by substituting these molecules for Erk-1 or Erk-2 in the following assay.
Specifically, assay plates are made by coating the wells of a 96-well ELISA plate with 0. lml of protein G (lug/ml) for 2 hr at room temp, (RT). The plates are then rinsed with PBS and blocked with 3% BSA/PBS for 1 hr at RT. The protein G plates are then treated with 2 commercial monoclonal antibodies (lOOng/well) against Erk-1 and Erk-2 (1 hr at RT) (Santa Cruz Biotechnology). (To detect other molecules, this step can easily be modified by substituting a monoclonal antibody detecting any of the above described molecules.) After 3-5 rinses with PBS, the plates are stored at 4°C until use.
A431 cells are seeded at 20,000/well in a 96-well Loprodyne filteφlate and cultured overnight in growth medium. The cells are then starved for 48 hr in basal medium (DMEM) and then treated with EGF (6ng/well) or 50 ul of the supematants obtained in Example 11 for 5-20 minutes. The cells are then solubilized and extracts filtered directly into the assay plate.
After incubation with the extract for 1 hr at RT, the wells are again rinsed. As a positive control, a commercial preparation of MAP kinase (lOng/well) is used in place of A431 extract. Plates are then treated with a commercial polyclonal (rabbit) antibody (lug/ml) which specifically recognizes the phosphorylated epitope of the Erk-1 and Erk-2 kinases (1 hr at RT). This antibody is biotinylated by standard procedures. The bound polyclonal antibody is then quantitated by successive incubations with Europium-streptavidin and Europium fluorescence enhancing reagent in the Wallac DELFIA instrument (time-resolved fluorescence). An increased fluorescent signal over background indicates a phosphorylation.
Example 21: Method of Determining Alterations in a Gene Corresponding to a Polynucleotide
RNA isolated from entire families or individual patients presenting with a phenotype of interest (such as a disease) is be isolated. cDNA is then generated from these RNA samples using protocols known in the art. (See, Sambrook.) The cDNA is then used as a template for PCR, employing primers surrounding regions of interest in
SEQ ID NO:X. Suggested PCR conditions consist of 35 cycles at 95°C for 30 seconds; 60-120 seconds at 52-58°C; and 60-120 seconds at 70°C, using buffer solutions described in Sidransky, D., et al., Science 252:706 (1991). PCR products is then sequenced using primers labeled at their 5' end with T4 polynucleotide kinase, employing SequiTherm Polymerase. (Epicentre Technologies). The intron-exon borders of selected exons is also determined and genomic PCR products analyzed to confirm the results. PCR products harboring suspected mutations is then cloned and sequenced to validate the results of the direct sequencing. PCR products is cloned into T-tailed vectors as described in Holton, T.A. and
Graham, M.W., Nucleic Acids Research, 19: 1 156 (1991) and sequenced with T7 polymerase (United States Biochemical). Affected individuals is identified by mutations not present in unaffected individuals.
Genomic rearrangements are also observed as a method of determining alterations in a gene corresponding to a polynucleotide. Genomic clones isolated according to Example 2 are nick-translated with digoxigenindeoxy-uridine 5'- triphosphate (Boehringer Manheim), and FISH performed as described in Johnson, Cg. et al., Methods Cell Biol. 35:73-99 (1991). Hybridization with the labeled probe is carried out using a vast excess of human cot-1 DNA for specific hybridization to the corresponding genomic locus.
Chromosomes are counterstained with 4,6-diamino-2-phenylidole and propidium iodide, producing a combination of C- and R-bands. Aligned images for precise mapping are obtained using a triple-band filter set (Chroma Technology, Brattleboro, NT) in combination with a cooled charge-coupled device camera (Photometries, Tucson, AZ) and variable excitation wavelength filters. (Johnson, Cv. et al., Genet. Anal. Tech. Appl., 8:75 (1991).) Image collection, analysis and chromosomal fractional length measurements are performed using the ISee Graphical Program System. (Inovision Coφoration, Durham, ΝC.) Chromosome alterations of the genomic region hybridized by the probe are identified as insertions, deletions, and translocations. These alterations are used as a diagnostic marker for an associated disease.
Example 22: Method of Detecting Abnormal Levels of a Polypeptide in a Biological Sample A polypeptide of the present invention can be detected in a biological sample, and if an increased or decreased level of the polypeptide is detected, this polypeptide is a marker for a particular phenotype. Methods of detection are numerous, and thus, it is understood that one skilled in the art can modify the following assay to fit their particular needs. For example, antibody-sandwich ELISAs are used to detect soluble polypeptides in a sample, preferably a biological sample. Wells of a microtiter plate are coated with specific antibodies, at a final concentration of 0.2 to 10 ug/ml. The antibodies are either monoclonal or polyclonal and are produced by the method described in Example 10. The wells are blocked so that non-specific binding of the polypeptide to the well is reduced.
The coated wells are then incubated for > 2 hours at RT with a sample containing the polypeptide. Preferably, serial dilutions of the sample should be used to validate results. The plates are then washed three times with deionized or distilled water to remove unbounded polypeptide. Next, 50 ul of specific antibody-alkaline phosphatase conjugate, at a concentration of 25-400 ng, is added and incubated for 2 hours at room temperature. The plates are again washed three times with deionized or distilled water to remove unbounded conjugate.
Add 75 ul of 4-methylumbelliferyl phosphate (MUP) or p-nitrophenyl phosphate (NPP) substrate solution to each well and incubate 1 hour at room temperature. Measure the reaction by a microtiter plate reader. Prepare a standard curve, using serial dilutions of a control sample, and plot polypeptide concentration on
the X-axis (log scale) and fluorescence or absorbance of the Y-axis (linear scale). Inteφolate the concentration of the polypeptide in the sample using the standard curve.
Example 23: Formulating a Polypeptide The secreted polypeptide composition will be formulated and dosed in a fashion consistent with good medical practice, taking into account the clinical condition of the individual patient (especially the side effects of treatment with the secreted polypeptide alone), the site of delivery, the method of administration, the scheduling of administration, and other factors known to practitioners. The "effective amount" for puφoses herein is thus determined by such considerations.
As a general proposition, the total pharmaceutically effective amount of secreted polypeptide administered parenterally per dose will be in the range of about 1 μg/kg/day to 10 mg/kg/day of patient body weight, although, as noted above, this will be subject to therapeutic discretion. More preferably, this dose is at least 0.01 mg/kg/day, and most preferably for humans between about 0.01 and 1 mg/kg day for the hormone. If given continuously, the secreted polypeptide is typically administered at a dose rate of about 1 μg/kg/hour to about 50 μg/kg/hour, either by 1-4 injections per day or by continuous subcutaneous infusions, for example, using a mini-pump. An intravenous bag solution may also be employed. The length of treatment needed to observe changes and the interval following treatment for responses to occur appears to vary depending on the desired effect.
Pharmaceutical compositions containing the secreted protein of the invention are administered orally, rectally, parenterally, intracistemally, intravaginally, intraperitoneally, topically (as by powders, ointments, gels, drops or transdermal patch), bucally, or as an oral or nasal spray. "Pharmaceutically acceptable carrier" refers to a non-toxic solid, semisolid or liquid filler, diluent, encapsulating material or formulation auxiliary of any type. The term "parenteral" as used herein refers to modes of administration which include intravenous, intramuscular, intraperitoneal, intrasternal, subcutaneous and intraarticular injection and infusion. The secreted polypeptide is also suitably administered by sustained-release systems. Suitable examples of sustained-release compositions include semi-permeable polymer matrices in the form of shaped articles, e.g., films, or mirocapsules. Sustained-release matrices include polylactides (U.S. Pat. No. 3,773,919, EP 58,481), copolymers of L-glutamic acid and gamma-ethyl-L-glutamate (Sidman, U. et al., Biopolymers 22:547-556 (1983)), poly (2- hydroxyethyl methacrylate) (R. Langer et al., J. Biomed. Mater. Res. 15: 167-277 (1981), and R. Langer, Chem. Tech. 12:98- 105 (1982)), ethylene vinyl acetate (R. Langer et al.) or poly-D- (-)-3-hydroxybutyric
acid (EP 133,988). Sustained-release compositions also include liposomally entrapped polypeptides. Liposomes containing the secreted polypeptide are prepared by methods known per se: DE 3,218,121; Epstein et al., Proc. Natl. Acad. Sci. USA 82:3688-3692 (1985); Hwang et al., Proc. Natl. Acad. Sci. USA 77:4030-4034 (1980); EP 52,322; EP 36,676; EP 88,046; EP 143,949; EP 142,641 ; Japanese Pat. Appl. 83-118008; U.S. Pat. Nos. 4,485,045 and 4,544,545; and EP 102,324. Ordinarily, the liposomes are of the small (about 200-800 Angstroms) unilamellar type in which the lipid content is greater than about 30 mol. percent cholesterol, the selected proportion being adjusted for the optimal secreted polypeptide therapy. For parenteral administration, in one embodiment, the secreted polypeptide is formulated generally by mixing it at the desired degree of purity, in a unit dosage injectable form (solution, suspension, or emulsion), with a pharmaceutically acceptable carrier, i.e., one that is non-toxic to recipients at the dosages and concentrations employed and is compatible with other ingredients of the formulation. For example, the formulation preferably does not include oxidizing agents and other compounds that are known to be deleterious to polypeptides.
Generally, the formulations are prepared by contacting the polypeptide uniformly and intimately with liquid carriers or finely divided solid carriers or both. Then, if necessary, the product is shaped into the desired formulation. Preferably the carrier is a parenteral carrier, more preferably a solution that is isotonic with the blood of the recipient. Examples of such carrier vehicles include water, saline, Ringer's solution, and dextrose solution. Non-aqueous vehicles such as fixed oils and ethyl oleate are also useful herein, as well as liposomes.
The carrier suitably contains minor amounts of additives such as substances that enhance isotonicity and chemical stability. Such materials are non-toxic to recipients at the dosages and concentrations employed, and include buffers such as phosphate, citrate, succinate, acetic acid, and other organic acids or their salts; antioxidants such as ascorbic acid; low molecular weight (less than about ten residues) polypeptides, e.g., polyarginine or tripeptides; proteins, such as semm albumin, gelatin, or immunoglobulins; hydrophilic polymers such as polyvinylpyrrolidone; amino acids, such as glycine, glutamic acid, aspartic acid, or arginine; monosaccharides, disaccharides, and other carbohydrates including cellulose or its derivatives, glucose, manose, or dextrins; chelating agents such as EDTA; sugar alcohols such as mannitol or sorbitol; counterions such as sodium; and/or nonionic surfactants such as polysorbates, poloxamers, or PEG.
The secreted polypeptide is typically formulated in such vehicles at a concentration of about 0.1 mg/ml to 100 mg/ml, preferably 1-10 mg/ml, at a pH of
about 3 to 8. It will be understood that the use of certain of the foregoing excipients, carriers, or stabilizers will result in the formation of polypeptide salts.
Any polypeptide to be used for therapeutic administration can be sterile. Sterility is readily accomplished by filtration through sterile filtration membranes (e.g., 0.2 micron membranes). Therapeutic polypeptide compositions generally are placed into a container having a sterile access port, for example, an intravenous solution bag or vial having a stopper pierceable by a hypodermic injection needle.
Polypeptides ordinarily will be stored in unit or multi-dose containers, for example, sealed ampoules or vials, as an aqueous solution or as a lyophilized formulation for reconstitution. As an example of a lyophilized formulation, 10-ml vials are filled with 5 ml of sterile-filtered 1% (w/v) aqueous polypeptide solution, and the resulting mixture is lyophilized. The infusion solution is prepared by reconstituting the lyophilized polypeptide using bacteriostatic Water-for-Injection.
The invention also provides a pharmaceutical pack or kit comprising one or more containers filled with one or more of the ingredients of the pharmaceutical compositions of the invention. Associated with such container(s) can be a notice in the form prescribed by a governmental agency regulating the manufacture, use or sale of pharmaceuticals or biological products, which notice reflects approval by the agency of manufacture, use or sale for human administration. In addition, the polypeptides of the present invention may be employed in conjunction with other therapeutic compounds.
Example 24: Method of Treating Decreased Levels of the Polypeptide
It will be appreciated that conditions caused by a decrease in the standard or normal expression level of a secreted protein in an individual can be treated by administering the polypeptide of the present invention, preferably in the secreted form. Thus, the invention also provides a method of treatment of an individual in need of an increased level of the polypeptide comprising administering to such an individual a pharmaceutical composition comprising an amount of the polypeptide to increase the activity level of the polypeptide in such an individual. For example, a patient with decreased levels of a polypeptide receives a daily dose 0.1-100 ug/kg of the polypeptide for six consecutive days. Preferably, the polypeptide is in the secreted form. The exact details of the dosing scheme, based on administration and formulation, are provided in Example 23.
Example 25: Method of Treating Increased Levels of the Polypeptide
Antisense technology is used to inhibit production of a polypeptide of the present invention. This technology is one example of a method of decreasing levels of a polypeptide, preferably a secreted form, due to a variety of etiologies, such as cancer. For example, a patient diagnosed with abnormally increased levels of a polypeptide is administered intravenously antisense polynucleotides at 0.5, 1.0, 1.5, 2.0 and 3.0 mg/kg day for 21 days. This treatment is repeated after a 7-day rest period if the treatment was well tolerated. The formulation of the antisense polynucleotide is provided in Example 23.
Example 26: Method of Treatment Using Gene Therapy
One method of gene therapy transplants fibroblasts, which are capable of expressing a polypeptide, onto a patient. Generally, fibroblasts are obtained from a subject by skin biopsy. The resulting tissue is placed in tissue-culture medium and separated into small pieces. Small chunks of the tissue are placed on a wet surface of a tissue culture flask, approximately ten pieces are placed in each flask. The flask is turned upside down, closed tight and left at room temperature over night. After 24 hours at room temperature, the flask is inverted and the chunks of tissue remain fixed to the bottom of the flask and fresh media (e.g., Ham's F12 media, with 10% FBS, penicillin and streptomycin, is added. The flasks are then incubated at 37°C for approximately one week.
At this time, fresh media is added and subsequently changed every several days. After an additional two weeks in culture, a monolayer of fibroblasts emerge. The monolayer is trypsinized and scaled into larger flasks. pMV-7 (Kirschmeier, P.T. et al., DNA, 7:219-25 (1988)), flanked by the long terminal repeats of the Moloney murine sarcoma vims, is digested with EcoRl and Hindlll and subsequently treated with calf intestinal phosphatase. The linear vector is fractionated on agarose gel and purified, using glass beads.
The cDNA encoding a polypeptide of the present invention can be amplified using PCR primers which correspond to the 5' and 3' end sequences respectively as set forth in Example 1. Preferably, the 5' primer contains an EcoRl site and the 3' primer includes a Hindlll site. Equal quantities of the Moloney murine sarcoma vims linear backbone and the amplified EcoRl and Hindlll fragment are added together, in the presence of T4 DNA ligase. The resulting mixture is maintained under conditions appropriate for ligation of the two fragments. The ligation mixture is then used to
transform bacteria HB 101, which are then plated onto agar containing kanamycin for the puφose of confirming that the vector has the gene of interest properly inserted. The amphotropic pA317 or GP+aml2 packaging cells are grown in tissue culture to confluent density in Dulbecco's Modified Eagles Medium (DMEM) with 10% calf semm (CS), penicillin and streptomycin. The MSV vector containing the gene is then added to the media and the packaging cells transduced with the vector. The packaging cells now produce infectious viral particles containing the gene (the packaging cells are now referred to as producer cells).
Fresh media is added to the transduced producer cells, and subsequently, the media is harvested from a 10 cm plate of confluent producer cells. The spent media, containing the infectious viral particles, is filtered through a millipore filter to remove detached producer cells and this media is then used to infect fibroblast cells. Media is removed from a sub-confluent plate of fibroblasts and quickly replaced with the media from the producer cells. This media is removed and replaced with fresh media. If the titer of vims is high, then virtually all fibroblasts will be infected and no selection is required. If the titer is very low, then it is necessary to use a retroviral vector that has a selectable marker, such as neo or his. Once the fibroblasts have been efficiently infected, the fibroblasts are analyzed to determine whether protein is being produced. The engineered fibroblasts are then transplanted onto the host, either alone or after having been grown to confluence on cytodex 3 microcarrier beads.
It will be clear that the invention may be practiced otherwise than as particularly described in the foregoing description and examples. Numerous modifications and variations of the present invention are possible in light of the above teachings and, therefore, are within the scope of the appended claims. The entire disclosure of each document cited (including patents, patent applications, journal articles, abstracts, laboratory manuals, books, or other disclosures) in the Background of the Invention, Detailed Description, and Examples is hereby incoφorated herein by reference.
(1) GENERAL INFORMATION:
(i) APPLICANT: Human Genome Sciences, Inc. et al . (ii) TITLE OF INVENTION: 186 Human Secreted Proteins (iii) NUMBER OF SEQUENCES: 644 (iv) CORRESPONDENCE ADDRESS:
(A) ADDRESSEE: Human Genome Sciences, Inc. (B) STREET: 9410 Key West Avenue
(C) CITY: Rockville
(D) STATE: Maryland
(E) COUNTRY: USA
(F) ZIP: 20850
(v) COMPUTER READABLE FORM:
(A) MEDIUM TYPE: Diskette, 3.50 inch, 1.4Mb storage
(B) COMPUTER: HP Vectra 486/33
(C) OPERATING SYSTEM: MSDOS version 6.2 (D) SOFTWARE: ASCII Text
(vi) CURRENT APPLICATION DATA:
(A) APPLICATION NUMBER:
(B) FILING DATE: March 6, 199! (C) CLASSIFICATION:
(vii) PRIOR APPLICATION DATA:
(A) APPLICATION NUMBER:
(B) FILING DATE:
(viii) ATTORNEY/AGENT INFORMATION:
(A) NAME: A. Anders Brookes, Esq.
(B) REGISTRATION NUMBER: 36,373
(C) REFERENCE/DOCKET NUMBER: PS002.PCT
(vi) TELECOMMUNICATION INFORMATION:
(A) TELEPHONE: (301) 309-8504
(B) TELEFAX: (301) 309-8439
(2) INFORMATION FOR SEQ ID NO: 1:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 733 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 1:
GGGATCCGGA GCCCAAATCT TCTGACAAAA CTCACACATG CCCACCGTGC CCAGCACCTG 60
AATTCGAGGG TGCACCGTCA GTCTTCCTCT TCCCCCCAAA ACCCAAGGAC ACCCTCATGA 120
TCTCCCGGAC TCCTGAGGTC ACATGCGTGG TGGTGGACGT AAGCCACGAA GACCCTGAGG 180
TCAAGTTCAA CTGGTACGTG GACGGCGTGG AGGTGCATAA TGCCAAGACA AAGCCGCGGG 240
AGGAGCAGTA CAACAGCACG TACCGTGTGG TCAGCGTCCT CACCGTCCTG CACCAGGACT 300
GGCTGAATGG CAAGGAGTAC AAGTGCAAGG TCTCCAACAA AGCCCTCCCA ACCCCCATCG 360
AGAAAACCAT CTCCAAAGCC AAAGGGCAGC CCCGAGAACC ACAGGTGTAC ACCCTGCCCC 420
CATCCCGGGA TGAGCTGACC AAGAACCAGG TCAGCCTGAC CTGCCTGGTC AAAGGCTTCT 480
ATCCAAGCGA CATCGCCGTG GAGTGGGAGA GCAATGGGCA GCCGGAGAAC AACTACAAGA 540
CCACGCCTCC CGTGCTGGAC TCCGACGGCT CCTTCTTCCT CTACAGCAAG CTCACCGTGG 600
ACAAGAGCAG GTGGCAGCAG GGGAACGTCT TCTCATGCTC CGTGATGCAT GAGGCTCTGC 660
ACAACCACTA CACGCAGAAG AGCCTCTCCC TGTCTCCGGG TAAATGAGTG CGACGGCCGC 720
GACTCTAGAG GAT 733
(2) INFORMATION FOR SEQ ID NO: 2:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 5 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 2:
Trp Ser Xaa Trp Ser 1 5
(2) INFORMATION FOR SEQ ID NO: 3:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 86 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 3:
GCGCCTCGAG ATTTCCCCGA AATCTAGATT TCCCCGAAAT GATTTCCCCG AAATGATTTC 60 CCCGAAATAT CTGCCATCTC AATTAG 86
(2) INFORMATION FOR SEQ ID NO: 4:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 4: GCGGCAAGCT TTTTGCAAAG CCTAGGC 27
(2) INFORMATION FOR SEQ ID NO: 5:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 271 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 5: CTCGAGATTT CCCCGAAATC TAGATTTCCC CGAAATGATT TCCCCGAAAT GATTTCCCCG 60
AAATATCTGC CATCTCAATT AGTCAGCAAC CATAGTCCCG CCCCTAACTC CGCCCATCCC 120
GCCCCTAACT CCGCCCAGTT CCGCCCATTC TCCGCCCCAT GGCTGACTAA TTTTTTTTAT 180 TTATGCAGAG GCCGAGGCCG CCTCGGCCTC TGAGCTATTC CAGAAGTAGT GAGGAGGCTT 240
TTTTGGAGGC CTAGGCTTTT GCAAAAAGCT T 271
(2) INFORMATION FOR SEQ ID NO: 6:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 32 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 6:
GCGCTCGAGG GATGACAGCG ATAGAACCCC GG 32
(2) INFORMATION FOR SEQ ID NO: 7:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 31 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 7:
GCGAAGCTTC GCGACTCCCC GGATCCGCCT C 31
(2) INFORMATION FOR SEQ ID NO: 8: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 12 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 8: GGGGACTTTC CC 12
(2) INFORMATION FOR SEQ ID NO: 9: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 73 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 9:
GCGGCCTCGA GGGGACTTTC CCGGGGACTT TCCGGGGACT TTCCGGGACT TTCCATCCTG 60 CCATCTCAAT TAG 73
(2) INFORMATION FOR SEQ ID NO: 10:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 256 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 10: CTCGAGGGGA CTTTCCCGGG GACTTTCCGG GGACTTTCCG GGACTTTCCA TCTGCCATCT 60
CAATTAGTCA GCAACCATAG TCCCGCCCCT AACTCCGCCC ATCCCGCCCC TAACTCCGCC 120
CAGTTCCGCC CATTCTCCGC CCCATGGCTG ACTAATTTTT TTTATTTATG CAGAGGCCGA 180
GGCCGCCTCG GCCTCTGAGC TATTCCAGAA GTAGTGAGGA GGCTTTTTTG GAGGCCTAGG 240
CTTTTGCAAA AAGCTT 256
(2) INFORMATION FOR SEQ ID NO: 11: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 582 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 11:
GGCACGAGGT AATTTCTACC AGAAATTTCC AGAGCATTAT GTAGGTAGAA AAAAATGCAA 60 GCAAGCTGTT AAAGATCTTG GATCCCATTA TATAGTATGT ATAGCTGAAA TCTGTAATTC 120
AATCACTTTT TCTCTTTTAT CCTCTAACCA AAAAATTGTT TAATTTTGCA TCCCAAATGT 180
TTTTAATCTT TGTATATTTT TTAAAAATCC TTTTCTCCTC ATCATTGCCT TTTTTGTGGT 240
TGTAAATAGA CTTACTTGCA CTTTGAAGAT GAGTTACTCC TTGTCATCTT ACAAATATGT 300
GATATGGTAA TTTTCATAAC AGATGTCAGT TTTGAACCAA GAATTGGTGA TTTGTTTATA 360 AGAAAAAAAC TGGCTTCATT TCTGTGAAAT TGCTCTTTGA AAATTTCTTT TTACACGTGT 420
AAGCCAACTG AGATACCGTG ATGGTGTTGA TTTCTTTCAA TGATGCTTAC CATCTATTTT 480
AGCCACTGAG CCTTTTATTA TTTGTCTATT TGTAAAGTTT ATTTGTCTTA ACTCATTTAA 540
TAAATATACT GTTTATCTGT TTCTGAAAAA AAAAAAAAAA AA 582
(2) INFORMATION FOR SEQ ID NO: 12:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 465 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 12:
GTTTGGGGGT GAGGCCGAGC TGCTGCGGGG CTTCGTCGCC GGCCAGGACA CAGCTACTCG 60
CACGGCGGCG GCGCCTGGCT ATGATGTTCC TCACCCAGGG CGGGCCTCTG CCCTCTACTC 120 GTGCCAGGCC CACTTGCCAG GCAGGAGCCC TCCCCAAGCC TTCAGGGCTG CTCGGAGTCA 180
CCTGTTGGAA TGGACTAAAA GGACCCTTGT GTGGGAACAG GTGCTCCCCA AACACCCTGC 240
TGCTGGCTGC CAGGCAGGCC CTCTGGAAGG GAAGGGGCAG GACTCATCAG GACCTCCCTG 300
GACCCCTGCA GGGCAGGCAG CTTGGGCCCG AGCCCAAGCA TTTGGCTCTG CTGCCCCCAA 360
GGGGACAGGA AGCCTCTTGG GCCTCTTCCC TTCCTGGACA AGGCCCCCTG CCTTTGCCTC 420 ACATAAACTG TACAGTATTT TCATTAAAAG CCTCTTTCAT AAAAA 465
( 2 ) INFORMATION FOR SEQ ID NO : 13 :
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 474 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 13: ATGCAATTCC TGCTCACAGC CTTTCTGTTG GTGCCACTTC TGGCTCTTTG TGATGTCCCC 60
ATATCCCTAG GCTTCTCCCC CTCCTAGAAG GGCTTCTTGA TAGATTAGAA AATAAGAATG 120
AGTGACATTT CCTATGTGCA TATAAGAAGG AGCCACAAGA CATGTCTTTT AAATAAAAGG 180
ACAGTGTCCA TCCTTTTAGC TGCCGAATAG AACCTTGGTC TCATCCTCCT GGAGCTAGGC 240
CTTTAAAACA GCTTCTGTGT TTCTCATTTG TCTCAGTGTT TTGCCAGGGT TTTATCGGAA 300 AGATAATGTT CCGTTTAAAA TATTTCCTAA TGAGGCCGGG CGTGGTGGCT CACGCCTGTA 360
ACCCTAGCAM TTGGGGGCTG AGCGGGTGGA TCACGAGGTC AGGAGATCGA GACCATCCTG 420 GSTAACATGG TGAAACCCCG TCTCTACTAA AAATACAAAA AAAAAAAAAA AAAA 474
(2) INFORMATION FOR SEQ ID NO: 14:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 314 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 14:
TTATGTTGGG GAGCAAGACC TGATAGCCAG CCTTTACATG GGAGTATAAT TCTGTCCTCC 60
ATCTCATAAG CCCCAGTACC TGAGCCAGAA TGATTATAAC CAACCACACT GTCTCTTTAT 120
CATGGATGGC TTTAGCAGTA GGTTATTTTC ATCATTGCCA TTTGTAGCTC TACAGTGGTT 180 TATAGTAATT TCTCATCTTT TAAGTCTCTC CCTCAGTGCC TGTTGTTATC AAACTCATTG 240
CTCTCTCANG CAGTTGAGCT CTGCATTCTC CCYTATGGGG GAGAGCTGTG TTGGAGAGAG 300
AGAATATNAC TTCC 314
(2) INFORMATION FOR SEQ ID NO: 15:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 613 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 15:
CTCATATTGC CGTCTGGCTA AAAGTGAACA TGCCATTGAT CAATCTGCTT TTATTATATT 60
ATGTTCCTAA TGGTGGCAAG CAAGACAAGA AGTAGAAAGA AAGATGGTGT AAGCTCAAGA 120
ACCCACTAAA TCTATCCTAT GGCCTGGGTT CACCCAGCCT GCTTTGTGGA TTTTGTCTCA 180 CTATAACAGA GCTCCCAAGG AGACTGCAGA GTCAGCTCCC TTAAGCACTG TAACTAAAGC 240
CTAACTCTTC CGTTCCACCC AACAATGTYC CCAGCTCATC CTCTTTCCCR AAGTCCCCTT 300
TCTGCCCCAG ATGCGAATTG CATTTAACTA ATCCTCAAGT GAAATGTCCA CACAGRATTC 360
CATTTTAATT AGCATACCAT AGTTTTTGTG CAAATTTGCT TTCAGARGAC TCCCATTGCA 420
GCTGCTCAGA GACGCTAAWG GCAGGGCCTC TTGAWGCTTT CCCGATAGCT TTCAGCTGCA 480 ATAGCTCTTA GGCAGAATGC CATGAGCGTC CTGCCCAACT GTATTACTGG GGAACACCTG 540
ATTGGCTAGA AGTTGATCCT CCTGTAACTT TTCTGAGTTC TTTACATTTA CTCGTGAAAC 600 CCAAATATGC CAC 613
(2) INFORMATION FOR SEQ ID NO: 16:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 356 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 16:
CCCCCCCCAT TGAACCCTGG GCTGTGAAAG TTTTTGCCTG TGTGGGTCGT TCTGTGTGGC 60
GCCTGGTGTG TGGKTCCCAA CTCCTGTTGC AAAGTGGCAG CAGCCAATCA TGAAGCGCCC 120
TTATTTTTAG TTGCAGATGA CCAGGTCTCC CCCCCACAGC CTCTGTCTGG TCCCTCATTG 180 GTGAGTGGTC TGCCTGCCCA AGGAGCCTGA TTGGTGGGAA ATGGCATCAT CTAATATGAT 240
GGGAAGGCAT TTGGTCCTGG TTATGTTTAT TACAACATCA TTGCACTCTG GGACTCCAGT 300
CCCTGAAAAC GTAATTTGTG GTGTTACCAA AGGACCACAG GGGAAAAAAA AAAAAA 356
(2) INFORMATION FOR SEQ ID NO: 17:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 414 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 17:
GAAACTANAT CCCGGGGCTT TTAACNGGTA CTTGGGAAAT AAGTATTGGG TAATCACTAA 60
GNGGACATTG ACTGCACCAA ACCAAAGCTA TAGAAAGAAA TGATTGACTT TTTAAAATAT 120
ATTCACATTA ACTGTCCTAG GATACTTCTC TTGAGGCTTT GGAAAACTTC TTCCTTGAAA 180 TTTGCATATC CACTCCAGTT CTGTCACCAA AGATTTTAAT CTTCAGATCG CAATTTCCTC 240
TCTCCCAGAA AAAAGTACTA CAACAGGCTC AAGGGATATG CTTTGGTGGT CAAGGGATTA 300
CACTATGGTT TTCCTTCTGT TCACAATGGT ATTTACAGGA GACCTTGTCA TCAGAGGACG 360
TACTGAACTA TCTTTATGAC TTTGGATTTG ATCAGAGGTT TAAAAAAAAA AAAA 414
(2) INFORMATION FOR SEQ ID NO: 18:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 469 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 18:
AATCACCATT GCAATACAAA TGATCTGCCT GGTGAATGYT GAGCTGTACC CCACATTCGT 60
CAGGAACYTC GGAGTGATGG TGTGTTCCTC CCTGTGTGAC ATAGGTGGGA TAATCACCCC 120 CTTCATAGTC TTCAGGCTGA GGGAGGTCTG GCAAGCCTTG CCCCTCATTT TGTTTGCGGT 180
GTTGGGCCTG CTTGCCGCGG GAGTGACGCT ACTTCTTCCA GAGACCAAGG GGGTCGCTTT 240
GCCAGAGACC ATGAAGGACG CCGAGAACCT TGGGAGAAAA GCAAAGCCCA AAGAAAACAC 300
GATTTACCTT AAGGTCCAAA CCTCAGAACC CTCGGGCACC TGAGAGAGAT GTTTTGCGGC 360
GATGTCGTGT TGGAGGGATG AAGATGGAGT TATCCTCTGC AGAAATTCCT AGACGCCTTC 420 ACTTCTCTGT ATTCTTCCTC ATACTTGCCT ACCCCCAAAT TAATATCAG 469
(2) INFORMATION FOR SEQ ID NO: 19:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 550 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 19: CCCCCCCCCC CCCCCACACT TTCAGGAGTC ACCCCCCAGC ATTTGGGGTT GGGTTGGCCC 60
TACTCCAGCC TGGAGCTCCC TGAGGGAGCC TGCACTCCCT GCTCCCAATC CCCGCTACTG 120
GTGCAGGGAT GCAGCCTGGA GCTGGCGTCC TTGTTCTGGG CCTGCTGCTG CCGCCACCCC 180
AGAGCCCCAG CCTGTCCTGA ATTGACATCA GTGCTTCCCT GAACTGCCTC CCCCACCCCT 240
GGGCATTATC CCAGGAAACT TTATGTTTTC TAGAAGCTAA GCAGCTGCTG GGACTCAGGG 300 ACTGGTGCAG GTAGGCTGAG TGGCAGCTCA GTCCTAGAAG GTCTCTGAAG ATCTGGACTG 360
AGGACCTTGC TACTCCCCAA GCCAGAGCCC ATCAGCCAGG CCTGCTGTGA GCCACCTGCC 420
TGTGGAGTGC TGAGCTCAAC CAAAGGCTGG CAAGCTCTGG GCCTCATTTA AGGGATTCTG 480
ATGAGCCGAT GGGCCCTGGA GGCAGCCCAT TAAAGCATCT GGCTCGTTTT TGGAAAAAAA 540 AAAAAAAAAA 550
(2) INFORMATION FOR SEQ ID NO: 20:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 741 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 20:
TCTTGAAGAG TGTACAGTAC AGGATTATTA TAATGAAAGT TTATATCAAC AGGGTTTCGT 60 TGGCTCTGCA TATATTATAA GCAAAAGAGA TTGGTAAAGT GCCACAGTAT TCCAGATAAC 120
TTTTCAGTTG CGGCCTTTCT TCTCGTTCTT TAATTTGAAA CCTAGATACA TGCAGTAAAA 180
ACTAGGAGAA TGACTTTTAC CCTTGGGGAC AGCCAAGTTT TGTTGATAAA CCTATTTCCT 240
AGCATGCCTT CAGGAAGTTG TGCCAGACCC TAGATTGTGA AGGACCCACT GTTCTTCTGT 300
TGTACGAGCT CCCTGAACCA TTGTTCAGAG GACCAATGTC ACATCGCTTC ATGGGCATGG 360 NCCATGGGAG CATCTGGGTG ATAYCTGTCT ACAGTATTGG CTCTTCTGCG AGGCTGATAC 420
ACAAGGCCTC TCTTCCACAT GATCATTTGC AAACCTCCCC CAGCCCCTAC CATCCAATGT 480
GGAAGGAAAA CAAGAACTGC CTGAAGAAGA GTCCAAGCTA CAGATACACA GCGTGTGCAT 540
TGCGGCTGTC ACCTTCCTCC TCCCACTTCT GTATCCTCAG AGATGCTGCG TGGATGTTTC 600
CTTAACCTCA GCTGACTTCC CTGTGAATGT CTAATGCTAG TTCAGGGCCT CCAGGCATTG 660 ATTTGTACAG TGGTAACTCC CAATGAGGCT TCTGTTATCA TTTGGTGTGC TTTYTCTGTC 720
ATTAAAAGAA ATGATTTTCC C 741
(2) INFORMATION FOR SEQ ID NO: 21:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 991 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 21:
GGCACGAGTC TCCCCTGGGG AAGTTTTTCT TTTTCAGGAG GGAGGAGGGC TTTCCCAGGT 60 AATGTGTCTA GAGTGTTGGG CAGAAAATCT GGGACCACAC CACACCAGTT CTCTCCTTAA 120
TCCACGTCAT TTGCCTTCTA TCCCAGCTAT GTTTCCAGTG TCCTCTGGGT GTTTCCAAGA 180
GCAACAAGAA ATGAATAAAT CTCTGGTGAG TTGTTTATTT GTTCTTCACT TTGTTTTACA 240 CTGTATTTTC TGAGTTTATG GGTGTCTGTG AATTAAAAAG GAAAAGTAGA AATAAGTAAA 300
ACTCAGGTTG AAGGAAATAT ACATAAATAA GATAAAGCTG ACCTGTAGAT ATAGCAGGTT 360
ATAAAGCTTA GAGTTGTCTA AGTTGAGTGC AAATTTTCCT CTGATCTTTC TGATGCCGAA 420
CAAAAAAGCA GTCATGTTTG TTATGTGATT GGAATGGAAC CCGAGAAGAG AGCATGCTGT 480
GTTCTTGTGG GACAGGAAAG CTTGCGTGCA CCAAGTCTGA ACCACCACCT TCATGGTGAC 540 ATAGATTATG TGCTGGAACA TATTTCACAC CGGCCTGGCA GTAAACACTT GTAGTGTTGT 600
GCAGTGGAAA CGGTCATCTT CCGCTAAAGC ACGGCGTGTT GTGCAGCGGA AATGGTCATC 660
TGCTGCTAAA ACACAGCTTC CATCGTAATG TATGCTCCTT ACTCAAAGAG TGTGGTCCCA 720
AACAGCCTTT GGGAGGTCCT CCTTGATTCA TGGATGAAAC CTGGAACATC TTGAGGACTG 780
AGTTAACCAT AGGTCCTTAA ATAACTCTCC ACACGTTTTT CTTAGTTTAT CTCTACATGC 840 AGGGTGTGCA GCAGCCTGTT CAAAGTCATA TTTTCTGGGA AATATTTCCA GTGTTTATTT 900
GCACTTTAGC CCACTCTGTG TAGCCTTATT TCTTCTAAAC TCACCATTAA TCTGAATAAT 960
AGTCAAATTT AGGGGGACTG TATTTGCCTT A 991
(2) INFORMATION FOR SEQ ID NO: 22:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 653 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 22:
CCACGCGTCC GGAATTCCCC TGAGGATCTT GGGCTATCTT TGACAGGGGA TTCTTGCAAG 60
TTGATGCTTT CTACAAGTGA ATATAGTCAG TCCCCAAAGA TGGAGAGCTT GAGTTCTCAC 120
AGAATTGATG AAGATGGAGA AAACACACAG ATTGAGGATA CGGAACCCAT GTCTCCAGTT 180 CTCAATTCTA AATTTGTTCC TGCTGAAAAT GATAGTATCC TGATGAATCC AGCACAGGAT 240
GGTGAAGTAC AACTGAGTCA GAATGATGAC AAAACAAAGG GAGATGATAC AGACACCAGG 300
GATGACATTA GTATTTTAGC CACTGGTTGC AAGGGCAGAG AAGAAACGGT AGCAGAAGAA 360
GTTTGTATTG ATCTCACTTG TGATTCGGGG AGTCAGGCAG TTCCGTCACC AGCTACTCGA 420
TCTGAGGCAC TTTCTAGTGT GTTAGATCAG GAGGAAGCTA TGGAAATTAA AGAACACCAT 480 CCAGAGGAGG GGTCTTCAGG GTCTGAGGTG GAAGAAATCC CTGAGACACC TTGTGAAAGT 540
CAAGGAGAGG AACTCAAAGA AGAAAATATG GAGAGTGTTC CGTTGCACCT TTCTCTGACT 600
GAAACTCAGT CCCAAGGGTT GTGTCTTCGG AGGCATCCAA AAAAAAAAAA AAA 653
(2) INFORMATION FOR SEQ ID NO: 23:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1486 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 23:
GGCAGGCTGA CGACCTGCAA GCCACAGTGG CTGCCCTGTG CGTGCTGCGA GGTGGGGGAC 60
CCTGGGCAGG AAGCTGGCTG AGCCCCAAGA CCCCGGGGGC CATGGGCGGG GATCTGGTGC 120
TTGGCCTGGG GGCCTTGAGA CGCCGAAAGC GCTTGCTGGA GCAGGAGAAG TCTCTRGCCG 180
GCTGGGCACT GGTGCTGGCA SGARCTGGCA TTGGACTCAT GGTGCTGCAT GCAGAGATGC 240
TGTGGTTCGG GGGGTGCTCG GCTGTCAATG CCACTGGGCA CCTTTCAGAC ACACTTTGGC 300
TGATCCCCAT CACATTCCTG ACCATCGGCT ATGGTGACGT GGTGCCGGGC ACCATGTGGG 360
GCAAGATCGT YTGCCTGTGC ACTGGAGTCA TGGGTGTCTG CTGCACAGCC CTGCTGGTGG 420
CCGTGGTGGC CCGGAAGCTG GAGTTTAACA AGGCAGAGAA GCACGTGCAC AACTTCATGA 480
TGGATATCCA GTATACCAAA GAGATGAAGG AGTCCGCTGC CCGAGTGCTA CAAGAAGCCT 540
GGATGTTCTA CAAACATACT CGCAGGAAGG AGTCTCATGC TGCCCGCANG CATCAGCGCA 600
ANCTGCTGGC CGCCATCAAC GCGTTCCGCC AGGTGCGGCT GAAACACCGG AAGCTCCGGG 660
AACAAGTGAA CTCCATGGTG GACATCTCCA AGATGCACAT GATCCTGTAT GACCTGCAGC 720
AGAATCTGAG CAGCTCACAC CGGGCCCTGG AGAAACAGAT TGACACGCTG GCGGGGAAGC 780
TGGATGCCCT GACTGAGCTG CTTAGCACTG CCCTGGGGCC GAGGCAGCTT CCAGAACCCA 840
GCCAGCAGTC CAAGTAGCTG GACCCACGAG GAGGAACCAG GCTACTTTCC CCAGTACTGA 900
GGTGGTGGAC ATCGTCTCTG CCACTCCTGA CCCAGCCCTG AACAAAGCAC CTCAAGTGCA 960
AGGACCAAAG GGGGCCCTGG CTTGGAGTGG GTTGGCTTGC TGATGGCTGC TGGAGGGGAC 1020
GCTGGCTAAA GTGGGKAGGC CTTGGCCCAC CTGAGGCCCC AGGTGGGAAC ATGGTCACCC 1080
CCACTCTGCA TACCCTCATC AAAAACACTC TCACTATGCT GCTATGGACG ACCTCCAGCT 1140
CTCAGTTACA AGTGCAGGCG ACTGGAGGCA GGACTCCTGG GTCCCTGGGA AAGAGGGTAC 1200
TAGGGGCCCG GATCCAGGAT TCTGGGAGGC TTCAGTTACC GCTGGCCGAG CTGAAGAACT 1260
GGGTATGAGG CTGGGGCGGG GCTGGAGGTG GCGCCCCCTG GTGGGACAAC AAAGAGGACA 1320
CCATTTTTCC AGAGCTGCAG AGAGCACCTG GTGGGGAGGA AGAAGTGTAA CTCACCAGCC 1380
TCTGCTCTTA TCTTTGTAAT AAATGTTAAA GCCAGAAAAA AATAAAAAAA AAAAAAAAAA 1440
AACTCGAGGG GGGCCCRKAC CCAATCWCCC TATAGTAKAC GTANNN 1486
(2) INFORMATION FOR SEQ ID NO: 24:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2323 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 24:
CTTCGCCGTT TCTCCTGCCA GGGGAGGTCC CGGCTTCCCG TGGAGGCTCC GGACCAAGCC 60
CCTTCAGCTT CTCCCTCCGG ATCGATGTGC TGCCGCCGCC GCCGCCGCCG TCCCGCGTCC 120
TTCGGTCTCT GCTCCCGGGA CCCGGCTCCG CGCAGCCAGC CAGCATGTCG GGGATCAAGA 180
AGCAAAAGAC GGAGAACCAG CAGAAATCCA CCAATGTAGT CTATCAGGCC CACCATGTGA 240
GCAGGAATAA GAGAGGGCAA GTGGTTGGAA CAAGGGGTGG GTTCCGAGGA TGTACCGTGT 300
GGCTAACAGG TCTCTCTGGT GCTGGGAAAA ACAACGATAA GTTTTGCCCT GGAGGAGTAC 360
TTGTCTCCCA TGCCATCCCT GTTAATTCCT GGATGGGGAC AATGTCCGTC ATGGCCTTAA 420
CAGAATCCCC CAGATGGCTT CATGGCCCCC AAAGCATGGA AGGTCCTGAC AGATTATTAC 480
AGGTCCCTGC AGAAGAACTA AGCCTTTGGT CCAGAGTTTC TTTCTGAAGT GCTCTTTGAT 540
TACCTTTTCT ATTTTTATGA TTAGATGCTT TGTATTAAAT TGCTTCTCAA TGATGCATTT 600
TAATCTTTTA TAATGAAGTA AAAGTTGTGT CTATAATTAA AAAAATATAT ATATATATAC 660
ACACACACAT ATACATACAA AGTCAAACTG AAGACCAAAT CTTAGCAGGT AAAAGCAATA 720
TTCTTATACA TTTCATAATA AAATTAGCTC TATGTATTTT CTACTGCACC TGAGCAGGCA 780
GGTCCCAGAT TTCTTAAGGC TTTGTTTGAC CATGTGTCTA GTTACTTGCT GAAAAGTGAA 840
TATATTTTCC AGCATGTCTT GACAACCTGT ACTCTTCCAA TGTCATTTAT CAGTTGTAAA 900
ATATATCAGA TGTGTCCTCT TCTGTACAAT TGACAAAAAA AAAAATTTTT TTTTCTCACT 960
CTAAAAGAGG TGTGGCTCAC ATCAAGATTC TTCCTGATAT TTTACCTCAT GCTGTACAAA 1020
GCCTTAATGT TGTAATCATA TCTTACGTGT TGAAGACCTG ACTGGAGAAA CAAAATGTGC 1080
AATAACGTGA ATTTTATCTT AGAGATCTGT GCAGCCTATT TCTGTCACAA AAGTTATATT 1140
GTCTAATAAG AGAAGTCTTA ATGGCCTCTG TGAATAATGT AACTCCAGTT ACACGGTGAC 1200
TTTTAATAGC ATACAGTGAT TTGATGAAAG GACGTCAAAC AATGTGGCGA TGTCGTGGAA 1260
AGTTATCTTT CCCGCTCTTT GCTGTGGTCA TTGTGTCTTG CAGAAAGGAT GGCCCTGATG 1320
CAGCAGCAGC GCCAGCTGTA ATAAAAAATA ATTCACACTA TCAGACTAGC AAGGCACTAG 1380 AACTGGAAAA GACCACAGAA AACAAAGAAT CCAACCCTTT CATCTTACAG GTGAACAAAC 1440 TGTGATGATG CACATGTATG TGTTTTGTAA GCTGTGAGCA CCGTAACAAA ATGTAAATTT 1500 GCCATTATTA GGAAGTGCTG GTGGCAGTGA AGAAGCACCC AGGCCACTTG ACTCCCAGTC 1560 TGGTGCCCTG TCTACACCAG ACAACACAGG AGCTGGGTCA GATTCCCCTC AGCTGCTTAA 1620 CAAAGTTCCT CGAACAGAAA GTGCTTACAA AGCTGCCTTC TCGGATACTG AAAGGTCGAG 1680 TTTTCTGAAC TGCACTGATT TTATTGCAGT TGAAAAAAAA AAAAAGCTAT TCCAAAGATT 1740 TCAAGCTGTT CTGAGACATC TTCTGATGGC TTTACTTCCT GAGAGGCAAT GTTTTTACTT 1800 TATGCATAAT TCATTGTTGC CAAGGAATAA AGTGAAGAAA CAGCACCTTT TAATATATAG 1860 GTCTCTCTGG AAGAGACCTA AATTAGAAAG AGAAAACTGT GACAATTTTC ATATTCTCAT 1920 TCTTAAAAAA CACTAATCTT AACTAACAAA AGTTCTTTTG AGAATAAGTT ACACACAATG 1980 GCCACAGCAG TTTGTCTTTA ATAGTATAGT GCCTATACTC ATGTAATCGG TTACTCACTA 2040 CTGCCTTTAA AAAAAAAAAC CAGCATATTT ATTGAAAACA TGAGACAGGA TTATAGTGCC 2100 TTAACCGATA TATTTTGTGA CTTAAAAAAT ACATTTAAAA CTGCTCTTCT GCTCTAGTAC 2160 CATGCTTAGT GCAAATGATT ATTTCTATGT ACAACTGATG CTTGTTCTTA TTTTAATAAA 2220 TTTATCAGAG TGAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA 2280 AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAA 2323
(2) INFORMATION FOR SEQ ID NO: 25:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 683 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 25: GGCACGAGCC TGTGTGGTCA TGTTCCTCGT GGTGCAGTAC CTGACATGAG CCAGCCACGC 60 TCAGTGGCTG AACAGCATTC CCACAGCCTG CAAGTGTGTG TGTGTGTGAA AGAGAGAGGG 120 GGGCCCAGAG CCGCCTTTTG AAATGTTTGC CTGTCTGAAC TGTGAAGACA CTTGGGAGTG 180 ATTGTGGTCT AATTTCCAAC CTGCTCTGTT TTCTGTGACA TCTTGGAGGG GAGCTAGTGC 240 CACACCATGC GCGGTGCTTA GAAATGAAAA AGTCCCGGGT CTGTCTCTCT CACTCTCGCT 300 CTCATGGGGG AGGGAAAGAA TGGCTTTGGT GGCTTTGTTC ACACAGCTGA TGCGTGCTGG 360 GAAGGTGTCC ACAGTGAGCC TGTGTGCAGG ACTGTCCACA CGGTTCACAC TTGTCACCAT 420
CAGGCCTTTC TGGTCCTGAT AGGGTGGAGC AAAAGTGGAA AGGAAAGGAA AGAGGCTTTT 480
CTCACAGCCA TTATATTAAA TAGTAGGTCG ATTCACATCT CGTGCTCCTG GCCACCTTCC 540
CCTGTGCCTC AGTGACATGT AGATGACTGA CTGCCAATAC TTGTCACCAT TCCCTGGAAG 600
CAGCTACCTA GGGGAAACAA GATGTAGTGC TATTGCCGAT AACAAGTAAG ATTTTCCACA 660 CTAAAAAAAA AAAAAAAAAA AAA 683
(2) INFORMATION FOR SEQ ID NO: 26:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2036 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 26: CTGAGAAAGG AAAGCATTCG GATCTGCTGC AAAAACACAT ATATCCATAA AGACTCATGT 60
TATTCAGAAA ACAGATTGTG AACACAATCA CATTCGCATG AATCCTTTAA AAGGAAGAAG 120
ACCTTAAAGT ATCTGCAAAT CTGAATTTCT ATTTATTCCT TCACTGAATA TAGAAACAAT 180
GGTTATCTGA TTATTAGAGA TATTATTTTG GATATGTTAC TTATTAACTT GCTATGGCTG 240
GTAACCATGA TAAAGTCTGT TATTAATAAC AACATAATTC TTTTTTTAAA GAAGAAAAGC 300 TTATTTTTCA TTGACAGTGT ATAGATTTAT CTACTTAGTT GTGTTTTGCT ATTAGTGTTT 360
TAATTTTTTT TTTAAGTTGA GTGTTTGATA AATTTTAAGA CCCTGTCCCC ACCTTGTTTT 420
GAGTCCTGTG TTGACTACAG GTATATAGCY CAWTTTAAAA ATCCTAAAGC AAAAGAATTT 480
TATTTATAAA AGAATCMAMC MGTTGCATGC ATGAGGCTGT GAAGTCAGAT ATTTAGTAAT 540
AAAAGCAGCA GTGCCTTTTT TTGTATTTAC CCATTGACCC CCACCAAATG CAACTGTTTT 600 ATATTAAGAA AATAGTAACA ATTTTAAAAT CTCAGAGTAA AATCTATTTC ACTACATGCT 660
TTTCCCCCCT TGTTCTGATT TAAGCAGTGT GTACTTGGCA TCTCTACATT GTCCTAGGGA 720
CAGTGGTGTT CTACAATATT ATCATGTATG ATGTTTTATT GGTGCTTTTT ATTCATAGTG 780
GCTTCTTACC AGAAACAGTA GGAAGAAACA CATGAACTGT GTACAAGACA TGAAACATTG 840
CTGCTGATAT GTTGTTTTTT CACATGCTTT TGAGTTTTCA CTTTTTAAAC GAGAGCCAGC 900 AAGCAAAATA GATGTGGCTG GGTCTGCCTG TCCGGGCGGC TYTTTGCACC GAGCTCTCAA 960
ATCCTGTGTA TTGAGGGTTC CTTTTTGGTA CTCAGGATTG GAGCTACAGC TGGGCCCCCC 1020
TCTCTCCCAT TCGTTTGAAG AGACACTGAG GGAAACAAGG GTTTCTTTTG AGGTGTCCTT 1080
GGCTGCCTTT TACGGGATGG GAGCCTTCTC CGGATCTTTT GTTCTTCTGC ACCTCTTGTA 1140
GCTACTGCCG GTGCAAGGTT GTAGATGTTA TTCCCCAGGA GCCTGGGCTK GGGGGCTGAG 1200
CTGGGCTGAA TGCAAAAGCA TGCAACCAGA AGGCGGGCAA GGGGAGGAAA AGCAGGCCTG 1260
GCCTCATTGG TCCCCTGGAG ATGTCTGTAG CAGTCAGCTC CAGCTTGGGC CTGGGGAAGC 1320
AGCCTGACCA AGGCGCTCAG GTGTGCCTGT TACAAGAAGA ACCTGCAGAA GGATAATTTG 1380
CACATGGAGC TGTGATAACA CTAATGTTGA TTTTTTTTTT TTTTACAAGT CATCAGRGAT 1440
GTTTGCAAAG TGAGTTTTAT TTTTTTGTAA TTCCTTTATC TTTACTTAAA GGTGAATGTG 1500
TATTCCTCTG GGAGGAATAG GAAGAAAACA GGAATGTTAA TAATGTCGAA CAGAAAACTT 1560
CCTCCCTTAT TAATATATAA TCYTCATGTA TTTATGCCNT AATGTAAGCT GACTTTTAAA 1620
AAGCTTTCTT TTGTTGCATG CCCTGTGCAG GCATCTGTAT TGTACATGCA TGCCTTTCGT 1680
CCTGTTTTCC TGTATAAAGT TAGTGAACAA AGAAATATTT TTGCCCTAGT TCATGTTGCC 1740
AAGCAATGCA TATTTTTTAA ATTTGTCATA TATGGAAAGA GCATGTTTGT TACATGTAAA 1800
AGCTTTACTG ATATACAGAT ATACTAATGT TTGAAGATGC TGTTCTTTGC AAGTGTACAG 1860
TTTTCAAATG TTGTTACCAG TGAAACACCC TTGTGGTTTA AACTTGCTAC AATGTATTTA 1920
TTATTCATTT CCTCCCATGT AACTAAGAAT CATGGCTATA TTTCATATCA ACGTTATATT 1980
GAAAGTGAAG GGAAATGATT AATACAAGGT TTTGTAACAA AAAAAAANAA ANNAAA 2036
(2) INFORMATION FOR SEQ ID NO: 27:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 717 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO : 27 : GGCACGAGAT AACATAGGCA CAATAATACT GTATGTCTAC TTCTAGGATT ATAAGGAATT 60 AACATTGAGA TGACATTTCC ATTTGAGAAG AAAATAGTTG CTTTCAGTGC CITTTATTTG 120 ATTCCTGGAG AGAGCAGACT CGCACCAACA TTCAACCCCA GCGCTGATAT GACAGTAATC 180 CTCAGAGGCA GAGCCCAGCA CAAAACAGCA ATGCTAGAAA GTTACAATTG GAAAGTTTCC 240 TGCCAGCTTC GGGAATGACA CTGCAAAGCT GATGCCAGAA ACTGCCAGAG TAATTCTCCT 300 CATTACTGCT CTACCCACCC ACTTTCAGCT CCCCAAATTA ACTAGTGCAG TTGACTAATC 360 CTCTTTACCT TTATCATTTA GGTGAGGCAT TGCACAAAAA CTCTCGACTT TGCCATATAA 420 GGGCTGTGGT TCTCTGTGGT CCTGGATAAG AGGCATCACC ATTATCTGGA AACATGCAGT 480
AAATGCAGAT TCTTCATCTT CTCCCCAGAC CTCCTGAGTT AGAAATTCAC AAGTTCTCCA 540
GGTGATCTCA TACATGCTAA AGTTTGAGAA CCATTGAGTA AAGTTAATGC ATTAAGAAGA 600
GATTAGATAG GGATGGTGGC GTATCTTCCT ACAGTTTCCC TGTTAACAAG AAAGTCAGAG 660
GTCAGTTGAT CAGACATTAG ATTATTTATT GCTAAAACTA AAAAAAATTA AAAAAAA 717
(2) INFORMATION FOR SEQ ID NO: 28: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 495 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS : double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 28:
GAATTCGGCA CGAGCAGCAT CCTAATTTTA GTTTGGAGAT GCATTCTAAA GGATCTTCTC 60 TATTGCTTTT TCTCCCACAA TTAATCTTGA TTCTGCCTGT CTGTGCACAT TTGCATGAGG 120
AACTGAACTG TTGTTTTCAT AGGTAAATGA GAGACTGAGT TTTTTCATTT CTGAAGAGAA 180
AGGGCATTTG CTCCTACAAG CTGAAAGGCA CCCCTGGGTG GCTGGGGCCC TCGTGGGAGT 240
TTCTGGGGGA TTGACCCTTA CAACATGCAG TGGCCCTACA GAAAAACCTG CAACTAAAAA 300
TTATTTTTTA AAAAGGCTCC TCCAGGAAAT GCATATAAGG GCTAATCACC CAGTATTTTG 360 ARGCTTCGAA GARGTAATAR AMCCCTGGAG AGAGAAACTG AGACATGTAA GAGGGTGGGA 420
ATGACTCAGT GGTGGCACAC TATGGAGTCC TGCCCACAAG TAGCACACAT CAACCCACTA 480
CACAGAAATC CTAGG 495
(2) INFORMATION FOR SEQ ID NO: 29:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 556 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 29:
AGCTTAACGT CATGATTCAT TAGGGGAATG CAAGGCAAAA CCATGATGAG AATGCCCCTA 60
GACACCTCTT AGAAGAGCTG CTAGAAAGGC AGACAGCACC AAGCGCTTAA ATGAGATGGG 120
GGCACTGGTG CTTCTTCTGT GCCTACTGGT AGGGGTGCAG CAGAGTGGTT CAGTCTGGGA 180 CAGTTAGCTG GACATCACGT GGACCCAACA CACGCATTTC CTGGGTTACT TACCAAGGAG 240
AATAGAAAGC AGGCAGATCT TTACAGCAGC TCTTACCTGW TTGCAAAACA ATGGAAATGC 300
CCACATGTCC ACAAACAAGT KTGTGGTCTG CCTGTGCCAT GAAGCACAGT GTGGCTGAGC 360
GTCAAGAGTC CCCACACTCA .AAGGAGGCAG CAGATACAGG GCTGCACACT GTGTGATTCC 420
ACACATGTGA CATTCTGGAC ACGGACATGC TGGATGGCAA AACGAGCATC GGGCTGAGAG 480 GACTGCTGAG AAGGGGAACG GGGCTGCTGG GATGTGGGTT GATTGTAGCA GTAGCTCATG 540
GAGATGTGAC CTCAAA 556
(2) INFORMATION FOR SEQ ID NO: 30:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 434 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 30:
CTAAATGGTG ACTGTGGCTT TGTCGAGACA GGCCCCAAAT GGTAGGTGTG AACACAACAT 60 GCACAGAATG AGGAGACATG CAGAGTGCTG AAATACTGTC CTGGACAGAT GTGTTACATG 120
ACTTTCTTTT CAGCTTATTT CTGTGGCCTG CCTTTGAAGA TAGAGCTTTG TTGATATTTA 180
CATTAAACCA AATTGTATAA YTATGTTCCA TTCTGACATG TTATTTAGCA AARGAAAAAR 240 GAGTAATTCT ACATCAGCAT CTTTAGTGCA TGCTAAAAGA TTAAAAATGT CTTTTGGGGA 300
A^TGTTTTG TATACATAAA TGTTTAGATA GAAATATTTA TAGAATNCTC TATGTGAGTA 360
TTNATCTCCC TATGTATATT TATATCTAGA TGTGTCAATC TTTGTATTGA TATGAAATGC 420
TATGAATAGT GAGA 434
(2) INFORMATION FOR SEQ ID NO: 31:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 715 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 31:
CCACGCGTCC GATCTCACAG CTCCGACACT ATTGCGAGCC ATACACAACC TGGTGTCAGG 60
AAACGTACTC CCAAACTAAG CCCAAGATGC AAAGTTTGGT TCAATGGGGG TTAGACAGCT 120 ATGACTATCT CCAAAATGCA CCTCCTGGAT TTTTTCCGAG ACTTGGTGTT ATTGGTTTTG 180
CTGGCCTTAT TGGACTCCTT TTGGCTAGAG GTTCAAAAAT AAAGAAGCTA GTGTATCCGC 240
CTGGTTTCAT GGGATTAGCT GCCTCCCTCT ATTATCCACA ACAAGCCATC GTGTTTGCCC 300
AGGTCAGTGG GGAGAGATTA TATGACTGGG GTTTACGAGG ATATATAGTC ATAGAAGATT 360
TGTGGAAGGA GAACTTTCAA AAGCCAGGAA ATGTGAAGAA TTCACCTGGA ACTAAGTAGA 420 AAACTCCATG CTCTGCCATC TTAATCAGTT ATAGGTAAAC ATTGGAACTC CATAGAATAA 480
ATCAGTATTT CTACAGAAAA ATGGCATAGA AGTCAGTATT GAATGTATTA AATTGGCTTT 540
CTTCTTCAGG AAAAACTAGA CCAGACCTCT GTTATCTTCT GTGAAATCAT CCTACAAGCA 600
AACTAACCTG GAATCCCTTC ACCTAGAGAT AATGTACAAG CCTTAGAACT CCTCATTCTC 660
ATGTTGCTAT TTATGTACCT AATTAAAACC CAAGTTAAAA AAAAAAAAAA AAAAA 715
(2) INFORMATION FOR SEQ ID NO: 32: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 486 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 32:
GAGCCAGTGC CGGCGAAAGG GGACCTTCCT CTACTTCCTG CCACAGACCC TGTCCCCACA 60 CACTTCCTGC CCCTGCTCTG CTGGGAGGCC ACTTCCTCCC CCAGTGCTGG ATTCCACCCC 120
CAGCTCACCC TCAAACATGG CCCCCTCTCT CCTCCTGCTT GCCCCTCTCT GCTCCCTGGA 180
GGCTGTTCTG TCCTCCCCTC TTGAAAAGCA ATGCCAGCTT CCTGGGATCT TCTGCCAACT 240
CCAGCTACCA TGCCCTTTGC TCCTGTCAGC TCAGCTCCTC AAGGGAATTG TCTAMCCTCG 300
GTGTCCTGCT TCCCTCCCTC AACCTCCTCA CCCTGCTCCA AGCTGGCATC TGCCCCTCCA 360 CTGCACAGAA CGGNTCCCCC ACCACCTGCC TTTACAGGGA GGAAGCAGCA ACATGGAAGA 420
ANCGAACTAT AGGGGCTACA ANGATGCTCA GCTCTGATCC CGAAGGCAAA AAGNATCTTT 480
GGGCAC 486
(2) INFORMATION FOR SEQ ID NO: 33:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 725 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 33:
GTTCCTCTGG TAATAATTAG GTTATTCCCA GAAGCACAGT GTCATTCTTT AAATAAAAGC 60
TTTCCTGTTT AAAGCTTTTC AAAGGAGCAG ACCACCTTGA AGATTCCCCC TAGGGTTGAT 120
ATGTGTCTAA TTCATTTTAT AAAAATTATT CTTGTCTTCA TTTTAAAGCT TTGGCTATAT 180 AGTCAGAAAT GTCCTAAATA ACAAACTATT TTGTATTTAA TTTAGGGAAG ACTAAAGGGA 240
AGAAAAATGA AAACTCAGTC TTTATGTAAG CTCCAAGGAT ATTAGGGCTT AAAGGGCTTT 300
TCTAGTTTTA TGAGAATTTG TACTACTGAT TTTTATATAT TCCTGTTTTT GATGAACAGA 360
TCTCTGGGGA AATTGTTGAG TTACAATGGC ATTTCACTGT GATCCCTCTC AAGCTCAGAT 420
CAGTTCTATA ACCCAATGAC AACCTGTCTC TTTGGTTTAC TGTCCTGTGA AATGTCAGCT 480 CAAGTTTCCC AGAAGTCGTG TGTTTATGAT GAGTCAGAGT GCTTTTCCTC GGTGGGACAG 540
TTGCTGGCCC TCTTAATTTT GGTGTATGTG CTTCCAAGTA TCTAAACCTC CAGTCTGATC 600
TGTATATGCT ATCCTAACTG TTAATTGTAT TATTGATTAT GTTGATTATC TTGCTTGAAG 660
GTTCATACTT TTCAATTTGA TAGAAATAAA GTTTTTTTCT GCTTATAAAA AAAAAAAAAA 720
AAAAA 725
(2) INFORMATION FOR SEQ ID NO: 34: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 437 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 34:
CACACAGCAT GCTGCCCTCA GACGTGTCCA TCCTGTACCA CATGAAAACG CTGCTGCTCC 60 TGCAAGATAC TGAGAGATTG AAGCATGCTC TGGAAATGTT CCCAGAACAT TGCACGATGC 120
CTCCTGCTTT TATTGGCTCT TGTCGAAATC AAATTGGAAG ATCTTCAGTC CCAGCTGCAC 180
CCAACGTGGA AAAGTATTCC AGGTCCATCC CCAAGGAACC AACACCGATG ACATGGACTC 240
AGGAATCTTA TAACCTACGT GGACTCTTTC CATCCGTACA TTGTCGTGCA CATGCCACTC 300
ATCACCTGGC GTGCCCAGAT CCTCGCARGG CAACACCCTG TGATAATTCC AGGTGATTCT 360 CTACATCTGC AGCTTGAGGT TAGCCTCATA TCACATTACA TTCTCACTAN AAACNAAAAA 420
AAAAAAAAAA AACTCNA 437
(2) INFORMATION FOR SEQ ID NO: 35:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 943 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS : double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 35:
GGCACGAGCT GGAACAGAGA CTAAATCCCA CGAAACTGAC ATTGTTAAAC ACACTAAAAC 60 AGAAGTACTT ACCTCTTGAA GATTTAATAT ATAATGGTTG ACATGATACA TGTACATGAT 120
GAATGACCAG ATGCTTATGG TCTACATTTT CCTTTATCCT GTTAGTATTA CCTTCCTTAA 180
TCTTTGTTCA TTAACATGCT AATTCCTCTT CAGTGTTTAT TTTCTAGTGA CAGAATGCTA 240 ACATTTCTTA CACCCTGGCA GAAGGGAGAG AAATGTGTTT TGGGGTGGGT AACTAAATTT 300
TTGAGTGAAA TATCATAAGA TGANAATGGA AANAAGGAGA CACAAANAGT TATNACAAAA 360
AAACAATGGT TTTTTTAGCC ATTTGACTGG CTCTTTAAAT AGTCTACAAG ACATTCACGT 420
TTAACATCAC TTTTAGTGAA ATAAAATGTG CCATACTAGT ATGTGCTTCA AAAGGGCAAA 480
TGTGCTTTAG TGCCCTAAGG CTAAATTTTG GTCATTTGAC ATCAGAGATG TTGTAAGTAT 540 TGCACTTAAT ACGCACCTAT TTNTCAATAG TGTTATTTTT TGGNTAGCAT TTTTTTTACC 600
ACTATNTTGT TGATAGCTTT TTGTTCTNTN AGGTTGNAAN ATGACAGTGC TNATNTCAAA 660
CAGATTACCC ATNTGCAGAA CTAAGGGAAG CNATTTATGT ATGAAAGNAA TTNTTGAATT 72
NGTCATTNTC AACCNTTGNA TTAAAGCTTA GACTAAATAG TAATATATNG TGGGNAGGAT 780
TTTGGTTTTG TGATATTTNT GTGNATTAAG GNATAGATGT TAACCNTTAT TTTGTAGNAA 840 AGTGANTTGT ATGTGGTTAA TTATAAATAA AACTGGTACC AGGNAAAAAA AAAAAAAAAN 900
NAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAA 943
(2) INFORMATION FOR SEQ ID NO: 36:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 604 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 36:
GGCACGAGAA ATCTTCATGC TGTAGTCACT CCAGACCATG GAGTGGCTTT CCAGCTGAAT 60 GAATCCTATG TCTCGCGTGC AGGTGGTTGG TTTTCAATGT TCTTGCTAAT TTTTTTTCTA 120
TTGGATCTTG GGAGTTTTCT TTGTTTGCTC CTGTGTTTGC CCAGCTTTAA TAAAACCAGG 180
CGCAAACAAA AACCATAGCA TTCTGAACAA TAGGGGGCCC ACATTGGACC CAGTATGTCA 240 CTTTAATGGA CTTCAAGAAA AAATCTGAAT GGGAAAAATG ACACTAGGAA TGTATACTCC 300
ACACATTTTA TGCCATATAA TGGTGTGTTT TCTTAATTTT GTTTCTTGTG GCGAAATGTG 360
GCTTTCAAAT TAAAATGACC TTTTCTTCTT TGAAACTTTT TGTTTTGACT TGTATAATTA 420
AGGGTTTGGA AAGATTCATA ATTCTGAGAG AGGTTTGCAA CCAGGAGATA CAAAGAAGTC 480
TCAGTAGTAA TCTTGTTCAT GTGCTTTTAC AGCCAGCTAC ATTTAAGGAT GTATTAGTTA 540 CAGAAATTAT ATGTCTGTGT ATGTGTCTCT ACTCAATAAA GTACATGCCT CCACAAAAAA 600
AAAA 604
(2) INFORMATION FOR SEQ ID NO: 37:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 349 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 37:
GTGAGTGCCC GGGAGCCCCG AGGCCCTGCC CCTAAGAAGG ATATCTYTRA CCGCTCCCTT 60 GTCCACACCC TAACCCCCCA GCTGCTCAGG CAGTGGGCAC ATGGCAGGGG CCTCACTGGG 120
GGCACATAGA GCATTTGGGG GACTGCGAGT GCTCACCTTT GACTTCCTGC AGGTCGGGGG 180
AAAACCAGAT CATGATGACC AAAGTYTACA TATTCTTGAT CTTCATGGTG CTGATCCTGC 240 CCTCCCTGGG TCTCACCAGG TATATGCCAC CΛCYTTCTGY TCTAAATTCA GAATAAGAGT 300
CACATCAGGA GAGCACTGTC CCCAGGANAA TGCAAACGGG TTGGCAGCA 349
(2) INFORMATION FOR SEQ ID NO: 38:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 672 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 38:
GTAGTCGTTG CGGTTGCCGG GATGGCGAAG ATCTCGCCGT TTGAAGTCGT AAAACGCACC 60 TCGGTACCGG TGCTTGTTGG TTTGGTGATT GTWATCGTTG CTACAGAGCT GATGGTGCCA 120
GGAACGGCAG CAGCGGTCAC AGGCAAGTAA ATAGTAATGC CGGAGCAAGT TTCCTCCGGC 180
TTTATCATGT CACCCACTGT GGTATATGCG TTGTGGTCTG CCAACTTTGC CGTGAACAAT 240 TTCAGCAATA ATCAGATGGC GGCTGGCGCA ATATTCAAGA TAACGCCTGG CAGTGGTGCG 300
GCTGATGGTT CAGTGCCTGC GSCACCGTTT YTGCCGTATG TTGCACACCA GGNTCTTTAA 360
ACAGTTTTCG SACCGCGTTT AGCGTCAAGG GTTCAATGCC GGTCGGTAGC TCGTCCTTAG 420
GTTCACCGCG AGCATAAGCA TTAAACATCT CATCAATTTG CTTCTGGCTG GCGCTATCAA 480
TACTTTCCAG CATATGTTTA CGCTGGCGGA AACGGGTTAG CGTTTGCCCC ARCMGWTCAT 540 AGGCAATGGG CTTAATGAGA TAATCAAATA CACCACAACG TACGGCTTCA GACACCGTTT 600
CCATATCGCT GGCTGCAGTG GTAAACACCA CGTCGCCGGG ATAATGCGCC TGCACCAGTT 660
CATGCAGTAA AT 672
(2) INFORMATION FOR SEQ ID NO: 39:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1908 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 39:
AGAGTTGATA TTTTTAGAAA CAGTAATTTT ACTTTTAAGG AAATTGGCTA GCTCTTTGAC 60
TNNAGAGCTG TAGGAAGCTC AACATTTCTT TGTAGAGAAC GTTGCTTTTT TTGGATTGTA 120
CAGGTATAAA AACATTGCTT TTGTTGAATT GTATAGGTGT AAAAAGGGAA TAACTGTATG 180 CAGGTTTGAA AAGGAAATGT GCTTTAGGCA TGAGTCATAA GATGCCATTG TACTTGTAGG 240
CATTTTATTT TCCTTTAGAA ATGGACATCA GCTCTTCTCT TCTGACTGGT AACACATAGC 300
CCCAAAGCAT GAGATTATTT TTCATTGGGT TTTTATTGTT GTTTAGTTTT GGTTTGTTAC 360
GCCAGCCCAG TCTGTCTGCG GAACACTGAC TCTGCTCTCT AATGAGAACA AAGTTAGAAA 420
TCTGCCGATA ACCTAAAATA ATTTAGAAAT GAATTAAAAA TGTGAAATCG GGTTAAAGTG 480 ATGATGATAA AATAGCATGC AAGAAACAAG CTCCTTCCAT CAGACTTGGC TACTGTTTTC 540
TTCTGGTACG ATTTGGTTTG GAAGAGCCTC TTGTTTCCTT CTCTTTGGGG TATGTCTTCG 600
TTTCTTAATA TGTTTGTAAC ATTATTGAGA TATAATTCAC ATACCTTACA ATTCACTTAT 660
TTTAAGGGTA CAATTTAGTG GTTTTTAGTG TATTCACAAA GTTGTGTAAC CGTGACCACA 720
GTCAATTTTA GAACATTTCG TTACCCCAAA AAGAAACCCT GTACCCTTGA GCAGTCACCT 780 CTCATTTTCT CCCAGTGCCC ACCCCATCCC CGAGCCCCKG GAACCACTAA TCTATTTCTC 840
TCTCTGTAGA TTTGCTTATT CTGGTCATTT CATATAAATG GAATTCTACA ATATTCGGTC 900 TTTTGGGACT GGCTTCCCAA ATATGATTTT CTATATGGAG TGAGAAAATT CTTCTCATCT 960 TGAGAACTCT TATTGCTGTG AAAGGGAGTG GTTGGTAAAA TCAATAGATT TCAGGCAAGA 1020 GGGCCAGATA CCTAACAGGT TTTTCTCCGT GAATCTTATG CTGAGTAGTT TTTCCTCATA 1080 ACCAAGCATT TATGATATAT TACTACTTAT AATACTGTGG CTAGTCTCTA GAATGGATGT 1140 TGAAATCTTT GCCTCCTCAG TCGGGAAGAG TCCTGCTAAA. AATCAGGCTA AAAATCAGGC 1200 CAAAAATCAG GCCAAATGAC TTGGCAAATA ATTGACAAAG TGGTTTTCAC GTGTGTCTAT 1260 CTTTGCTAGC AGCTTGTATA CCTCAGGCCA GGTGAGCTCC CCAAATTTCT TTTTTCATTT 1320 ACTCCAGTGA GTTTCTGCTG TCTTTTTCAA GTATGTACCA TAGGACTTAA AGGTGATTTG 1380 GATGCGTTGT AACACTGCTA AATATGCTAA GTACAGAATT TTATCTACAG TACTGTGAGA 1440 CAGTCAATTA TTGCCTAGGG TAGTTCAAAA ATATGATGTG AGCTAGTTAA GCCTTTGCTT 1500 GACTGATTTC AGTGATATTC AGAAGTGTGT ACCAATCAAG GCTCTTTAAA ATACGGAACG 1560 ACTCACTTAA TAACCAGGGA ACCAGCCAAA TACTGTGCAG CCGCAGAATA TGCATATCAA 1620 TGAGTTGGAG GTGATTATTC TCTGTAACTC CCTAATGATT GTTTTCTAAG CATTGTGGCT 1680 TCTCAGTGGC TTGACAGCAT CTTCCTGGTT GTATGTGGCC TGTTTACATG ATGTATTGAA 1740 TAATGTTGTT TGTTGTGAGC ATCAATGCCT GTAACACCAA ACTAAACACG TGTTTTTGGG 1800 ATATGTTTCC AATCTTTAAA TGACCTTGCC CTGTCCAATA AATAAATGAT TGTCTCACCC 1860 TGTTAAAAAA AAAAAAAATT AAAAAAACTG GGNGGGGGGC CCGGTACN 1908
(2) INFORMATION FOR SEQ ID NO: 40:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 458 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 40: CCTCAAAAAA AAAAANGAAA GGAAAGAGGT CTCTACACAA GCCCGTGATT CTTCATGGCA 60 AGGGATAACA TCAGAAATGT TTCATTTYCK GCTATTAGTT TCCATTCCTT TCCCCATCCA 120 GGCATAAAGA GAAACAAAAG ACAATGATGG TATTCTCTGT GTCCTCAGCT TTGGCACTTT 180 TGTTGATGTT GCTAAGGAGC AGTGACCTTG CTAAAAAGAC TGAATAATCC ACCCACTGAA 240 TAGCTAACCT GGGGAGGAAA TGAAAATTTC CTTTGTGGAT CTCCCCAAAT CCATTGTTGT 300
CACCAGGCCC TCCCAGAACC TCCTCAGTTC CTTCACAGTG CAACCCTGTG TACTTGGCCC 360
GCAACCCAAT AGTATTGTGC CTCACTTCAC CTTCCATGGG CAACTGCCCT CCCTTCTGGA 420 CATAAAACCT CATATTTTAA ATNAAGTTGA AATTTGAA 458
(2) INFORMATION FOR SEQ ID NO: 41:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1153 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 41:
GGCACAGAGC CTCCGACCCA GGTGGTCTGG AGCCTGCCGG GAGAGTGGTG GCATCTGAGA 60
GGCTGGTCGT GGACTGTGGT TGGGGGAGGT GGGAGCTGTT TTAACCGTGT GCCCCCTCTC 120
CTGTGCCGGC GTGGGCATCC CCCGGGGCAG TGGAACCCGG GCGCTCCTCC AGCTTCCGAG 180
TCCAGCCAGC CTGGGCGCGG GGCGCGCCCC GAGACACCCG AGGAGTCCGT TCCTCCCTGG 240
TTACGTGGAC TGTGGAGCTG GTCTCTTGTG GCTCAGCGCC GTGCGGAGGT TGAAGCGTAC 300
CTGCGGAGGT CGCACCAGGG CGTGAGGAGG AGGAGGAAGG GCATGAGCCG AGCTTGAGGA 360
ATCCGTGCTC CAAACTCTAC ACTCAAGGAT GCACTGCGCA ACTCTGGTGG CGATGGGCTG 420
GGGCAGATGT CCTTGGAGTT CTACCAGAAG AAGAAGTCTC GCTGGCCATT CTCAGACGAG 480
TGCATCCCAT GGGAAGTGTG GACGGTCAAG GTGCATGTGG TAGCCCTGGC CACGGAGCAG 540
GAGCGGCAGA TCTGCCGGGA GAAGGTGGGT GAGAAACTCT GCGAGAAGAT CATCAACATC 600
GTGGAGGTGA TGAATCGGCA TGAGTACTTG CCCAAGATGC CCACACAGTC GGAGGTGGAT 660
AACGTGTTTG ACACAGGCTT GCGGGACGTG CAGCCCTACC TGTACAAGAT CTCCTTCCAG 720
ATCACTGATG CCCTGGGCAC CTCAGTCACC ACCACCATGC GCAGGCTCAT CAAAGACACC 780
CTGCCCTCTG AGCGTCGCTG GATCTCTGGG AGCTCCTTGA TGGCTCCCAG ACCTTGGCTT 840
TTGGGAATTG CACTTTTGGG CCTTTGGGCT CTGGAACCTG CTCTGGGTCA TTGGTGAGAC 900
TTGGAAGGGG CAGCCCCCGC TGGCTTCTTG GTTTTGTGGT TGCCAGCCTC AGGTCATCCT 960
TTTAATCTTT GCTGACGGTT CAGTCCTGCC TCTACTGTCT CTCCATAGCC CTGGTGGGGT 1020
CCCCCTTCTT TCTCCACTGT ACAGAAGAGC CACCACTGGG ATGGGGAATA AAGTTGAGAA 1080
CATGAGTTTG GGCTGAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA 1140
AAAAAAAAAA AAA 1153
(2) INFORMATION FOR SEQ ID NO: 42:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1983 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 42:
GGCACGAGAG GGGCCGAGCC GACAAGATGT TCTTGCTGCC TCTTCCGGCT GCGGGGCGAG 60
TAGTCGTCCG ACGTCTGGCC GTGAGACGTT TCGGGAGCCG GAGTCTCTCC ACCGCAGACA 120
TGACGAAGGG CCTTGTTTTA GGAATCTATT CCAAAGAAAA AGAAGATGAT GTGCCACAGT 180
TCACAAGTGC AGGAGAGAAT TTTGATAAAT TGTTAGCTGG AAAGCTGAGA GAGACTTTGA 240
ACATATCTGG ACCACCTCTG AAGGCAGGGA AGACTCGAAC CTTTTATGGT CTGCATCAGG 300
ACTTCCCCAG CGTGGTGCTA GTTGGCCTCG GCAAAAAGGC AGCTGGAATC GACGAACAGG 360
AAAACTGGCA TGAAGGCAAA GAAAACATCA GAGCTGCTGT TGCAGCGGGG TGCAGGCAGA 420
TTCAAGACCT GGAGCTCTCG TCTGTGGARG TGGATCCCTG TGGAGACGCT CAGGCTGCTG 480
CGGAGGGAGC GGTGCTTGGT CTCTATGAAT ACGATGACCT AAAGCAAAAA AAGAAGATGG 540
CTGTGTCGGC AAAGCTCTAT GGAAGTGGGG ATCAGGAGGC CTGGCAGAAA GGAGTCCTGT 600
TTGCTTCTGG GCAGAACTTG GCACGCCAAT TGATGGAGAC GCCAGCCAAT GAGATGACGC 660
CAACCAGATT TGCCGAAATT ATTGAGAAGA ATCTCAAAAG TGCTAGTAGT AAAACCGAGG 720
TCCATATCAG ACCCAAGTCT TGGATTGAGG AACAGGCAAT GGGATCATTC CTCAGTGTGG 780
CCAAAGGATC TGACGAGCCC CCAGTCTTCT TGGAAATTCA CTACAAAGGC AGCCCCAATG 840
CAAACGAACC ACCCCTGGTG TTTGTTGGGA AAGGAATTAC CTTTGACAGT GGTGGTATCT 900
CCATCAAGGC TTCTGCAAAT ATGGACCTCA TGAGGGCTGA CATGGGAGGA GCTGCAACTA 960
TATGCTCAGC CATCGTGTCT GCTGCAAAGC TTAATTTGCC CATTAATATT ATAGGTCTGG 1020
CCCCTCTTTG TGAAAATATG CCCAGCGGCA AGGCCAACAA GCCGGGGGAT GTTGTTAGAG 1080
CCAAAAACGG GAAGACCATC CAGGTTGATA ACACTGATGC TGAGGGGAGG CTCATACTGG 1140
CTGATGCGCT CTGTTACGCA CACACGTTTA ACCCGAAGNT CATCCTCAAT GCCGCCACCT 1200
TAACAGGTGC CATGGATGTA GCTTTGGGAT CAGGTGCCAC TGGGGTCTTT ACCAATTCAT 1260
CCTGGCTCTG GAACAAACTC TTCGAGGCCA GCATTGAAAC AGGGGACCGT GTCTGGAGGA 1320
TGCCTCTCTT CGAACATTAT ACAAGACAGG TTGTAGATTG CCAGCTTGCT GATGTTAACA 1380
ACATTGGAAA ATACAGATCT GCAGGAGCAT GTACAGCTGC AGCATTCCTG AAAGAATTCG 1440
TAACTCATCC TAAGTGGGCA CATTTAGACA TAGCAGGCGT GATGACCAAC AAAGATGAAG 1500
TTCCCTATCT ACGGAAAGGC ATGACTGGGA GGCCCACAAG GACTCTCATT GAGTTCTTAC 1560
TTCGTTTCAG TCAAGACAAT GCTTAGTTCA GATACTCAAA AATGTCTTCA CTCTGTCTTA 1620
AATTGGACAG TTGAACTTAA AAGGTTTTTG AATAAATGGA TGAAAATCTT TTAACGGAGA 1680
CAAAGGATGG TATTTAAAAA TGTAGAACAC AATGAAATTT GTATGCCTTG ATTTTTTTTT 1740
CATTTCACAC AAAGATTTAT AAAGGTAAAG TTAATATCTT ACTTGATAAG GATTTTTAAG 1800
ATACTCTATA AATGATTAAA ATTTTTAGAA CTTCCTAATC ACTTTTCAGA GTATATGTTT 1860 TTCATTGAGA AGCAAAATTG TAACTCAGAT TTGTGATGCT AGGAACATGA GCAAACTGAA " 1920
AATTACTATG CACTTGTCAG AAACAATAAA TGCAACTTGT TGTGCAAAAA AAAAAAAAAA 1980
AAA 1983
(2) INFORMATION FOR SEQ ID NO: 43:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1406 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 43:
ATGATGATGA CTTTGAAGAC GATTTTATTC CTCTTCCTCC AGCTAAGCGC CTTGAGGTTA 60
ATAGTTGGAA AAGACTCTAT AGATATTGAC ATTTCTTCAA GGAGAAGAGA AGATCAGTCT 120
TTAAGGCTTA ATGCCTAAGC NCTTGGTCTT AACTTGACCT GGGATAACTA CTTTAAAGAA 180
ATAAAAAATT CCAGTCAATT ATTCCTCAAC TGAAAGTTTA GTGGCAGCAC TTCTATTGTC 240
CCTTCACTTA TCAGCATACT ATTGTAGAAA GTGTACAGCA TACTGACTCA ATTCTTAAGT 300
CTGATTTGTG CAAATTTTTA TCGTACTTTT TAAATAGCCT TCTTACGTGC AATTCTGAGT 360
TAGAGGTAAA GCCCTGTTGT AAAATAAAGG CTCAAGCAAA ATTGTACAGT GATAGCAACT 420
TTCCACACAG GACGTTGAAA ACAGTAATGT GGCTACACAG TTTTTTTAAC TGTAAGAGCA 480
TCAGCTGGCT CTTTAATATA TGACTAAACA ATAATTTAAA ACAAATCATA GTAGCAGCAT 540
ATTAAGGGTT TCTAGTATGC TAATATCACC AGCAATGATC TTTGGCTTTT TGATTTATTT 600
GCTAGATGTT TCCCCCTTGG AGTTTTGTCA GTTTCACACT GTTTGCTGGC CCAGGTGTAC 660
TGTTTGTGGC CTTTGTTAAT ATCGCAAACC ATTGGTTGGG AGTCAGATTG GTTTCTTAAA 720
AAAAAAAAAA AAAACGACAT ACGTGACAGC TCACTTTTCA GTTCATTATA TGTACCGAGG 780
GTAGCAGTGT GTGGGATGAG GTTCGATACA GNCGTATTTA TTGCTTGTCA TGTAAATTAA 840
AAACCTTGTA TTTAACTCTT TTCAATCCTT TTAGATAAAA TTGTTCTTTG CAAGAATGAT 900
TGGTGCTTAT TTTTTCAAAA ATTTGCTGTG AACAACGTGA TGACAACAAG CAACATTTAT 960
CTAATGAACT ACAGCTATCT TAATTTGGTT CTTCAAGTTT TCTGKTGCAC TTGTAAAATG 1020
CTACAAGGAA TATTAAAAAA ATCTATTCAC TTTAACTTAT AATAGTTTAT GAAATAAAAA 1080
CATGAGTCAC AGCTTTTGTT CTGTGGTAAC CTATAAAAAA AGTTTGTCTT TGAGATTCAA 1140
TGTAAAGAAC TGAAAACAAT GTATATGTTG TAAATATTTG TGTGTTGTGA GAAATTTTTG 1200
TCATAAGAAA TTAAAAGAAC TTACCAGGAA GGTTTTTAAG TTAGAAATAT TCCATGCCAA 1260
TAAAATAGGA AATTATAAAT ATATAGTTTT AAGCCTGCAT CAGTGGGAGT CTTGGCTATG 1320
TAGTTATGTA GTTATTATGN AACCACCAAG ATTTTTTTGG CTATTTACCG TAACCAAAGG 1380
GGCCGATTAA NTGGTTTGAA GNCTTG 1406
(2) INFORMATION FOR SEQ ID NO: 44:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1391 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 44: GGGCCTGAAG GCGGCRCGCC AGTCCCGAGC AGTGCTCGCT CCTGCTCGGG GCGCTGCGGC 60 CCCGGGCGTC GCCATGACCA GTGAGCTGGA CATCTTCGTG GGGAACACGA CCCTTATCGA 120 CGAGGACGTG TATCGCCTCT GGCTCGATGG TTACTCGGTG ACCGACGCGG TGGCCCTGCG 180 GGTGCGCTCG GGAATCCTGG AGCAGACTGG CGCCACGGCA GCGGTGCTGC AGAGCGACAC 240 CATGGACCAT TACCGCACCT TCCACATGCT CGAGCGGCTG CTGCATGCGC CGCCCAAGCT 300 ACTGCACCAG CTCATCTTCC AGATTCCGCC CTCCCGGCAG GCACTACTCA TCGAGAGGTA 360 CTATGCCTTT GATGAGGCCT TTGTTCGGGA GGTGCTGGGC AAGAAGCTGT CCAAAGGCAC 420 CAAGAAAGAC CTGGATGACA TCAGCACCAA AACAGGCATC ACCCTCAAGA GCTGCCGGAG 480 ACAGTTTGAC AACTTTAAAC GGGTCTTCAA GGTGGTAGAG GAAATGCGGG GCTCCCTGGT 540 GGACAATATT CAGCAACACT TCCTCCTCTC TGACCGGTTG GCCAGGGACT ATGCAGCCAT 600 CGTCTTCTTT GCTAACAACC GCTTTGAGAC AGGGAAGAAA AAACTGCAGT ATCTGAGCTT 660 CGGTGACTTT GCCTTCTGCG CTGAGCTCAT GATCCAAAAC TGGACCCTTG GACCCGTCGA 720 CTCACAGATG GATGACATGG ACATGGACTT AGACAGGAAT TTCTCCAGGA CTTGAAGGAG 780
CTCAAGGTGC TAGTGGCTGA CAAGGACCTT CTGGACCTGC ACAAGAGCCT GGTGTGCACT 840
GCTCTCCGGG AAAGCTGGGC GTCTTCTCTG AGATGGAAGC CAACTTCAAG AACCTGTCCC 900
GGGGGCTGGT GAACGTGCCG CCAAGCTGAC CCACAATAAA GATGTCAGAG ACCTGTTTGT 960
GGACCTCGTG GAGAAGTTTG TGGAACCCTG CCGCTCCGAC CACTGGCCAC TCAGCGACGT 1020
GCGGTTCTTC CTGAATCAGT ATTCAGCGTC TGTCCAATCC CTCGATGGCT TCCGACACCA 1080
GGCCCTCTGG GACCGCTACA TGGGCACCCT CCGCGGCTGC CTCCTGCGCC TGTATCATGA 1140
CTGAGGTGCC TCCCAACGTC CGCCCACGCT GACAATAAAG TTGCTCTGAG TTTGGAGACT 1200
GGTCCTCGCT CCGGGGAGCA AGTGGGGGGC GTGCAGATGT GCCTGTGTCT GTCTCTGAGC 1260
ACCTGGTGTC CGTGTACAAG GATGGATGTG TNCNGTGGCT CCTTGGGAAC TGAGACATAT 1320
CTCAGGGAAT GGTGTCTGTG CTCAGCCCAT CCACCAGAAG AGTCTGCTCA CAAAAAAAAA 1380
AAAAAAAAAA A 1391
(2) INFORMATION FOR SEQ ID NO: 45:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1569 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 45:
GGCACGAGTG GAGATGGCTG CGGCCGTGGC GGGGATGCTG CGAGGGGGTC TCCTGCCCCA 60
GGCGGGCCGG CTGCCTACCC TCCAGACTGT CCGCTATGGC TCCAAGGCTG TTACCCGCCA 120
CCGTCGTGTG ATGCACTTTC AGCGGCAGAA GCTGATGGCT GTGACTGAAT ATATCCCCCC 180
GAAACCAGCC ATCCACCCAT CATGCCTGCC ATCTCCTCCC AGCCCCCCAC AGGAGGAGAT 240
AGGCCTCATC AGGCTTCTCC GCCGGGAGAT AGCAGCAGTT TTCCAGGACA ACCGAATGAT 300
AGCCGTCTGC CAGAATGTGG CTCTGAGTGC AGAGGACAAG CTTCTTATTG CGACACCAGC 360
TGCGGAAACA CAAGATCCTG ATGAAGGTCT TCCCCAACCA GGTCCTGAAA GCCCTTCCTG 420
GAGGATTCCA AGTACCAAAA TCTGCTGCCC CTTTTTGTGG GGCACAACAT GCTGCTGGTC 480
AGTGAAGAGC CCAAGGTCAA GGAGATGGTA CGGATCTTAA GGGACTGTGC CATTCCTGCC 540
GCTGCTAGGT GGCTGCATTG ATGACACCAT CCTCAGCAGG CAGGGCTTTA TCAACTACTC 600
CAAGCTCCCC AGCCTGCCCC TGGTGCAGGG GGAGCTTGTA GGAGGCCTCA CCTGCCTCAC 660
AGCCCAGACC CACTCCCTGC TCCAGCACCA GCCCCTCCAG CTGACCACCC TGTTGGACCA 720
GTACATCAGA GAGCAACGCG AGRAAGGATT CTGTCATGTC GGCCAATGGG AAGCCAGATC 780
CTGACACTGT TCCGGACTCG TAGCCAGCCT GTTTAGCCAG CCCTGCGCAT AAATACACTC 840 TGCGTTATTG GCTGTGCTCT CCTCAATGGG ACATGTGGAA GAACTTGGGG TCGGGGAGTG 900 TGTTTGTCAC TTGGTTTTCA CTAGTAATGA TATTGTCAGG TATAGGGCCA CTTGGAGATG 960 CAGAGGATTC CATTTCAGAT GTCAGTCACC GGCTTCGTCC TTAGTTTTCC CAACTTGGGA 1020 CGTGATAGGA GCAAAGTCTC TCCATTCTCC AGGTCCAAGG CAGAGATCCT GAAAAGATAG 1080 GGCTATTGTC CCCTGCCTCC TTGGTCACTG CCTCTTGCTG CACGGGCTCC TGAGCCCACC 1140 CCCTTGGGGC ACAACCTGCC ACTGCCACAG TAGCTCAACC AAGCAGTTGT GCTGAGAATG 1200 GCACCTGGTG AGAGCCTGCT GTGTGCCAGG CTTTGTGCTG AGTGCTGTTA CATGTATTAG 1260 TTCCTTTACT GCTGACCACA TTGTACCCAT TTCACAGAGA AGGAGCAGAG AAATTAAGTG 1320 GCTTGCTCAA GGTCATGCAG TTAGTAAGTG GCAGAACAGG GACTTGAACC AAGCCCTCTG 1380 CTCTGAAGAC CGCGTCCTGA ATTTCTTCAC TAGAGCTTCC TCATCAGGTT ACCCAGAAGT 1440 GGGTCCCATC CACCATCCAG GTGTGCTTGG ATGTTAGTTC TCCACCCTCG AGGTGTACGC 1500 TGTGAAAAGT TTGGGAGCAC TGCTTTATAA TAAAATGAAA TATATTCTAA AAAAAAAAAA 1560 AAAAAAAAA 1569
(2) INFORMATION FOR SEQ ID NO: 46:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1924 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 46: GGGCCCCCCC WCGWKTTTTT TTTTTTTTTT TTTAATTAGG ATAATGCCTT TATTAACGAG 60 AATGAAACGT TCATTCCTCC TTCCACTCCT TCTCGTTGGT TTTCTGGACA CAGCTCACCT 120 GATCCTGCTA GAAACGTTGT CAGTCTGCTT GTGGCTTCCC TCCTTGATTG ACTCACGCTG 180 TGTGATGTCT TGAGAAGTAT CTATCCACTT CATGTGAATG AGCACTCCAA TATCAGCCAA 240 CATCAATCAT TCTTACCTAA AGAATAATAA GAAAAAGTTA ATATAAAAGA CAAGGGTATA 300 AAATAAAGGT TTGAAAATGC TAGTCAACTT CAAAATTTAA AGAGTAAAAA TCCAGAGATA 360 AAGATTGGGG GTAAGTTACA GCATAAAAAA ATAGGAAGAA ACTTCATGGT GGGGGGGAAA 420 TCTAAAATTA TTCTTACATA AAATAAGTAG ACACCTGAAT TAGAATGAAA ACTGTATTTT 480 CTTTAAAATG TAAAAGCCTG ACTCTCAGTT TCACCAGTCT GAGCACAAGT TTGACTGCAA 540
CCCAAAATAT ACTATCCCTT ATGTGAAGGT ATGTGACAAC GTTGACCTCA CCAAATGAGT 600
TTTAACATCA GCTCTTTTTT. CATATGAAAG CACATACCCT GCTCCCCATT CAAGTATGTC 660
TTCCATTGTC AGGCAGGCTG ACCACCTTCA GCAGGAGTCC TCCAAGAGTG CCCAACTCCC 720
CTTCCCACAG TACACAACGC TGTAGTTGTT GTCCTGCAAT CCTTTGTATT TACCTCATTC 780
TTTCCCATCT AAGTCCTCAC TGAGTTTTAA AGTTAGGGCT GGAAAAGCTA TGCCTTACTG 840
GGACAGCAAG GAACCAATTT TTTTCTGAGG GAGAAGACAT TCACCTTCAC TATATGCCTG 900
GCAGGGCCAC AGTGCACAAA ACAAAGATCA GCCTTCATTC AAGTTCCAGG TTTTTCTTCC 960
TCCCTGAATG ATTACTGCAA AGGGTATATG AAGTAAGAGT TCCCTGTTGC ACATGTACCA 1020
TCCATAAGGG ATACTATATC GTTTTGCATT CTTCCCCCCA TTCTCCACAT TGTCCTATCT 1080
TAAGTCCAAG CCCTTTTCAC TCTCAAAAAA AAAAAAAAAA TATTTTTTTC AGCACTGGTG 1140
TTCAAAAGCA ACGTTTTTAT GGTTAATGGT TTACCAGCAA CTGTTGAGAT TTCCAGTTGA 1200
GTCTTAAAAA TTGCCAATCA TTATCTAGCA GCAATGACAG ATGATTAGGA GCAGTCAAAT 1260
CCTCTGAATT CTTTCCCTAA TAGGCAGCCA TTTGAGAACT GCACTAGCTG ACATCACTAA 1320
AACATTATCA GCTAAAGCCA AAACCAAATA AAGGCCCAGA CCAACATCCT GGCTCTCTAA 1380
AACCTGTCCA AAATCATTAA GTGAAAGGCA GTAAATGCAG GACTGTGGAT CATGTCACTG 1440
CAGCTGACAA TGATTAACAA TAGGAGACAT GCAACCCCCA TTAAGGTTAA AAGTCCAAAA 1500
CTAGTCACAC GCATCTCTTT ATTGGGGAAA AGTGAGACTA TTATGCATTC TTGGTAGGTT 1560
TGCAACCTTG CATGAAGAGC ACCCATTGCA TTTCTTTCAT CTTTCAGAAA GCACCGGTAT 1620
CTGTTCCAAG GGCCTAACAG TACGAAAATA CATTCTGGCA TCACACCTCT GAACCCAAGA 1680
CTGTTCTCAT TAAAAATAAT TTTGGTTTGT AACAAAATTA TGAAATACAA TGCAAGCACC 1740
TCGGTATAGC ATTATTACTG AAACCACTTA ATTCCCAGCT TTTTGAGTTT TTTAAAAAAA 1800
CCCACTGCAC TAAGATTCAC AATTCATTGC TACATACAAA TTAAAGCTAG TAAGAACACA 1860
CTAACGTCAC AAGTTTCTCA TTCTAAAGTG CAAAAGCCTA ATCATCTGAA AGTGAACAGG 1920
GTAA 1924
(2) INFORMATION FOR SEQ ID NO: 47:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 475 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 47:
TGGTGTGGGG CCCAGAAAMC AAGGGACCAG TGAAAACAMC CCCAGAGACT TGTATCCGCC 60
AGGAAAGCCA TTGCCAMTYC TGAGCCCTTG AAGGGCAAGG AGGGAAACAG TGTTACCAGA 120
GCCCAGTAAG AACTGCTGTC ATGAAGGAGG GGCCACCTTG TAAGAGACAT CATTACTACC 180
AGAACTGTGG TGCCAAATTG CTGGTGTCTC TCTTTGGAGA AACCAACCAG ATACATCTGC 240 TGGAGACCCA GGTGGGCACA GAGAAGGGTG GAGAGAGAAT CTGGGAAGAG AAATGGAGAA 300
TAAGCAGCAC AGTGTTATTC ATTTCTGTAA ATTCCTATGT AGAAGGCTCA GTGTTAGAAA 360
TAAAGTTATT CTACTAGTTG CAAGTTAAGT GTTTCTGTTT GTTCTGCTTT CCTGTTAGCA 420
TAAGTAAACT CCCTTTGGAA CTACACAGGT ATGTCTCTCC TTCAACATGT GTGAA 475
(2) INFORMATION FOR SEQ ID NO: 48:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 346 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 48:
AAGGGACAGA GACCTGGATT CAGATCTCAT TTTACAATGA AGACCCCAAT GCAGAAAGTC 60
ATGTCTGAAA TTCTGAGCTT ACTCTTCTGC CTGCTGGGAC CTGCTCTGGA TGAGAGAAGG 120 GAGGAAAAGG ACTAATCAGA GGAGCCAATG AAGTCACTCC ATGAGTTTCC TGAACCCTGC 180
CCAGCTAGAG ATTAACGTYT GACCWTCAAC GTAGGACACT GTGCAGATGG CTACTTGCTG 240
GCGCACATGA AGACCAAAGC CAGGACCAAG CCCCMASCCT GCTWAACACG GCAGARTCTT 300
GCCCAGCCMA CYTCTGTGAR AATCTGCTTC CCTCCACAGC TGACCC 346
(2) INFORMATION FOR SEQ ID NO: 49:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1366 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 49:
TAGGTGTCAG CCGCCACCCC CCCCCCATAT GCAGATTTAC TSGGCATGGT AGTGGCCAGC 60
TTCTAACACA GCTGGTATTT CAAGTCTCCT GGGACCTCAC TCAGGAATGA TACCCCCTCA 120 GTAGAAGCAG CAGGTGATCT TAACTCCTTT CAAAGAGCAG GCCTGTCTGG GAAGCCATGT 180
CCTCAGCAGG CACAGCAACC CCTCTGGAAA TGGATCACAA ACTCACTTCT CAGCCAGGCA 240
GGCCAAGCTT CTATTGTAAC AGTAGGCACA GTATAGTCGG ATCATCACAT CAGCTGGGTT 300
TTTGGTTTAG TCATCTAGAG TCGTCTGGAC TAAAGGTCTT TCAGGTCTCC TTGCCCTGTG 360
AGTGCGTGAA CCTCCCCACC CGAATTGCCT CAGTTGTCCT GAGCCTCATG TCTCTCCTGG 420
TGGTGGGCCA GGCCCCTGCA TGGGAAGGGA GCCTGCTGCG GGGCAGGCCA GCTGGGGGTG 480
CTCACCTATG CGCAATGANA GTTATTGAAG GACTGGTTGT TGATGTTGGT GAGCGTATCC 540
TTCATGGCCA GCGCGAAGTC GGCCAGGTCA GCCAGGTGCT GCCAGCGCTC TCTCTCGGAC 600
TTGTCTTCCT GTGCCAGGGG ACCGTGGAGA AAGTGTCAGG GGCCGCTCAC TGCAGCAGCC 660
TGCTCTGCTG CCTTCCCTGG CAGTGTTCTG GGGGTGGATT CCCTACAMCT AGATGTTCAA 720
GGCCTTACTT TTCCTCCCAC AAAGGAGTCG CAGCCACGCT AGCTCTGACT TGCCACTGTG 780
ACAAAGTTCA CGTAGCAGGT CTAGGCAAAG ACTGGGCAAT TGAGCAGAGG AGACGGACCT 840
GTGAGTCTGA CCRYGAGSCG GRCCCCTTCA CCTTGGCTGG GCTGGTCCTG GTCCTTAGGT 900
TTTGTCAGGT TGTCCTTGTT TGGATCCCTC AACTAGGTGA TAAGCACTGG AGGGGGATGA 960
CCCGCCTTGG ACGTGTTTCT TTAACCTCAT CCATATAATA GGGCCGTGGG ATGGTTGTAG 1020
AGGTAAAGCA GGATGATGGT GTTTTAAGAC CAGAGCTTGG GACCAGGGCT CCTACACCTA 1080
ATTTTCTCTC CTGGTAGCTG AACAAAGGTC TAAATTAGCT TAACAAAAGA ACAGGCTGCC 1140
GTCAGCCAGA GTTCTGAAGG CCATGCTTTC AGTTTCCCTT GTTGACAATT GCTCTCCAGT 1200
TCCTATGAAA GCACAGAGCC TTAGGGGGCC TGGCCACAGA ACACAACCAT CTTAGGCCTG 1260
AGCTGTGAAC AGCAGGGGGT TGTGTGTCTG TTCTGTTTCT CTGCTTGCCG AACTTTCTCA 1320
ATAAACCCTA TTTCTTATTT ATAAAAAAAA AAAAAAAAAA AAAAAA 1366
(2) INFORMATION FOR SEQ ID NO: 50:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1405 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 50: GCAGTAATTC CTGTTAGCCA CTGCATCCAC CAAAACTAGT TTATTTTTCC CCTCAAATTC 60 ATGATTTTTA CGTCTGTTAC AAAGGGAATT TTGCTGATAG CTCTTTGGGT CCCACTGTTC 120 CATTTTATGC TAATAGATTC CATTCTAGGG CCCAGCCGTC TCTTGACTGA TGGTGTTCCC 180
TTTAACCCTT GGCATGTATA ATAGAATTTT GGTGAATGAA AGAACCCAAA TAGGCCAGAT 240
AGTCCCCCCA GGCCCTGATA TCCATAAAAG GCTTGGGAAT GCATTATGTA ATTGTCCTTA 300
GTCTTTTTGT TGTTTTAGAA AAAAAAAACA AGATGGGCTC AGATGGATGC CTACGTAAAA 360
ATGGTTCCTA GCTGTGTACT CATAACTTTT CTTTGAATTG AGTAGTGAAA GGAAGGAGGA 420
GGAAAGGAAA TTAAATGTCC TTCTAGTATT CTCTGGACTC AAGTCTGACA TATGAGATAA 480
TAACCTATAT TGAAATGCCA AGAATTGTAT CTGAAACAAG AGAACAGTTT GACACATTTA 540
TCATGCCTTC ATATTACATA TTAACTGAAA CCAATTAATA AACATATGAA ATATCCATTG 600
CACAAGGCAA AGGCACCTAA ACCTTTTGTT TCTTTTTCTA CATAGCAGAA ATTGATTTTT 660
TTTTTATTTT TTTAGGGGAA CCTATATAAT TATGACCCAG TGATGTCTTT TGGTGACTTA 720
AGCTTATGAA TTCAGGTTAC AATTGAGTTG ATTCTAGATG GTTACTACCT TGAAAAGGAT 780
GTTGGTGCCT TATGTGACAC GAGCCAGAGC CTGCTGGGGA ATAAACAAAG CAGGTTTCAT 840
GCCAACACCA ACTCGTAGCT TTAGTGGGCA GATGGGGAGT GGTTCACAGA CTTCCCAAAA 900
TGTGGGGGCT TTGGGATTTT CCACACCATC CCACGTGTGT TGTTCATTCT TCCTCTTTTC 960
ACACTCTTGG ATGGATWATT TGRAAATGGT GRAAWYMMCY YYKRAATTTG CCCAATAGCC 1020
WTGRGCCACC ATTCTTWATG ACACCATAAC CAAATAGTTC CWTAATGTTG AAATATTAGA 1080
AACCTGTTAC CAGCCYKSMA KTWACCCWWA WTTTTCCCAT GTTTGTGGAA TTGATATTGA 1140
AATAGCAGGG CTAAGGAATT ACTGGCAAGT TTTAGCCTGT GGGTAATACC TTAGGGTTAT 1200
TTAAATATTT GTAATTTTAT TTAAATGTTC ATGAATGTTT GAAAGGAACA AAATTATCAG 1260
GGATGGCTCT TTGCCATGGG TCTTATTTTC ACCCTCTTTT CTGTAAGAAA AAAGAACAAT 1320
GTCTTAATGT ATTTTTAAAG TTTTTGGTAT AGTTTCTAAT TCCAATTTTA ATAAAAGTTT 1380
TWTRTAAAAA AAAAAAAAAA AAAAA 1405
(2) INFORMATION FOR SEQ ID NO: 51:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 504 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 51: CGGATTTTCT AGGACCCCAA AAAAAAAAAA AGGGNAAAAA AAACCCNCAA AACCANCCAA 60 AACCCCAAAA AAAAAAAAAA TCCACAAAAA CAAAAAAACT ATAAAAAAGA AAGAATTAAA 120 AACTTTCAGA GAATTACTAT TTACTTTATT AACTTACGGA TTTATTATAT AAATATATAT 180
TCACCTAGCA ACATATCTCT GCCGTCTCTC CTGCTCTCAT AATGAAGACA TAGCCGATTC 240
TCTGCCCGGG CCCCTTGCTG ATGCTCCTCC GGGTCTGCGT CGGGCGTGGG TCTCTGGGGA 300
CCCTCCAGAG GTGGAGGTGG GCTGATGGCC TGGCTGCCTG GTGGTTGATG GTTTTGCTCC 360
CCCTACCTTT TTTTTTTGAG TTTATTCTGA TTGATTTTTT TTCTTGGTTT CTGGATAAAC 420 CACCCTCTGG GGACAGGATA ATAAAACATG TAATATTTTT AAGAAGGAAA AAAAAAAAAA 480
AAAAAACTNG GGGGGGGCCC CGAA 504
(2) INFORMATION FOR SEQ ID NO: 52:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 777 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 52:
NAAGTATCTT GGCCAGTTTA TTACAGAGGA CGATAAATGA TTCCATGTGG ATAGGGCATA 60 ACATACAGAG AATGAGACTA TGCCAGAAAT GGGAGGAGGC ATTTGAAACA ACATGAGTAT 120
CTCAGGGACA GATGGATTGA TTCTGCTATT GGTAGGCCTG GAAGCAANGG TCAGAAGTAG 180
CAAAAAATGG ATACCAAAAG CACTATTWGT CACCCAAGCT AAGTGGAATA GCTGGCCCAG 240 TAGGAGAAAT G<^∞TTTTG CTCTACACTA AGTTCTCCAA CTCTTGATAA GCCTCCAAAA 300
ACAAATGTTA GGGGAAAAAA ACGCAGCTGG TTATGAAAAG ATATATCTCA TTTCATTAAA 360
AAATCAATGT CAATGCTGTT AATAGAATCC TTTTATCTTC AGGACAGAGG CAATGCCCTA 420
AACAAACACC AGCTCAAGAG CCTCTGATGC CAACCTAGAG GGTACCCAAA CACAAACTTA 480
GCATAGAGGT AAGAATCTCT ATGTCTTTTG GTGGAGGCAA AGCCATTTGG TTGGTACTTC 540 ACAGGAACAT CTTTCTACCA AGTCTTCATC ATATGGTATG TGCCACGAGT CTCCAGTTGT 600
TTGCACCACT GTGTCATAGC TGAGAATACG CTGAAAGGTT AGTTTTGATC CTGGAAACCT 660
ATTTACAATT GCCAGCTGAT GTCCCTGCTG CCACTTAAAA AAGGCTTGGG TCTGGCATAG 720
GCAGAMAGGC CTGTGGTCCC CTCGTGCCGA TTCTNGGCTC GAGGCCAATT NCCTTAT 777
(2) INFORMATION FOR SEQ ID NO: 53:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 602 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 53:
ATGACTACAG TGTTATACCC TCCAATCTTT GCAGGTGGGC ATGGAACACT GCTTGTATCA 60
CTCTGTGCAC GGTATAAATC CATATATCCA CAAAAACACA CATCCATCCA TCAACATATA 120 CATGGTTTGG GATGAGCAGG TCAATAGTTT TGAGAGGGAG TTTGTTCCTT TTTTTTTTCT 180
CATTATACTC TTAAATTGTT GTCAGTTATC AAACAAACAA ACAGAAAAAT TGTTTGGAAA 240
AACCTTGCAT ACGCCTTTTC TATCAAGTGC TTTAAAATAT AGACTAAATA CACACATCCT 300
GCCAGTTTTT TCTTACAGTG ACAGTATCCT TACCTGCCAT TTAATATTAG CCTCGTATTT 360
TTCTCACGTA TATTTACCTG TGACTTGTAT TTGTTATTTA AACAGGAAAA AAAACATTCA 420 AAAAAAGAAA AATTAACTGT AGCGCTTCAT TATACTATTA TATTATTATT ATTATTGTGA 480
CATTTTGGAA TACTGTGGAA GTTTTATCTC TTGCATATAC TTTATACGGA AGTATTACGC 540
CTTAAAAATA CGAAAATAAA TTTTACAAGG TTCCGGTTTT GGTGGTGGAA AGAGTAAATT 600
GA 602
(2) INFORMATION FOR SEQ ID NO: 54:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1749 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 54:
AGTCACTGAC TTGGAGCCGC TCGGGGGAAG TCCCGCCCAG ACAGGCGGTG GGTGGGAATG 60
CCTCACTTCA GTTTGAAGAG GGTCCGGATC CAAAGGGGTT AAAACGAGCG AACCCCGATC 120 CCCGACCACA CTTCCCGCCT CCCTAAAACG CACACCCCGC TAGCCATGGG CAGCCGCGAC 180
CACCTGTTCA AAGTGCTGGT GGTGGGGGAC GCCGCAGTGG GCAAGACGTC GCTGGTGCAG 240
GATTATTCCC AGGACAGCTT CAGCAAACAC TACAAGTCCA CGGTGGGAGT GGATTTTGCT 300
CTGAAGGTTC TCCAGTGGTC TGACTACGAG ATAGTGCGGC TTCAGCTGTG GGATATTGCA 360
GGGCAGGAGC GCTTCACCTC TATGACACGA TTGTATTATC GGGATGCCTC TGCCTGTGTT 420 ATTATGTTTG ACGTTACCAA TGCCACTACC TTCAGCAACA GCCAGAGGTG GAAACAGGAC 480
CTAGACAGCA AGCTCACACT ACCCAATGGA GAGCCGGTGC CCTGCCTGCT CTTGGCCAAC 540
AAGTGTGATC TGTCCCCTTG GGCAGTGAGC CGGGACCAGA TTGACCGGTT CAGTAAAGAG 600
AACGGTTTCA CAGGTTGGAC AGAAACATCA GTCAAGGAGA ACAAAAATAT TAATGAGGCT 660
ATGAGAGTCC TCATTGAAAA GATGATGAGA AATTCCACAG AAGATATCAT GTCTTTGTCC 720
ACCCAAGGGG ACTACATCAA TCTACAAACC AAGTCCTCCA GCTGGTCCTG CTGCTAGTAG 780
TGTTTGGCTT ATTTTCCATC CCAGTTCTGG GAGGTCTTTT AAGTCTCTTC CCTTTGGTTG 840
CCCACCTGAC CATTTTATTA AGTACATTTG AATTGTCTCC TGACTACTGT CCAGTAAGGA 900
GGGCCCATTG TCACTTAGAA AAGACACCTG GAACCCATGT GCATTTCTGC ATCTCCTGGA 960
TTAGCCTTTC ACATGTTGCT GRCTCACATT AGTGCCAGTT AGTGCCTTCG GTGTAAGATC 1020
TTCTCATCAG CCCTCAATTT GTGATCCGGA ATTTTGTGAG AAGGATTAGA AATCAGCACC 1080
TGCGTTTTAG AGATCATAAT TCTCACCTAC TTCTGAGCTT ATTTTTCCAT TTGATATTCA 1140
TTGATATCAT GACTTCCAAT TGAGAGGAAA ATGAGATCAA ATGTCATTTC CCAAATTTCT 1200
TGTAGGCCGT TGTTTCAGAT TCTTTCTGTC TTGGAATGTA AACATCTGAT TCTGGAATGC 1260
AGAAGGAGGG GTCTGGGCAT CTGTGGATTT TTGGCTACTA GAAGTGTCCC AGAAGTCACT 1320
GTATTTTTGA AACTTCTAAC GTCATAATTA AGTTTCTCTT GTCTTGGCAT CAAGAATAGT 1380
CAAGTTTTTT GGCCGGGCAT GGTGGCTCAT GCCKGTAATC CCAGCACTTG GGGAGGCCAA 1440
GGCAGGCGGA TCACATGAGG CCAGGAATTC GAGACCAACC TGGTCAGCAT GGCAAAACCC 1500
CGTCTCTACT AAAAGTACAA AAATTAGCCA GGCGTGATGG CACGTGTCTG TAATCCCAGC 1560
TACTCTGGAG ACTGAGGTGG GAGAATCGCT TGAGACTGGG AGGCAGAGGT TGCAGTGAAC 1620
CGAGATCATG CCACCGCACT TCAGCCTGGG TGACAGAGAA GGACTCCGTC TCAAAAAAAA 1680
AAAAAAAAAA AAAACTCGAG GGGGGGCCCG GTACCCAAAT CGCCSTGATA GTGATCGTAW 1740
ACAATCNAA 1749
(2) INFORMATION FOR SEQ ID NO: 55:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1896 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 55: AAAGAGATGG GCTCTTTATT TTCTCGAAAA ACCAATTTGG AGTTACTCAT TTTTCCATAA 60 CATTAAATTT CTTACAGTGA ACTACATATT GTCCATAAGT GCTTCATCAG GACTCATCGC 120 CCTCCTGTCT ACTGGCTCCA AATAGACCAT GTCAGCTTCA CCCCCTGGCT TTGTGTCTAT 180 GGGTGGCCTG TGGTATATGG AAAAGTAGCA GGGTGGTCAG GGTGGGAGAC ACAAGATGTT 240
TTTATAGTCT AGAGCCTTTA AAAAACCCAG CAGAATGTAA TTCAGTATTT GTTTATTGGC 300
TGTTTTTTGA CAGATTGTTG AAATTAAATG AATTGAAAGG GAAACTCAGA GTACTAGGAC 360
GTTTATTAAA AGGAAAAAAA TGTCTTGCAA TGTGCTGTAA TCACAAGAGG AGAAAATAAC 420
TTGTTTCCTT GATCTGTCAG AGGTCACAGT AACCTGGGCC GAGCTGTTAT TATTTATTAT 480
ATAATAGTAG TAGGAAGTTA ATAACTGGTT CTCTGTGTTC CAAGCACAAT ATTACAACTT 540
CTTTTGAACC GTAAATATCA GAATGAATCC TCTTCCCAGG GGATTGAACA GAAGCTTAAT 600
GTTTACAAGT GTTTGAATTT GTGATCTGAA ATAACACAAA ATTAAAAACA TGATTTCTCT 660
AATTTTCCAA CTAGAGGAAG AGAAACTTGT GGAAAAGTTC TTTTTTTTTC TTTTTTTTTT 720
CTTAAAGAAG GGCAGCCAAG GTAGTAACCT AAAAATAGTG CCCAGGCATA TGAGAGTTGT 780
CCTACGAGGT TAAAGAACAC ACTGTTCCAC TGTATGGCTT TGGCCCTGAG TGGCCAGGGA 840
GGTCAACTTG ACCCTGCCAT GTTGGTTTGA CTTACTAAGA CACAGGAATC ATTGTTTTCC 900
TTGACCAGGG TCTCACACCC TGGAGGAATG TTAAGTAAGA GAAAGAACCT CTTTCCTGAA 960
TATTGACATG TAAAAGACCA AAGTAATTTT TCTGAACTTC TGCAATTCTG AGAACTCTCC 1020
AAGGAATTTA CAGTGATTTT AGTGCTTGTC AGCATTTTTC CATGAGGACT TTCATACATT 1080
TGACTCTTTA GTTCACAGGT TCCCATTGAT TGTGAGCAAG ATATTTATCT CTTTAGCCCT 1140
TGGGGATCCA GCTGAGAGCA ATCTCTTGCA TTTTTTTACC CGTGTATGTA CAGATATCAT 1200
TTCTTGTGTA TGCCATGACT TGAAAAAGTT TGGGAAGCTC TTTAGCAATA TCAGCTAAAA 1260
GGATATGAAA TCACAGGTGA TAGCAGTTGT CATTCAGTAA TTTCCTACAA GCAGCACCCC 1320
AAAGGAAATA TAGTCCTAAT CTTTACTATC CACTTCTAAA TTTAATGTGA ATTTCATACA 1380
TGTTATTAGT TGTTTTCTTT ATAATTTTAT AAAAATTATT CATCGGGAGT TTAACTTCCA 1440
CTTCCATGCT ATCGGATGTG TTGGGCTCCA TGCAAGAACT TGGAAGAAAA ACAGGCAGGA 1500
ATGCATTTGC ATAATGACCC AGATCATCAT TTTCTGCAAC TGAGAATTAT ATTTCATCAT 1560
TGCTTCTAGA AGTCTGCAAT TCTTTACTTT TCTTTGGTGC ATTATTATCT AGGTGCCATC 1620
ACTGGATAAT GTGGAGTGAC TAGAGAAGTC AYATATCACT GTAAGGTACA GTTAGGGGTA 1680
ACACTTTAGA GGTTTATTAT TTTTAAAAAA CTTTTCTTGA ACTCCTGGGC CAACATGGGT 1740
GAAACCCCGT CTTCTTACTT AAAAATACCC AAAATTAGGC CAGGGGCGTG GATGGGTGGG 1800
GTGCCTGTTA ATCTTCAGCT ACTTNGGGGA GGGCTTGAAG CCAGGGAGGA ACTGCCCTGG 1860
ANCCCCGGGG NGGGCCAGNA GGTTTGCCAG TTGAGT 1896
(2) INFORMATION FOR SEQ ID NO: 56:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1753 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 56:
TCTTTTTAAA ATAGACATTT GTGGGGCTCA CACAATATAT GAAATAGTAC CCTCTAAAAA 60
AGAGAAAAAA AAAATCAGGC GGTCAAACTT AGAGCAACAT TGTCTTATTA AAGCATAGTT 120
TATTTCACTA GAAAAAATTT AATATCAAGG ACTATTACAT ACTTCATTAC TAGGAAGTTC 180
TTTTTAAAAT GACACTTAAA ACAATCACTG AAAACTTGAT CCACATCACA CCCTGTTTAT 240
TTTCCTTAAA CATCTTGGAA GCCTAAGCTT CTGAGAATCA TGTGGCAAGT GTGATGGGCA 300
GTAAAATACC AGAGAAGATG TTTAGTAGCA ATTAAAGGCT GTTTGCACCT TTAAGGACCA 360
GCTGGGCTGT AGTGATTCCT GGGGCCAGAG TGGCATTATG TTTTTACAAA ATAATGACAT 420
ATGTCACATG TTTGCATGTT TGTTTGCTTG TTGAATTTTT GAACAGCCAG TTGACCAATC 480
ATAGAAAGTA TTACTTTCTT TCATATGGTT TTTGGTTCAC TGGCTTAAGA GGTTTCTCAG 540
AATATCTATG GCCACAGCAG CATACCAGTT TCCATCCTAA TAGGAATGAA ATTAATTTTG 600
TATCTACTGA TAACAGAATC TGGGTCACAT GAAAAAAAAT CATTTTATCC GTCTTTTAAG 660
TATATGTTTA AAATAATAAT TTATGTGTCT GCATATTGCA GAACAGCTCT GAGAGCAACA 720
GTTTCCCATT AACTCTTTCT GACCAATAGT GCTGGCACCG TTGCTTCCTC TTTGGGAAGA 780
GGAAAGGGTG TGTGAACATG GCTAACAATC TTCAAATACC CAAATTGTGA TAGCATAAAT 840
AAAGTATTTA TTTTATGCCT CAGTATATTA TTATTTAATT TTTTAGGTAA TGCCTATCTC 900
TTGGTCTATT AAGGAAAGAA GCAATCAGTA GAGAATTCAG GATAGTTTTG TTTAAATTCT 960
TGCAGATTAC ATGTTTTTAC AGTGGCCTGC TATTGAGGAA AGGTATTCTT CYATACAACT 1020
TGTTTTAACC TTTGAGAACA TTGACAGAAA TTATGCAATG GTTTGTTGAG ATACGGACTT 1080
GATGGTGCTG TTTAATCAGT TTGCTTCCAA AGTGGCCTAC TCAAGAGGCC CTAAGACTGG 1140
TAGAAATTAA AAGGATTTCA AAAACTTTCT ATTCCTTTCT TAAACCTACC AGGAAACTAG 1200
GATTGTGATA GCAATGAATG GTATGATGAA GAAAGTTTGA CCAAATTTGT TTTTTTGTTG 1260
TTGTTGTTGT TTTGAATTTG AAATCATTCT TATTCCCTTT AAGAATGTTT ATGTATGAGT 1320
GTGAAGATGC TAGCGAACCT ATGCTCAGAT ATTCATCGTA AGTCTCCCTT CACCTGTTAC 1380
AGAGTTTCAG ATCGGTCACT GATAGTATGT ATTTCTTTAG TAAGAATGTG TTAAAATTAC 1440
AATGATCTTT TAAAAAGATG ATGCAGTTCT GTATTTATTG TGCTGTGTCT GGTCCTAAGT 1500
GGAGCCAATT AAACAAGTTT CATATGTATT TTTCCAGTGT TGAATCTCAC ACACTGTACT 1560
TTGAAAATTT CCTTCCATCC TGAATAACGA ATAGAAGAGG CCATATATAT TGCCTCCTTA 1620
TCCTTGAGAT TTCACTACCT TTATGTTAAA AGTTGTGTAT AATTGTTAAA ATCTGTGAAA 1680
GAATAAAAAG TGGATTTAAA TTAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA 1740
AAAAAAAAGG GGG 1753
(2) INFORMATION FOR SEQ ID NO: 57:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1220 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 57:
GCGGAAGTTA CTGCAGCCGC GGTGTTGTGC TGTGGGGAAG GGAGAAGGAT TTGTAAACCC 60
CGGAGCGAGG TTCTGCTTAC CCGAGGCCGC TGCTGTGCGG AGACCCCCGG GTGAAGCCAC 120
CGTCATCATG TCTGACCAGG AGGCAAAACC TTCAACTGAG GACTTGGGGG ATAAGAAGGA 180
AGGTGAATAT ATTAAACTCA AAGTCATTGG ACAGGATAGC AGTGAGATTC ACTTCAAAGT 240
GAAAATGACA ACACATCTCA AGAAACTCAA AGAATCATAC TGTCAAAGAC AGGGTGTTCC 300
AATGAATTCA CTCAGGTTTC TCTTTGAGGG TCAGAGAATT GCTGATAATC ATACTCCAAA 360
AGAACTGGGA ATGGAGGAAG AAGATGTGAT TGAAGTTTAT CAGGAACAAA CGGGGGGTCA 420
TTCAACAGTT TAGATATTCT TTTTATTTTT TTTCTTTTCC CTCAATCCTT TTTTATTTTT 480
AAAAATAGTT CTTTTGTAAT GTGGTGTTCA AAACGGAATT GAAAACTGGC ACCCCATCTC 540
TTTGAAACAT CTGGTAATTT GAATTCTAGT GCTCATTATT CATTATTGTT TGTTTTCATT 600
GTGCTGATTT TTGGTGATCA AGCCTCAGTC CCCTTCATAT TACCCTCTCC TTTTTAAAAA 660
TTACGTGTGC ACAGAGAGGT CACCTTTTTC AGGACATTGC ATTTTCAGGC TTGTGGTGAT 720
AAATAAGATC GACCAATGCA AGTGTTCATA ATGACTTTCC AATTGGCCCT GATGTTCTAG 780
CATGTGATTA CTTCACTCCT GGACTGTGAC TTTCAGTGGG AGATGGAAGT TTTTCAGAGA 840
ACTGAACTGT GGAAAAATGA CCTTTCCTTA ACTTGAAGCT ACTTTTAAAA TTTGAGGGTC 900
TGGACCAAAA GAAGAGGAAT ATCAGGTTGA AGTCAAGATG ACAGATAAGG TGAGAGTAAT 960
GACTAACTCC AAAGATGGCT TCACTGAAGA AAAGGCATTT TAAGATTTTT TAAAAATCTT 1020
GTCAGAAGAT CCCAGAAAAG TTCTAATTTT CATTAGCAAT TAATAAAGCT ATACATGCAG 1080
AAATGAATAC AACAGAACAC TGCTCTTTTT GATTTTATTT GTACTTTTTG GCCTGGGATA 1140
TGGGTTTTAA ATGGACATTG TCTGTACCAG CTTCATTAAA ATAAACAATA TTTGTAAAAA 1200 TCAWAAAAAA AAAAAAAAAA 1220
(2) INFORMATION FOR SEQ ID NO: 58:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1049 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 58:
TCGCGCCTGC AGACACAGCA TCTACTCAGC GTGGGTCACC TCTGTGAACA TCACTGACTG 60
CAAGCCTCCC TCAATTTCTG GTGCAGCCCA TCAGGGACCC ACAGCGCCTG GGAGGATGGT 120
GCGGATCTTG GCCAATGGGG AAATCGTGCA GGACGACGAC CCCCGAGTGA GGACCACTAC 180
CCAGCCACCA AGAGGTAGCA TTCCTCGACA GAGCTTCTTC AATAGGGGCC ATGGTGCTCC 240
CCCAGGGGGT CCTGGCCCCC GCCAGCAGCA GGCAGGTGCC AGGCTGGGTG CTGCTCAGTC 300
CCCCTTCAAT GACCTCAACC GGCAGCTGGT GAACATGGGC TTTCCGCAGT GGCATCTCGG 360
CAACCATGCT GTGGAGCCGG TGACCTCCAT CCTGCTCCTC TTCCTGCTCA TGATGCTTGG 420
TGTTCGTGGC CTCCTCCTGG TTGGCCTTGT CTACCTGGTG TCCCACCTGA GTCAGCGGTG 480
ACCTCTGAGG GCTGATAGGG GTGGGTTTGT TGAGAGGGAC TTGCTGGGCC TTGGTGTGAG 540
AGCAGGCATA TTTGGAGGGG ATCTGGTGGT GCCTTGAAGG TATGATCAGA GAGGGGACCA 600
CAGGTGTGTG TTTCCCCTTT GTGTTAAGCG TGAGGCAGAG GGAGACGTTA GTCCCAGCAT 660
TTCCCAAAGT GTGGGTGGGT CCGTTGGTTC CCGAGATACT TTTAGGTGGT ATGGGGCCTG 720
CATTAAGTGG CACAAAATCA GAGCAAGAAA GCGATGCCCT TCCCAATTCT CTCAATCCTT 780
TTATGCCGAG AAGATCTCAG CTGGATGCCA ACATGTTCCG ATGCCTGTGG AAGACATGCC 840
GACGTCTCCT CTGCCTAGGG AGCAGGACTT GGGCTTAGGG CAGGTGGAAA AAATTCCAGA 900
CTTTTTTAGC ACTGTTTTTG TTTTAATGGT ATATTTTTAT TGGCTACTTT ATTGTTTAGG 960
ACAAGTGGTA GTGGCATTCT ATTTATTGTG ACCTTTTCAA TAAATAGATT TAAGTAAAAA 1020
AAAAAAAAAA AAAACTCGAG GGGGGGCCC 1049
(2) INFORMATION FOR SEQ ID NO: 59: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1776 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 59:
AAAGAGGATG TGMAGCTAGA GGTCCCCGAT GGCTGGTCGG ATGGGAAGCA CAAGGCTGAG 60
GGACTGGATT GTAAAGGCAC TAAGTCGTTC TGCGGTGAGA ATCAGACATG GGGGACCTCT 120
AGCTTCACAT CCTCTTTCCT TGCAGSTCTG GACATCCTGA GCCCAAGTCC CCCACACTCA 180
GTGCAGTGAT GAGTGCGGAA GTGAAGGTGA CAGGGCAGAA CCAGGAGCAA TTTCTGCTCC 240
TAGCCAAGTC GGCCAAGGGG GCAGCGCTGG CCACACTCAT CCATCAGGTG CTGGAGGCCC 300
CTGGTGTCTA CGTGTTTGGA GAACTGCTGG ACATGCCCAA TGTTAGAGAG CTGGCTGAGA 360
GTGACTTTGC CTCTACCTTC CGGCTGCTCA CAGTGTTTGC TTATGGGACA TACGCTGACT 420
ACTTAGCTGA AGCCCGGAAT CTTCCTCCAC TAACAGAGGC TCAGAAGAAT AAGCTTCGAC 480
ACCTCTCAGT TGTCACCCTG GCTGCTAAAG TAAAGTGTAT CCCATATGCA GTGTTGCTGG 540
AGGCTCTTGC CCTGCGTAAT GTGCGGCAGC TGGAAGACCT TGTGATTGAG GCTGTGTATG 600
CTGACGTGCT TCGTGGCTCC CTGGACCAGC GCAACCAGCG GCTCGAGGTT GACTACAGCA 660
TCGGGCGGGA CATCCAGCGC CAGGACCTCA GTGCCATTGC CCGAACCCTK AANAAAAACC 720
ATTAAAGTTA CGACGGCAGC AGCAGCCGCA GCCACATCTC AGGACCCTGA GCAACACCTG 780
ACTGAGCTGA GGGAACCAGC TCCTGGCACC AACCAGCGCC ASCCAGCAAG AAAGCCTCAA 840
AGGGCAAGGG GCTCCGAGGG ANCGCCAAGA TTTGGTCCAA GTCGAATTGA AAGRACTGTC 900
GTTTCCTCCC TGGGGATGTG GGGTCCCAGC TGCCTGCCTG CCTCTTAGGA GTCCTCAGAG 960
AGCCTTCTGT GCCCCTGGCC AGCTGATAAT CCTAGGTTCA TGACCCTTCA CCTCCCCTAA 1020
CCCCAAACAT AGATCACACC TTCTCTAGGG AGGAGKCAAA TGTAGGTCAT GTTTTTGTTG 1080
GTACTTTCTG TTTTTTGTGA CTTCATGTGT TCCATTGCTC CCCGCTGCCA TGCTCTCTCC 1140
CTTGTTTCCT TAAGAGCTCA GCATCTGTCC CTGTTCATTA CATGTCATTG AGTAGGTGGG 1200
TAGCCCTGAT GGGGGTCGCT CTGTCTGGAG CATAACCCAC AGGCGTTTTT TCTGCCACCC 1260
CATCCCTGCA TGCCTGATCC CCAGTTCCTA TACCCTACCC CTGACCTATT GAGCAGCCTC 1320
TGAAGAGCCA TAGGGCCCCC ACCTTTACTC ACACCCTGAG AATTCTGGGA GCCAGTCTGC 1380
CATGCCAGGA GTCACTGGAC ATGTTCATCC TAGAATCCTG TCACACTACA GTCATTTCTT 1440
TTCCTCTCTC TGGCCCTTGG GTCCTGGGAA TGCTGCTGCT TCAACCCCAG AGCCTAAGAA 1500
TGGCAGCCGT TTCTTAACAT GTTGAGAGAT GATTCTTTCT TGGCCCTGGC CATCTCGGGA 1560
AGCTTGATGG CAATCCTGGA AGGGTTTAAT CTCCTTTTGT GAGTTTGGTG GGGAAGGGAA 1620
GGGTATATAG ATTGTATTAA AAAAAAAAAG GTATATATGC ATATATCTAT ATATAATATG 1680 ACGCAGAAAT AAATCTATGA GAAATCTATC TACAAAMWAA AAAAAAAAAA AAAAAAAAAA 1740 AGGAATTCGA TNTCAAGCTT ATCGATACCG TCNACC 1776
(2) INFORMATION FOR SEQ ID NO: 60:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 443 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO : 60: ACAGATAAAT AAATAAATAA TAAATTAAAT TAAATAAAAA ATCTGAGCTA ATCTGAATAA 60 ATTGAGAGAT TTCACATGAA AGCCAGGATT TCTGGCTTCC CAGGAACAGT CAGAAGAGCT 120 AGCTAGCAAC ACTGGTCTGC TTGGCTACCT TCTTTGGAAC AACATGAAAT CTAGCTCCCT 180 TTTTTTTTTT TTTTTGGCCC ACTTCATCCA TTCACATGAC CTGCCTGGCC TCTGCAGGTA 240 AGTGAGTATG CAACAAAAAT GTAGCACAGG TTTTGTCGCT GAACTACGTG GTTTCAGGTC 300 CAGCTCTGCC ACTTGCTAGC ATGACCTCGT GCCGAATTCC NGCACGAAGT TTTTTTTTTT 360 TTTTTCAGTG CTCCAGTCCC CCTATTGGAG AATCCTGCCC CCCCCTGGGA CAGAATGTTC 420 ACCCTGGCCC CGCGANTCCC TGA 443
(2) INFORMATION FOR SEQ ID NO: 61:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2888 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 61: TTAATGTTGT CAATAACCAC CAGGCCAAAC AGAATTTATA TGACCTGGAT GAAGATGATG 60 ATGGTATAGC TTCCGTTCCT ACTAAACAGA TGAAGTTTGC AGCCTCAGGC GNCTTTCTCC 120 ACCACATGGC TGGGCTAAGC AGTTCCAAGC TTTCCATGTC CAAGGCCCTC CCTCTCACCA 180 AAGTGGTTCA GAATGATGCA TACACAGCTC CTGCTCTCCC TTCCTCTATT CGAACAAAAG 240 CCTTGACCAA CATGTCCCGG ACACTGGTGA ACAAGGAAGA ACCCCCCAAA GAGCTGCCAG 300 CTGCTGAGCC TGTTCTCAGC CCATTGGAAG GCACCAAGAT GACTGTGAAT AATCTGCACC 360
CTCGAGTCAC TGAGGAGGAC ATTGTTGAGC TTTTCTGTGT GTGTGGGGCC CTCAAGCGAG 420
CTCGACTGGT CCATCCTGGG GTAGCGGAGG TGGTGTTTGT GAAAAAGGAC GATGCCATCA 480
CCGCATATAA GAAGTACAAC AACCGGTGTC TGGACGGGCA GCCGATGAAG TGCAACCTTC 540
ACATGAATGG GAATGTTATC ACCTCAGACC AGCCCATCCT GCTGCGGCTG AGTGACAGCC 600
CATCAATGAA AAAGGAGAGC GAGCTGCCTC GCAGGGTGAA CTCTGCCTCC TCCTCCAACC 660
CCCCTGCYGA AGTGGACCCT GACACCATCC TGAAGGCACT CTTCAAGTCC TCAGGGGCCT 720
CTKTGACCAC GCAGCCCACA GAATTCAAAA TCAAGCTTTG AGCAGGGGAG TGAGGCAGCC 780
AGAAGTGGGG GCAGAGGAGG GTGGCTCTGT TTCCCCAAGG CAAAGCTTAT GACCAATGGG 840
CCATCGGACT GGAGACCCCT GATTGTGGGA AGGGTTGCCA GGGATAAAGA GCTTCCTCAC 900
TGGATGGGAC CCGCCTTTCT GTGTTGTGTT CTGCCCTGTG CTCTTCTCTC TACGTTAACG 960
TTTCCTGTAG TATGTTTCTT CATCTCATCG CCAAGGTAGG CTTGTGTTTT TCAGTGTGTG 1020
CCTCCCCGAG CCTCAGCCCC AAGCTGATTT CTTATCTGGA AATGGTACAC TGAATTCTCT 1080
GGGTGGCTTT CTTGTGGCCC CATGGGATGC AGCGTGGGGG CTGTCTGAAG GACCCTGCTT 1140
TTTCCAGGGG CCGAGGGGCT GCCTTTCCTT TGTGTGTATT AAGCTTTTCA AACAATGGAG 1200
GGGATGGAGA GCCCTGGTGT CCTGACGGGA GCCAGGTCGG CCTGAGAGCT GTGCCGCTCC 1260
TCTGTCTTGT CAGTGGAGGT GCCTGGGTGG GGAGCAGGTC TCAGGCCTCT TGTCCTCTCC 1320
CCAGTGGCTC CAGGCCTCAC TAGTGGCAAG GGCAGGATGA GGCTGCACCG CTGGGAAGAG 1380
TCTATCTAAG YTCTTGGCTT GGAGTCCCGT GTCGTCTCCR CCCAGAGGAA GTTCTCCAGA 1440
GTTCACCTTT CCCTTTTCCT TGAGTTGTGC TGAATGCCCC ACCCCAGCTC TCTTTCCCTT 1500
CTGGGTGTCT TTGCTGGGAG GGGGCTGTGT TGTGAGCCCT CCCGGTTCTC ACCTCGCCTG 1560
GCACTTAACC ACACCCTGGT TTTGTGTAGC CGCCAGCTCT CTTCTGGTTG GGCCTTTGAA 1620
AGGCTCAGCC TCCCATTGTG CAGTGCTTGG GTTTGGAGCT TATTTGAATG GAAGAGGTCA 1680
GTTTGTTCCT GGCTCTCCAT TTCTGGCCTC AGTTGTCTAC AGGACAGTGG TCAGGGATGC 1740
CTGGAGGCAT ATATCCAGCT GCCACCAAGG GGCACTGTTT GTTCCCACTT ATGTGAGTGA 1800
CCCCATCCAT CCATGACCAG AGGATTATTT TCCTGCCTTG GCAGAGGAGG AGGAGTCAAG 1860
GGAGCAGGGC AGCTCTACCA GGCAAGGTGT TTCCCCAGCA TAGGCGCAGA CAGTTGGGAC 1920
GAAACTTCAG AGCCCAGGCA GTCCCTGAAT GACCAGGCCA GTGTTGTCAC TGAGTGGTCC 1980
CCTGCTGGTT GGGAGTGAAG AGAATCCAGG CTGGCAGAGC TGGAGCCAGT TGGGGAGCAC 2040
GGTTCTGGGA GCTCTGCAAA ATCAGTAGCA AGTGCTGGAA AAGGCACATG CCGAAGATAC 2100
TCAAGAGCTC CCAAGATTTG CTTGAGGCTA GCCCAGTGAA RAAAACCAGA GACTCATGTT 2160
TCCAGGGGTC AGTCTGTCAG GCAGGAAGGA CCCAGGATTT GAACCCAGCT TCAGTGTGCA 2220
GGCTCTGAGG CTGCCCAGGA CGGGAAAGTC CAAGGAAGGG GCCTGGTGGT GCTCCACTTG 2280
CAGTTCTTTA AAGAATGCTG CTTTTTATTC TCCTAACCCT TTCAAGTGGG TGCAGACTTC 2340
TCGTTAGCAG CTGGAAGACA TTCCTCCCAC ACTTTTCCCT TCCTGGCCCA AGAGAGCATC 2400
CAGAAGGCAG TAGGACCTGG TTTTTCAGGT ACTGGGAGCC GGGGGCTCAC TGCTTGCACT 2460
GTGCTTAGGG TAGGGATGGT AAATATCCTC CCTGCATGGC TTTATCCTCC CTCTCATCCC 2520
AAAGCAGGTA TCTTCTGGTT GTCACAGAGT TTCATTGAGT CCAGCTGCAG CCACGTGGCC 2580
ATCTGGAGCT GGTGCTATAG GTGACCATCT GGTACATTGA GGGGACCTGT TTGCCTCCTC 2640
CACTCTATAA GCAGTCATCT TGGGAGACCG GGAGGAGAAG GTGGTGGGCT AGTCCTGTGT 2700
CCTCCTCCAC TTCCCATGCC TCTATGTTAC CCATCTGTGT CTCCTGTGCA GAAGGAGAGG 2760
AAGGGGCATT AAGAGATGAA GGGTGATTAT GTATTACTTA TCCATTTCTG AATAAACATT 2820
TGTTATTCCT AAAAAAAAAA AAAAAAAACT CGAGGGGGGG CCCGGWACCC AWATCGCCSK 2880
AAAGTGAG 2888
(2) INFORMATION FOR SEQ ID NO: 62:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1851 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 62:
CACTAGTATA ATTTATAATT ATAACCTATT CTGATTTCTT TTCAAATATT AGGTGTCCTA 60
GTTGCCTATG AAGGTTTGCC ACTTCATCTT GCACTGTTCC CCAAACTTTG GACTGAGCTA 120
TGCCAGACTC AGTCTGCTAT GTCAAAAAAC TGCATCAAGC TTTTGTGTGA AGATCCTGTT 180
TTCGCAGAAT ATATTAAATG TATCCTAATG GATGAAAGAA CTTTTTTAAA CAACAACATT 240
GTCTACACGT TCATGACACA TTTCCTTCTA AAGGTTCAAA GTCAAGTGTT TTCTGAAGCA 300
AACTGTGCCA ATTTGATCAG CACTCTTATT ACAAACTTGA TAAGCCAGTA TCAGAACCTA 360
CAGTCTGATT TCTCCAACCG AGTTGAAATT TCCAAAGCAA GTGCTTCTTT AAATGGGGAC 420
CTGAGGGCAC TCGCTTTGCT CCTGTCAGTA CACACTCCCA AACAGTTAAA CCCAGCTCTA 480
ATTCCAACTC TGCAAGAGCT TTTAAGCAAA TGCAGGACTT GTCTGCAACA GAGAAACTCA 540
CTCCAAGAGC AAGAAGCCAA AGAAAGAAAA ACTAAAGATG ATGAAGGAGC AACTCCCATT 600
AAAAGGCGGC GTGTTAGCAG TGATGAGGAG CACACTGTAG ACAGCTGCAT CAGTGACATG 660
AAAACAGAAA CCAGGGAGGT CCTGACCCCA ACGAGCACTT CTGACAATGA GACCAGAGAC 720
TCCTCAATTA TTGATCCAGG AACTGAGCAA GATCTTCCTT CCCCTGAAAA TAGTTCTGTT 780
AAAGAATACC GAATGGAAGT TCCATCTTCG TTTTCAGAAG ACATGTCAAA TATCAGGTCA 840
CAGCATGCAG AAGAACAGTC CAACAATGGT AGATATGACG ATTGTAAAGA ATTTAAAGAC 900
CTCCACTGTT CCAAGGATTC TACCCTAGCC GAGGAAGAAT CTGAGTTCCC TTCTACTTCT 960
ATCTCTGCAG TTCTGTCTGA CTTAGCTGAC TTGAGAAGCT GTGATGGCCA AGCTTTGCCC 1020
TCCCAGGACC CTGAGGTTGC TTTATCTCTC AGTTGTGGCC ATTCCAGAGG ACTCTTTAGT 1080
CATATGCAGC AACATGACAT TTTAGATACC CTGTGTAGGA CCATTGAATC TACAATCCAT 1140
GTCGTCACAA GGATATCTGG CAAAGGAAAC CAAGCTGCTT CTTGACATTA GGTGTAGCAT 1200
GTCTACTTTT AAGTCCCTCA CCCCCAACCC CCATGCTGTT TGTATAAGTT TTGCTTATTT 1260
GTTTTTGTGC TTCAGTTTGT CCAGTGCTCT CTGCTTGAAT GGCAAGATAG ATTTATAGGC 1320
TTAATTCTTG GTCAGGCAGA ACTCCAGATG AAAAAAACTT GCATCTTCAG TATACTTCCT 1380
AAAGGGCAAT CAGATAATGG ATATGTTTTA TGTAATTAAG AGTTCACTTT AGTGGCTTTC 1440
ATTTAATATG GCTGTCTGGG AAGAACAGGG TTGCCTAGCC CTGTACAATG TAATTTAAAC 1500
TTACAGCATT TTTACTGTGT ATGATATGGT GTCCTCTGTG CCAGTTTTGT ACCTTATAGA 1560
GGCAGATTGC CTCCGATCGC TGTGGTTCTT ATTATCAAAA TTAAGTTTAC TTGTATACGG 1620
AACAACCACA AGAAATTTGA TTCTGTAAAG AATCCTCTTT AGCTGTGGCC TGGCAGTATA 1680
TAAATGGTGC TTTATTTAAC AGAATACCTG TGGAGGAAAT AAAGCACACT TGATGTAAAA 1740
ATAATTGTTT TATTTTTATT GACATGACTG ATTGATTGCT ATTCTGTGCA CTTAATTAAA 1800
CTGATTGTGA TGACTTWWAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA A 1851
(2) INFORMATION FOR SEQ ID NO: 63:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 3542 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 63: TCCAATGCTG ATGAGCGTCT TCGCTGGCAG GCCAGCTCCT TGCCTGCTGA TGACCTTTGC 60 ACAGAAAATG CCATCATGCT GAAACGATTC AATAGGTATC CGCTGATCAT TGACCCCTCT 120 GGACAGGCCA CAGAATTCAT TATGAATGAA TATAAGGWTC GTAAGATCAC ACGGACCAGC 180
TTCCTGGATG ACGCCTTCAG AAAGAACTTA GAGAGTGCAC TGAGATTCGG TAACCCCCTT 240
CTGGTCCAGG ATGTGGAAAG CTACGATCCA GTTTTGAACC CGGTGCTGAA CCGTGAAGTG 300
CGGCGAACAG GGGGGAGAGT GCTGATCACT CTCGGGGACC AGGACATAGA CCTGTCGCCA 360
TCGTTTGTCA TCTTCCTGTC CACCCGGGAT CCAACTGTCG AGTTCCCACC AGATCTCTGT 420
TCCCGGGTTA CTTTTGTAAA CTTCACAGTT ACCCGTAGCA GTTTACAAAG CCAGTGTCTA 480
AATGAAGTAC TTAAAGCAGA AAGACCTGAT GTGGACGAGA AACGATCTGA TCTTCTTAAA 540
CTTCAAGGGG AATTTCAGCT CCGTTTGCGT CAGCTGGAAA AATCTCTACT ACAAGCTCTG 600
AACGAGGTGA AAGGGCGCAT TTTGGATGAC GACACGATCA TAACCACTCT GGAGAACCTG 660
AAGAGAGAGG CTGCAGAGGT CACCAGGAAA GTTGAGGAGA CGGACATTGT CATGCAGGAG 720
GTGGAGACCG TGTCCCAGCA GTACCTCCCG CTCTCCACCG CCTGCAGCAG CATCTACTTC 780
ACCATGGAGT CCCTCAAGCA GATACACTTC TTGTACCAGT ACTCCCTCCA GTTTTTCCTG 840
GACATTTATC ACAACGTCCT ATACGAGAAC CCGAACCTGA AGGGTGTCAC CGACCACACA 900
CAGCGCCTGT CCATTATAAC AAAGGACCTC TTCCAGGTGG CGTTTAACCG AGTGGCTCGA 960
GGCATGCTGC ATCAGGACCA CATTACCTTT GCCATGCTGC TGGCAAGAAT CAAACTGAAG 1020
GGCACCGTGG GGGAGCCCAC CTACGATGCA GAATTCCAGC ACTTCTTGAG AGGAAATGAG 1080
ATTGTCCTGA GTGCTGGCTC CACCCCCAGG ATCCAGGGCC TGACTGTGGA GCAGGCGGAG 1140
GCGGTGGTGA GGCTGAGCTG CCTTCCCGCG TTTAAGGACT TGATTGCAAA GGTTCAGGCA 1200
GACGAGCAAT TTGGCATCTG GCTGGACAGC AGCTCCCCGG AGCAGACTGT GCCCTACCTC 1260
TGGAGTGAAG AAACACCTGC AACACCCATT GGCCAGGCCA TCCACCGCCT GCTCCTGATC 1320
CAGGCTTTCC GGCCCGATCG CCTGTTGGCC ATGGCCCACA TGTTTGTTTC AACAAACCTT 1380
GGGGAGTCTT TCATGTCCAT CATGGAGCAG CCGCTCGACC TGACCCACAT TGTGGSCACA 1440
GAGGTGAAGC CCAACACTCC TGTCTTAATG TGCTCTGTGC CTGGTTATGA TGCCAGTGGA 1500
CATGTCGAGG ACCTTGCAGC CGAGCAGAAC ACGCAGATCA CTTCAATTGC AATCGGCTCT 1560
GCAGAAGGCT TTAACCAAGC AGATAAGGCA ATAAACACCG CTGTAAAGTC GGGCAGGTGG 1620
GTGATGCTGA AGAATGTGCA TCTGGCCCCA GGGTGGCTGA TGCAGCTGGA GAAGAAGTTG 1680
CATTCCCTGC AGCCGCATGC CTGCTTCCGA CTCTTCCTCA CCATGGAGAT CAACCCCAAG 1740
GTGCCTGTGA ATCTGCTCCG TGCGGGCCGC ATCTTTGTGT TCGAGCCACC GCCAGGGKTG 1800
AAGGCCAACA TGCTGAGGAC GTTCAGCAGC ATTCCCGTCT CACGGATATG CAAGTCTCCC 1860
AACGAGCGTG CCCGCTTGTA CTTCCTGCTG GCCTGGTTTC ATGCGATCAT CCAAGAACGC 1920
TTACGATACG CACCACTGGG GTGGTCAAAG AAGTATGAAT TTGGAGAGTC TGACCTGCGG 1980
TCANYTTGCG ATACGGTGGA CACGTGGCTG GATGACACGG CCAAGGGCAG GCAGAACATC 2040
TCACCGGATA AGATCCCGTG GTCTGCACTA AAGACCTTAA TGGCCCAGTC CATTTATGGC 2100
GGGCGCGTGG ACAACGAGTT TGACCAGCGT CTGCTCAACA CCTTCCTGGA GCGCCTGTTC 2160
ACAACCAGGA GTTTCGACAG TGAGTTTAAG CTGGCATGCA AGGTCGACGG ACATAAAGAC 2220
ATTCAAATGC CAGATGGCAT GCAGGCGAGA GGAGTTTGTG CAGTGGGTGG AGTTGCTCCC 2280
CGACACCCAG ACGCCCTCCT GGCTGGGCCT GCCCAACAAC GCCGAGAGAG TCCTCCTTAC 2340
CACACAGGGT GTGGACATGA TCAGTAAAAT GCTGAAGATG CAGATGTTGG AGGATGAGGA 2400
CGACCTGGCC TACGCAGAGA CTGAGAAGAA GACGAGGACA GACTCCACGT CCGACGGGCG 2460
CCCTGCCTGG ATGCGGACAC TGCACACCAC CGCGTCCAAC TGGCTGCACC TCATCCCCCA 2520
GACGCTGAGC CACCTCAAGC GCACCGTGGA GAATATCAAG GATCCTTTGT TCAGGTTCTT 2580
TGAGAGAGAA GTGAAGATGG GCGCAAAGCT GCTTCAGGAC GTTCGCCAGG ACCTTGCAGA 2640
TGTCGTCCAG GTGTGCGAAG GAAAGAAGAA GCAGACCAAC TACTTGCGCA CGCTGATCAA 2700
CGAGCTAGTG AAAGGGATCT TGCCTCGGAG CTGGTCCCAC TACACGGTGC CTGCCGGCAT 2760
GACCGTCATC CAGTGGGTGT CCGACTTCAG CGAGAGGATC AAACAGCTGC AGAACATCTC 2820
ACTGGCAGCT GCATCTGGTG GCGCCAAGGA GCTAAAGAAC ATCCACGTGT GCCTGGGTGG 2880
CCTGTTCGTG CCTGAGGCGT ACATCACTGC CACCAGGCAG TATGTGGCCC AGGCCAACAG 2940
CTGGTCCCTG GAGGAGCTCT GCCTGGAAGT CAACGTCACC ACCTCACAGG GCGCCACCCT 3000
TGACGCTTGC AGCTTCGGAG TCACGGGTTT GAAACTTCAA GGGGCCACGT GCAACAACAA 3060
CAAGCTGTCA CTGTCCAATG CCATCTCAAC CGCCCTTCCC CTGACGCAGC TGCGCTGGGT 3120
CAAGCAGACA AACACCGAGA AGAAGGCCAG TGTGGTAACC TTACCTGTCT ACCTGAACTT 3180
CACCCGTGCA GACCTCATCT TCACCGTGGA CTTCGAAATT GCTACAAAGG AGGATCCTCG 3240
CAGCTTCTAC GAGCGGGGTG TCGCAGTCTT GTGCACAGAG TAAACTTTTC TAGCTGCCCC 3300
TTTCTGTAAT AGTGAAAGTT GGTATTTAAC ATTTATTCAT TTTTAAAATA TTTGGAAGGT 3360
CTGAGCTTGT GAAAAGAAAG TGGTTGGTCT GAGGTTGGAG GAAGCTGAAT GGAATCTGAC 3420
GGTTGGGAGT GGTGGAAATT GGAAGGATAC CAGGAGGTAT TTGGGAAGGC CAATGGCGTG 3480
GCTCCTTTGA GGAAATAAAA CACTAAGCAT GAAAAAAAAA AAAAAACTTA CAANCCNCAA 3540
GG 3542
( 2 ) INFORMATION FOR SEQ ID NO : 64 :
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 883 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 64:
AGGTGATTTT AATGATAGGT GTCATATATA GGACGGATAA TCTGTTTACA TTCTGTTCTT 60
CTCGATGCAC TCACAAGCGG GTAACTAGGT GACAAGAAAA CAAAGATCTT ATTCAAAAGA 120
GGTCTTACAG CAACCCAACG TCTCATCTTC CCATAGTAAA GATGACGGCG CCTTGAGGTA 180 AGCTACAGGC AACACCACTT CCGCGTTTCT CTTGCGCCCT GGTCCAAGAT GGCGGATGAA 240
GCCACGCGAC GTGTTGTGTC TGAGATCCCG GTGCTGAAGA CTAACGCCGG ACCCCGAGAT 300
CGTGAGTTGT GGGTGCAGCG ACTGAAGGAG GAATATCAGT CCCTTATCCG GTATGTGGAG 360
AACAACAAGA ATGCTGACAA CGATTGGTTC CGACTGGAGT CCAACAAGGA AGGAACTCGG 420
TGGTTTGGAA AATGCTGGTA TATCCATGAC CTCCTGAAAT ATGAGTTTGA CATCGAGTTT 480 GACATTCCTA TCACATATCC TACTACTGCC CCAGAAATTG CAGTTCCTGA GCTGGATGGA 540
AAGACAGCAA AGATGTACAG GGGTGGCAAA ATATGCCTGA CGGATCATTT CAAACCTTTG 600
TGGGGCCAGG AATGTGCCCA AATTTGGACT AGCTCATCTC ATGGCTCTGG GGCTGGGTCC 660
ATGGSTGGCA GTGGAAATCC CTGATCTGAT TCAGAAGGGC GTCATCCAAC ACAAAGAGAA 720
ATGCAACCAA TGAAGAATCA AGCCACTGAG GCAGGGCAGA GGGACCTTTG ATAGGCTACG 780 ATACTAWTTT CCTGTGCATC ACACTTAACT CATCTAACTG TTCCCCGGAC ANCCTCCACT 840
CTAGTTGTTA CTAAGTANTG CAGTAGCATT NTGGGGAAGA ACA 883
(2) INFORMATION FOR SEQ ID NO: 65:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 1541 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 65:
GGCACGAGGT GGCCTCTACC CTGGGCTCAT CTGGCTACAC AGGGACTCTA AACGCTTCCA 60 GATTCCCTGG AAACATGCCA CCCGGCATAG CCCTCAACAA GAAGAGGAAA ATACCATTTT 120
TAAGGCCTGG GCTGTAGAGA CAGGGAAGTA CCAGGAAGGG GTGGATGACC CTGACCCAGC 180
TAAATGGAAG GCCCAGCTGC GCTGTGCTCT CAATAAGAGC AGAGAATTCA ACCTGATGTA 240 TGATGGCACC AAGGAGGTGC CCATGAACCC AGTGAAGATA TATCAAGTGT GTGACATCCC 300
TCAGCCCCAG GGCTCGATCA TTAACCCAGG ATCCACAGGG TCTGCTCCCT GGGATGAGAA 360
GGATAATGAT GTGGATGAAG AAGATGAGGA AGATGAGCTG GATCAGTCGC AGCACCATGT 420
TCCCATCCAG GACACCTTCC CCTTCCTGAA CATCAATGGT TCTCCCATGG CGCCAGCCAG 480
TGTGGGCAAT TGCAGTGTGG GCAACTGCAG CCCGGAGGCA GTGTGGCCCA AAACTGAACC 540
CCTGGAGATG GAAGTACCCC AGGCACCTAT ACAGCCCTTC TATAGCTCTC CAGAACTGTG 600
GATCAGCTCT CTCCCAATGA CTGACCTGGA CATCAAGTTT CAGTACCGTG GGAAGGAGTA 660
CGGGCAGACC ATGACCGTGA GCAACCCTCA GGGCTGCCGA CTCTTCTATG GGGACCTGGG 720
TCCCATGCCT GACCAGGAGG AGCTCTTTGG TCCCGTCAGN CTGGAGCAGG TCAAATTCCC 780
AGGTCCTGAG CATATTACCA ATGAGAAGCA GAAGCTGTTC ACTAGCAAGC TGCTGGACGT 840
CATGGACAGA GGACTGATCC TGGAGGTCAG CGGTCATGCC ATTTATGCCA TCAGGCTGTG 900
CCAGTGCAAG GTGTACTGGT CTGGGCCATG TGCCCCATCA CTTGTTGCTC CCAACCTGAT 960
TGAGAGACAA AAGAAGGTCA AGCTATTTTG TCTGGAAACA TTCCTTAGCG ATCTCATTGC 1020
CCACCAGAAA GGACAGATAG AGAAGCAGCC ACCGTTTGAG ATCTACTTAT GCTTTGGGGA 1080
AGAATGGCCA GATGGGAAAC CATTGGAAAG GAAACTCATC TTGGTTCAGG TCATTCCAGT 1140
AGTGGCTCGG ATGATCTACG AGATGTTTTC TGGTGATTTC ACACGATCCT TTGATAGTGG 1200
CAGTGTCCGC CTGCAGATCT CAACCCCAGA CATCAAGGAT AACATCGTTG CTCAGCTGAA 1260
GCAGCTGTAC CGCATCCTTC AAACCCAGGA GAGCTGGCAG CCCATGCAGC CCACCCCCAG 1320
CATGCAACTG CCCCCTGCCC TGCCTCCCCA GTAATTGTGA ATGCCATCTT CTTCCTTCTC 1380
TTTTTTATAA TATTGTACAT ATGGATTTTT TTATTGTTTA GATTTAACCA GCTTTTAAAT 1440
CTCTGTTTTC TGTGACAGTG TTAGAAGTTT GTGATTCTCC AAATATGCCT AGATTTAAAG 1500
CTGATTTAAT TTATGGAAAA AAAAAAAAAA AAAAAAAAAA A 1541
(2) INFORMATION FOR SEQ ID NO: 66:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 732 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 66: AGAAAATGAA TGTTAGAAGG TGCCTGCCGA GGCGGGACAG AGTGTTTGCT CGCGCTGGAG 60 AAGGCTCTGC TCAGCCCTGA GAGTCCCTTC CTGCCCCACC GATACTGGCA CTTTAAAAAG 120
GAAGCTGACC GCACAGTGTC CAGACGAATT GGCCCCCAGA AGATGGGGAG TTCTGTCCTG 180
CCCTTCTGTG TCTGCGTGAC CTCACCCAGC CTAGGAGGGA GGTGCATTCA GGGTAGATTT 240 GCCTCTCATT CAAAGTTCTG GGGCTTTGGG CGGAAAACAG CCAGCTTTGG CGCTGTTGGG 300
GAGACTCCTC CAGACCAGGA ACCCCAGAAG GAGACAGAGC CTGCCACATC CTCCCACGCC 360
AGGCCCTGGG CCAGGGTGAT TGGACTGAGA ATTTGGCCAC AACCAAATTG ATGCTGGCTG 420
GAACCAGAGG CCAGAAAGCC TGGCCTTGTC CCCATGTGGG AGCCCTGTCC TCAGCCCTCT 480
TGTCCCCTTG AGCTCAGTGA ATTCCCACCA GGTGCCCACA GCTCCTGGAC TTCAAATTCT 540 ATATATTGAG AGAGTTGGAG AGTATATCAG AGATATTTTT GGAAAGGAGT TGGTCTATGC 600
AATGTCAGTT TGGAATCTTC TTGAAAGTTT AATGTTTTTA TTAGGAGATT TAAAGAAAAT 660
AAAGGTCTAC AATATCAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA 720
AAAAAAAAAA AA 732
(2) INFORMATION FOR SEQ ID NO: 67:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 629 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 67:
TTAAGGAATT CGGCMCGATC CCGGCAAGTA ACATGACTAA AAAGAAGCGG GAGAATCTGG 60
GCGTCGCTCT AGAGATCGAT GGGCTAGAGG AGAAGCTGTC CCAGTGTCGG AGAGACCTGG 120 AGGCCGTGAA CTCCAGACTC CACAGCCGGG AGCTGAGCCC AGAGGCCAGG AGGTCCCTGG 180
AGAAGGAGAA AAACAGCCTA ATGAACAAAG CCTCCAACTA CGAGAAGGAA CTGAAGTTTC 240
TTCGGCAAGA GAACCGGAAG AACATGCTGC TCTCTGTGGC CATCTTTATC CTCCTGACGC 300
TCGTCTATGC CTACTGGACC ATGTGAGCCT GGCACTTCCC CACAACCAGC ACAGGCTTCC 360
ACTTGGCCCC TTGGTCAGGA TCAAGCAGGC ACTTCAAGCC TCAATAGGAC CAAGGTGCTG 420 GGGTGTTCCC CTCCCAACCT AGTGTTCAAG CATGGCTTCC TGGCGGCCCA GGCCTTGCCT 480
CCCTGGCCTG CTGGGGGGTT CCGGGTCTCC AGAAGGACAT GGTGCTGGTC CCTCCCTTAG 540
CCCAAGGGAG AGGCAATAAA GAACACAAAG CTGAAAAAAA AAAAAAAAAA AACTCGTAGG 600
GGGGGCCCGT ACCCAATCGC CCTNTCGTG 629
(2) INFORMATION FOR SEQ ID NO: 68:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1751 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS : double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 68:
CTGCTAGCCG GCCGGCGCAG GCTGCCGAGC GGGTGAGCGC GCAGGCCAGG CCAAAGCCCT 60
GGTACCCGCG CGGTGCGGGC CTCAGTCTGC GGCCATGGGG GCGTCCGCGC GGCTGCTGCG 120
AGCGGTGATC ATGGGGGCCC CGGGCTCGGG CAAGGGCACC GTGTCGTCGC GCATCACTAC 180
ACACTTCGAG CTGAAGCACC TCTCCAGCGG GGACCTGCTC CGGGACAACA TGCTGCGGGG 240
CACAGAAATT GGCGTGTTAG CCAAGGCTTT CATTGACCAA GGGAAACTCA TCCCAGATGA 300
TGTCATGACT CGGCTGGCCC TTCATGAGCT GAAAAATCTC ACCCAGTATA GCTGGCTGTT 360
GGATGGTTTT CCAAGGACAC TTCCACAGGC AGAAGCCCTA GATAGAGCTT ATCAGATCGA 420
CACAGTGATT AACCTGAATG TGCCCTTTGA GGTCATTAAA CAACGCCTTA CTGCTCGCTG 480
GATTCATCCC GCCAGTGGCC GAGTCTATAA CATTGAATTC AACCCTCCCA AAACTGTGGG 540
CATTGATGAC CTGACTGGGG AGCCTCTCAT TCAGCGTGAG GATGATAAAC CAGAGACGGT 600
TATCAAGAGA CTAAAGGCTT ATGAAGACCA AACAAAGCCA GTCCTGGAAT ATTACCAGAA 660
AAAAGGGGTG CTGGAAACAT TCTCCGGAAC AGAAACCAAC AAGATTTGGC CCTATGTATA 720
TGCTTTCCTA CAAACTAAAG TTCCACAAAG AAGCCAGAAA GCTTCAGTTA CTCCATGAGG 780
AGAAATGTGT GTAACTATTA ATAGTAAGAT GGGCAAACCT CCTAGTCCTT GCATTTAGAA 840
GCrcCTTTTC CTAAGACTTC TAGTATGTAT GAATTCTTTG AAAATTATAT TACTTTTATT 900
TCTACTGATT TTATTTTGGA TACTAAGGAT GTGCCAAATG ATTCGGATAC TAAGATGCAT 960
CGTTTGAAAT CATCTAGTGT GTTGTATGCA GTTATCCTCA AAAACATCAG CGATGTCTGA 1020
ACCTTTAAAA CATCTGTTAG AGCAAAATTA AAAGAGCATT TGGTAGTAAT CTAACTTTTT 1080
GTTCAGTTAA TAAGTGGTTG ATAAAGTTTC CATATTTTTC TGGAAAAGTT AAAAAAAGTT 1140
ACATGTCATT TGGAGAAAAT ACGTAATCAG AAATTTGTGC ATAGATTGAT GCCAAAAAAG 1200
ACATTTCCAG CATTGTGGAA CATGGTGAGA CACTATATAA AATTCCAGAA AGAAAGCAAC 1260
TGGATTTACA GATTTATTGT GAGACACAAA TTCACTGCTG CCTTTACACT AAGAAATGTA 1320
TATGTTAACC ATATATGCTG TATTTATTTT GTCGTTAAGC ATACTTTCAG TTTACTCAGA 1380
ATTTTCAATT TGCTATAAAG ATGTATCAAT TAGCATATAG AAAAATATTA CTTTAAGATG 1440
ACTTGTTTCC TTTGAAAATA CCTGTGTACT GAGGGTTATG ATTTGTGTCA AAAATTGACA 1500
TAAGTGCTTT TACAAGCACC AAAGTTGAAT GAATTTTCAA CAAAATGTAA TTAAAGTCTA 1560
TGTTTTCAGT TATGACTCAG GTTAAGAAAT GTGTTTTAGG ATCTACTTGC TGGTTTTTCT 1620
TTTTGATCCA AATGTGTGAT CTGCCCTGAT AAATAACAAG TTATNGTACC ATCTCCCCCG 1680
CCAATAAAAA AAAAAAAAAA AAAAAAAAAC TCGAGGGGGG GCCCGGTACC CAATTCTCCG 1740
NAATAGGNAG T 1751
(2) INFORMATION FOR SEQ ID NO: 69:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 508 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO 69: GGCACGAGAT TATGTATTAA AATGTTTTTG AATTGTGAAA TATTAGAATA TTGTTACTAT 60 TTGACCCAAC TCAAAATCTC CATGGGAAAA TACCTGTCGA TACCCACAGT ATTGTTGAAA 120 ATAATCAGAT GCAGTATCAC AGCTGTGTCA GACTCTAGTA CCAGTTGGGC AATCAAGGCA 180 CAGCTAAAAA TTGAAAACAA AGATCTGGAC AACAAAACAG CCAAAGGTGG GGGTCAAGAA 240 GCTCTGACGT GTACCTAGCT GTAGAATGCT ATGCACACGT GCCAGGTGTA GTGTGCATAT 300 CCAGGAAAAA CTGCAGAGAG CCCCAGTCTT CACCTCTGGT TGACCATGAG CTCTGTGTAA 360 GCAGGAAGTG AAGGCTAAGG CAGATTTAAG CTCTGAAAGC ATTCCACAAC ATACACACAA 420 ATCGTGCAAA GCATTAAGGA AATCTTGTTA CTGCTAAGTG TTGCTGACCC AGGAACAACT 480 CCTACTCAGC TGGACTTAAA AATAAAAA 508
(2) INFORMATION FOR SEQ ID NO: 70:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 245 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 70: TACATAGAGC AAAGAGAAAT TTCCAGAATT TCTARAATTC TGGAAAGAGA ATTTTCCTGA 60 GATTGCAGAT TTGCTTGTGT CCTCAGGTGA TGATGAGGGC TGTTTTCCCC TGTTGTCCTT 120 TCCTCACACT CATGCTTCCT CTCCTAGAGT GTCTGGTTGG CATGATCATG TGCTACCTAG 180
GCATTTCTTT CACTGATACA AGGAAAACTG CAGGGTTAAA AAAAAAAAAA AAAAAAAAAA 240 NCNCG 245
(2) INFORMATION FOR SEQ ID NO: 71: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 361 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 71:
ATGTTCCTCA TGAGGATGCA CTTGTGCTTC TGCAAGTATT GCTGCAGCTT CATAGTGACT 60 CCCACCAGCA CCAGCAATAC AGCTAGCTAC CTGTGGCCTT GGATCTCAGC CAGCATGGCT 120
GGGAGAGGGA GCAGCTGGGC ATGTACCCTA AATGCTGTTA CCAGGGAAGG ACTCCCAGAG 180
TGAAGACAAG TAGGGACTTC CTGCAGAGGT GGTACATGTG CTCTCTGTAT CCATACTTTT 240
TTTTTTTTTT TTTTGAGATA GAGTTTCACC CTTGTTGCCC TGGCTGGAGT GCAATGGTGC 300
GATCTCAGCT CACTGCAACC TCTCTGCCTC CCGGGTTCAA GTGATTCTCC TGCCTCAGCC 360 T 361
(2) INFORMATION FOR SEQ ID NO: 72:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 713 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 72: AGGATCACAC AATAGAGAAC ACTGTAGTAA CATTTCGGTC TGCTCACAAG ACCCAGAACA 60
TTGATCAGTT TTTGTTGTTG GTTTATTATT TTTCTGTTAA AAAATTGTGA AAAGTTTGTT 120
TTAGCTAGAT GATATTTTAA TAGCTGCGAG TGCTTTGGAA CTATAAAGAT GTCACTACTT 180
AACACACATA CCTTATGTTT TGTTTTGTTT TGTTTTACAC TCAGTATAAA TCAGGAGAAG 240
TTAGCCAACC ATCTAGCATT TAGAATCCTC TTTTTTATTG TCTTCTAAGG ATATGGATGT 300 TCCCATAACA GCAACAAAAC AGCAACAAAA ACATTTCATA AATATCACTT GATAGACTGT 360
AAGCACCTGC TTAACTTTGT GTCCCAAATA TTTAGTGTGT ATATATATAT ATATATATAC 420
ACACACACAC ACATATATAT TCAACAAATA AAGCAAAATA TAACATGCAT TTCACATTTT 480
GTCTTTCCCT GTTACGATTT TAATAGCAGA ACTGTATGAC AAGTTTAGGT GATCCTAGCA 540
TATGTTAAAT TCAAATTAAT GTAAAACAGA TTAACAACAA CAAAGAAACT GTCTATTTGA 600 GTGAAGTCAT GCTTTCTATT ATAATAACTT GGCTTCGGTT ATCCATCAAA TGCACACTTA 660
TACTGTTATC TGATTGTTTA TAATAAAGAA TACTGTACTT ATAAAAAAAA AAA 713
(2) INFORMATION FOR SEQ ID NO: 73:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 862 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 73:
GAAAGTCAGA GCTGTCCAAT CCCTCAGCAC CTTTTAGATT TGCTCCAAAT TAGAAACGTG 60 GGGACTATGT GTTCTGGGCA ATCACAGGTC TGGAAAATGG CTCTGCAGGC TCTTGATAGT 120
GAGACAGTGG TCATCTTACC AGACATGCAT CTGATTTTAA GCCTCAGGCT AATCCACAAT 180
GCTCGGCCAT GCCTATGATT AACAAACAAA AGCAAAATCT GCTTTTATAG TTTAGGAAAC 240 CTGGATAGAA CAGTATTTTT CAGCATTCTT GGATAAAGCA GTTCTGCATT TTTAAATTGG 300
GACTGCAGAA GTGACTGTCT ATAGTTGTGA AATACAAAAA ATGGTATGTT TGATCAGAAA 360
AGGAAGCCCG TGCCTGGCAC TTGGAAAGAT ACTGAGCATC ATAACCCTAA TGAGAAAATG 420
TAGGCTCTGT GAATGTTAAC TACAAATCAG GTTAGGAAAG CATATGACAC CCTTTGTCAA 480
ACTAAGCTTC ACTAGGAGGA CCTGTGCTCA TAGAAGAATA TGCTTTAAAA GTATCAATTT 540 TCCACAGTCG ATGATGGAGA AAAGTTCATT TGCACCAGAA TGCTGATAGT CACAATACAC 600
AGCCTGACAT ATATAACAAT ACAGTTTTCT GTAAACAGAA GTTCTTCCTC TTCCAATTCA 660
GGAGTCAGTC AGAGCATAAA TATTGCATGT TTCACTTTAG AAACTGATTC ATTTTAGAAA 720
GCAGATCTGG ATTATTTTGC AGGGTAGAAA TGAAGGCTAT TTCTGGCATT CTTGCTCAAA 780
AAGTCAATAT ATGTACATTA AGTATAAAAA AGGGTCTCTT TCACCTCTTT TGTTTCGTAG 840 CATTGGCTAC ATAACTCGTG CC 862
(2) INFORMATION FOR SEQ ID NO: 74:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 4602 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 74:
GCGAGGGGGC GKGGGGAGCA GCGCCGARGC CGCCGCCTCC GCCTCCGCCG CCTAGGACTA 60
GGGGGTGGGG GACGGACAAG CCCCGATGCC GGGGGAKACG GAAGAGCCGA GACCCCCGGA 120
GCAGCAGGAC CAGGAAGGGG GAGAGGCGGC CAAGGCGGCT CCGGAGGACC CGCAACAACG 180
GCCCCCTGAG GCGGTCGCGG CGGCGCCTGC AGGGACCACT AGCAGCCGCG TGCTGAGGGG 240
AGGTCGGGAC CGAGGCCGGG CCGCTGCGRC CGCCGCGCMG CAGCTGTGTC CCGCCGGAGA 300
AGGCCGAGTA TCCCCGCCGG CGAGGAGCAG CCCCAGCGCC AGGCCTCCCG ACGTCCCCGG 360
GCAGCAGCCC AGGCCGCGAA GTCCCCGTCT CCAGTTCAGG GCAAGAAGAG TCCGCGACTC 420
CTATGCATAG AAAAAGTAAC AACTGATAAA GATCCCAAGG AAGAAAAAGA GGAAGAAGAC 480
GATTCTGCCC TCCCTCAGGA AGTTTCCATT GCTGCATCTA GACCTAGCCG GGGCTGGCGT 540
AGTAGTAGGA (^TCTGTTTC TCGCCATCGT GATACAGAGA ACACCCGAAG CTCTCGGTCC 600
AAGACCGGTT CATTGCAGCT CATTTGCAAG TCAGAACCAA ATACAGACCA ACTTGATTAT 660
GATGTTGGAG AAGAGCATCA GTCTCCAGGT GGCATTAGTA GTGAAGAGGA AGAGGAGGAG 720
GAAGAAGAGA TGTTAATCAG TGAAGAGGAG ATACCATTCA AAGATGATCC AAGAGATGAG 780
ACCTACAAAC CCCACTTAGA AAGGGAAACC CCAAAGCCAC GGAGAAAATC AGGGAAGGTA 840
AAAGAAGAGA AGGAGAAGAA GGAAATTAAA GTGGAAGTAG AGGTGGAGGT GAAAGAAGAG 900
GAGAATGAAA TTAGAGAGGA TGAGGAACCT CCAAGGAAGA GAGGAAGAAG ACGAAAAGAT 960
GACAAAAGTC CACGTTTACC CAAAAGGAGA AAAAAGCCTC CAATCCAGTA TGTCCGTTGT 1020
GAGATGGAAG GATGTGGAAC TGTCCTTGCC CATCCTCGCT ATTTGCAGCA CCACATTAAA 1080
TACCAGCATT TGCTGAAGAA GAAATATGTA TGTCCCCATC CCTCCTGTGG ACGACTCTTC 1140
AGGCTTCAGA AGCAACTTCT GCGACATGCC AAACATCATA CAGATCAAAG GGATTATATC 1200
TGTGAATATT GTGCTCGGGC CTTCAAGAGT TCCCACAATC TGGCAGTGCA CCGGATGATT 1260
CACACTGGCG AGAAGCATTA CAATGTGAGA TCTGTGGATT TACTTGTCGA CAAAAGGCAT 1320
CTCTTAATTG GCACATGAAG AAACATGATG CAGACTCCTT CTACCAGTTT TCTTGCAATA 1380
TCTGTGGCAA AAAATTTGAG AAGAAGGACA GCGTAGTGGC ACACAAGGCA AAAAGCCACC 1440
CTGAGGTGCT GATTGCAGAA GCTCTGGCTG CCAATGCAGG CGCCCTCATC ACCAGCACAG 1500
ATATCTTGGG CACTAACCCA GAGTCCCTGA CGCAGCCTTC AGATGGTCAG GGTCTTCCTC 1560
TTCTTCCTGA GCCCTTGGGA AACTCAACCT CTGGAGAGTG CCTACTGTTA GAAGCTGAAG 1620
GGATGTCAAA GTCATACTGC AGTGGGACGG AACGGGTGAG CCTGATGGCT GATGGGAAGA 1680
TCTTTGTGGG AAGCGGCAGC AGTGGAGGCA CTGAAGGGCT GGTTATGAAC TCAGATATAC 1740
TCGGTGCTAC CACAGAGGTT CTGATTGAAG ATTCAGACTC TGCCGGACCT TAGTGGACAG 1800
GAAGACTTGG GGCATGGGAC AGCTCAGACT TTGTATTTAA AAGTTAAAAA GGACAAAAAA 1860
AAAATCTAAA GCATTTAAAA TCTAGTGAAA TAACTGAAGG GCCTGCTCTT TCCATTGTGG 1920
ATCACAGCAC ACACATACAT ACACCCTCCA CCTCCCCATC CCCTGTTCTC CCrCTGTTGC 1980
TCCCCTTATA AAATTGATGT TGTCTTTACC AGAAAGGTAG ACAAAAAAGA AGCAGCAGCA 2040
GCTCTTAAAG TGAGGGTTAT TCTCATACTC GGTTCCAGCC ATCAGCAGAC TTCCTGCTCA 2100
TCGGCAGATC CCCCTTTCCA ACCTGTAACT CTGATGTGCT CTGGATCAGC TTTTAACTTT 2160
TAATCATATA TTACTGTCTT CTAAATCCCT TCTCCTCCTC TACTGCTGCC CTATGGTTCT 2220
GGCTCCTACC CCCTGCGGCA CACTTATCTT CAAATACCAT AGAATTCTAA TCTCTGAAAT 2280
CATAGCTCTC CAGTGGCTTT TAAAGAAAGC TGGTCCTCAG CACTAACAAA ATCACTACAA 2340
TAGCCTAGTG CTTTTTTGGA AGCCTTTTTA GGGAAGAATG TTAGGTTCAT GGTAACTAGT 2400
ATGCTCTTTG AGATTTTTAC AGTGTTGAAA CTTAAGAATT TTGAGAGGGT GAGGAGGGTT 2460
GTTCAGAATC TAAATTACAG ATAGATGATT GTTTCTTGTG AATTTGTTTC TTTTCCTTTT 2520
TTTTTGTCCC TACCATTTCC TTACATTTCC CTTGGGGCCC ATCTCTGGCT CCTTGCTTTT 2580
TGTTTCTTGC TTTGCTTTAT CAGTTCATTC CAGCTCCCTG TTAGTGAAGG ACACTGCTGT 2640
TAGTGAAGGA ACAAAGTCTA TGAGTCCTAA AATTTTAAGT CAAAGAAAAC TGCTCTGTTT 2700
CCCCTTTAGT AACACTTCTG AAGAGGAAAA ACTTCAATAG CCAAAGTTAA TAATCCTATA 2760
TAATAATTGC TTTGGCTTTC ACCTAAAATT CTGGGCATCA CAATTTCCTT GGGATAGAGG 2820
TTGTGTTGGG GAATAGATTG CTTATTGCTG TTCACTGGAG AGAAAAGGTA GTGTTTTTGT 2880
ACAAGGTCAT ACCGCCAGAA GCCCCAAATC CTATTTTGGC TCATCTTCAG GTAAAGAGTA 2940
ATTCCTATCC TGTGTGCCTC AGAAGCTAGA ATCGAAGGCT TACCCTATTC ATTGTTTATT 3000
GTCAGAAATG CATGATGGCT CTTGGAAAGA ATGACGTTTT GCTGGAAAAA AAAAAAARAA 3060
CMGTTTGTGT TTCACAAACA TGGCTTATCA ATTTTTTCAA AGAATTCTTT TTTCCCAAAA 3120
AGAGGAGTAA CAAAATGTCA TTTCTGAAAG AGGCTTACTT TATACCAACT AGTGTCAGCA 3180
TTTGGGATGC CAGGGAACAG AGAGTGAGAC ACCTACAATC ACCAGTCTCA AATGCGCTAT 3240
TGTTTCTTTT CAGAGTGTTG CAGATTTGCC ATTTCTCCAT AATATGGGGA TAGAAAATGG 3300
AATAAAGATA GAAGGGATGT AGAATATGCT TTCCTGCCAA CATCrøTTTGG AGTCGACTTT 3360
GGTATATTGA CTAGATTTGA AAATACAAGA TTGATTAGAT GAATCTACAA AAAAGTTGTC 3420
CTCCTCTCAG GTCCCTTTTA CACTTTTTGA CTAACTAGCA TCTATATTCC ACACTTAGCT 3480
TTTTTGTCAC ACTTATCCTT TGTCTCCGTA AATTTCATTT GCAGTGGTTA GTCATCAGAT 3540
ATTTTAGCCA CCTACACAAA AGCAAACTGC ATTTTTAAAA ATCTTTCTGA GATGGGAGAA 3600
AATGTATTCT CCTTTCCTAT ACCGCTCTCC CAACAAAAAA ACAACTAGTT AGTTCTACTA 3660
ATTAGAAACT TGCTGTACTT TTTCTTTTCT TTTAGGGGTC AAGGACCCTC TTTATAGCTA 3720
CCATTTGCCT ACAATAAATT ATTGCAGCAG TTTGCAATAC TAAAATATTT TTTATAGACT 3780
TTATATTTTT CCTTTTGATA AAGGGATGCT GCATAGTAGA GTTGGTGTAA TTAAACTATC 3840
TCAGCCGTTT CCCTGCTTTC CCTTCTGCTC CATATGCCTC ATTGTCCTTC CAGGGAGCTC 3900
TTTTAATCTT AAAGTTCTAC ATTTCATGCT CTTAGTCAAA TTCTGTTACC TTTTTAATAA 3960
CTCTTCCCAC TGCATATTTC CATCTTGAAT TGGTGGTTCT AAATTCTGAA ACTGTAGTTG 4020
AGATACAGCT ATTTAATATT TCTGGGAGAT GTGCATCCCT CTTCTTTGTG GTTGCCCAAG 4080
GTTGTTTTGC GTAACTGAGA CTCCTTGATA TGCTTCAGAG AATTTAGGCA AACACTGGCC 4140
ATGGCCGTGG GAGTACTGGG AGTAAAATAA AAATATCGAG GTATAGACTA GCATCCACAT 4200
AGAGCACTTG AACCTCCTTT GTACCTGTTT GGGGAAAAAG TATAATGAGT GTACTACCAA 4260
TCTAACTAAG ATTATTATAG TCTGGTTGTT TGAAATACCA TTTTTTTCTC CTTTTGTGTT 4320
TTTCCCACTT TCCAATGTAC TCAAGAAAAT TGAACAAATG TAATGGATCA ATTTAAAATA 4380
TTTTATTTCT TAAAAGCCTT TTTTGCCTGT TGTAATGTGC AGGACCCTTC TCCTTTCATG 4440
GGAGAGACAG GTAGTTACCT GAATATAGGT TGAAAAGGTT ATGTAAAAAG AAATTATAAT 4500
AAAAGGGATA CTTTGCTTTT CAAATCTTTG TTTTCTCTTA TTCTAGGTAA GGCATATTAA 4560
AAATAAATAT GTAAAGAAGA AAAATAAAAG TTGTCTTCAT GG 4602
(2) INFORMATION FOR SEQ ID NO: 75:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1255 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 75: CGCGCCCCGG GCCGGCGGGT TTCTCTAACA AATAAACAGA ACCCGCACTG CCCAGGCGAG 60 CGTTGCCACT TTCAAAGTGG TCCCCTGGGG GAGCTCAGCC TCATCCTGAT GATGCTGCCA 120 AGGCGCACTT TTTATTTTTA TTTTATTTTT ATTTTTTTTT TAGCATCCTT TTGGGGCTTC 180 ACTCTCAGAG CCAGTTTTTA AGGGACACCA GAGCCGCAGC CTGCTCTGAT TCTATGGCTT 240 GGTTGTTACT ATAAGAGTAA TTGCCTAACT TGATTTTTCA TCTCTTTAAC CAAACTTGTG 300
GCCAAAAGAT ATTTGACCGT TTCCAAAATT CAGATTCTGC CTCTGCGGAT AAATATTTGC 360 CACGAATGAG TAACTCCTGT CACCACTCTG AAGGTCCAGA CAGAAGGTTT TGACACATTC 420 TTAGCACTGA ACTCCTCTGT GATCTAGGAT GATCTGTTCC CCCTCTGGAT GAACATCCTC 480 TGATGATCAA GGCTCCCAGC AGGCTACTTT GAAGGGAACA ATCAGATGCA AAAGCTCTTG 540 GGTGTTTATT TAAAATACTA GTGTCACTTT CTGAGTACCC GCCGCTTCAC AGGCTGAGTC 600 CAGGCCTGTG TGCTTTGTAG AGCCAGCTGC TTGCTCACAG CCACATTTCC ATTTGCATCA 660 TTACTGCCTT CACCTGCATA GTCACTCTTT TGATGCTGGG GAACCAAAAT GGTGATGATA 720 TATAGACTTT ATGTATAGCC ACAGTTCATC CCCAACCCTA GTCTTCGAAA TGTTAATATT 780 TGATAAATCT AGAAAATGCA TTCATACAAT TACAGAATTC AAATATTGCA AAAGGATGTG 840 TGTCTTTCTC CCCGAGCTCC CCTGTTCCCC TTCATTGAAA ACCACCACGG TGCCATCTCT 900 TGTGTATGCA GGGCTATGCA CCTGCAGGCA CGTGTGTATG CACTCCCCGC TTGTGTTTAC 960 ACAAGCTGTG GGGTGTTACG CATGCCTGCT TTTTTCACTT AATAATACAG CTTGGAGAGA 1020 TTTTTGTATC ACATTATAAA TCCCACTCGC TCTTTTTGAT GGCCACATAA TAACTACTGC 1080 ATAATATGGA TACGCCTTAT TTGATTTAAC TAGTTCCCTA ATGATGGACT TTTAAGTTGT 1140 TTCCTTTTTT TTTCTTTTTT GCTACTGCAA ACGATGCTAT AATAAATGTC CTTATCAAAA 1200 AAAAAAAAAA AAAAAAAAAA AAAAAANCCC NGGGGGGGGG CCCCGGGAAC NCAAT 1255
(2) INFORMATION FOR SEQ ID NO: 76:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 475 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO : 76: GGCACGAGAG AAATGTTTGA TTCTCTTTCC TATTTTAAGG GATCTTCTCT CTTGTTGATG 60 TTGAAAACTT ACCTTAGTGA AGATGTGTTT CAACATGCTG TTGTCCTTTA CCTGCATAAT 120 CACAGCTATG CATCTATTCA AAGTGATGAT CTGTGGGATA GTTTTAATGA GGTCACAAAC 180 CAAACACTAG ATGTAAAGAG AATGATGAAA ACCTGGACCC TGCAGAAAGG ATTTCCTTTA 240 GTGACTGTTC AAAAGAAAGG AAAGGAACTT TTTATACAAC AAGAGAGATT CTTTTTAAAT 300 ATGAAGCCTG AAATTCAGCC TTCAGATACA AGGTACATGC CCTCTTTCTT TTCATGCCAT 360 CTCTTTTGCA CTCTCAGGTG GAAATATTTT GAAGTGTTTT ATAATCATAA GTTCTTGTGA 420
AACCTAACAA GATTATCCCT TCCTAAGAAT ACTTAACCTT CCTACCAAAT TAAAA 475
(2) INFORMATION FOR SEQ ID NO: 77:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 465 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 77:
TTCTCTCTGC TCTTCGACTG CACCGCACTC GCGCGTGACC CTGACTCCCC CTAGTCAGCT 60
CAGCGGTGCT GCCATGGCGT GGCGGCGGCG CGAACCRGCG TCGGGGCTCG CGGCGTGTTG 120 GCTCTGGCGT TGCTCGCCCT GGCCCTGTGC GTGCCCGGGG CCCGGGGCCG GGCTCTCGAG 180
TGGTTCTCGG CCGTGGTAAA CATCGAGTAC GTGGACCCGC AGACCAACCT GACGGTGTGG 240
AGCGTCTCGG AGAGTGGCCG CTTCGGCGAC AGCTCGCCCA AGGAGGGCGC GCATGGCCTG 300
GTGGGCGTCC CGTGGGCGCC CGGCGGAGAM CTCGARGGCT KCGCGCCCGA CACGCGCTTC 360
TTCGTGCCCG AGCCCGGCGG CCGAGGGGCC GCGCCCTGGG TCGCCCTGGT GGTCGTGGGG 420 GCTGCACCTT TCAAGGACAA AGTGCTGGTG GCGGCGCNGA ANGAA 465
(2) INFORMATION FOR SEQ ID NO: 78:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1907 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 78: ACATGCAGCC CAACTACAGA TTCTTATGGA ATTCCTCAAG GTTGCAAGAA GAAATAAGAG 60
AGAGCAACTG GAACAGATCC AGAAGGAGCT AAGTGTTTTG GAAGAGGATA TTAAGAGAGT 120
GGAAGAAATG AGTGGCTTAT ACTCTCCTGT CAGTGAGGAT AGCACAGTGC CTCAATTTGA 180
AGCTCCTTCT CCATCACACA GTAGTATTAT TGATTCCACA GAATACAGCC AACCTCCAGG 240
TTTCAGTGGC AGTTCTCAGA CAAAGAAACA GCCTTGGTAT AATAGCACGT TAGCATCAAG 300 ACGAAAACGA CTTACTGCTC ATTTTGAAGA CTTGGAGCAG TGTTACTTTT CTACAAGGAT 360
GTCTCGTATC TCAGATGACA GTCGAACTGC AAGCCAGTTG GATGAATTTC AGGAATGCTT 420
GTCCAAGTTT ACTCGATATA ATTCAGTACG ACCTTTAGCC ACATTGTCAT ATGCTAGTGA 480
TCTCTATAAT GGTTCCAGTA TAGTCTCTAG TATTGAATTT GACCGGGATT GTGACTATTT 540
TGCGATTGCT GGAGTTACAA AGAAGATTAA AGTCTATGAA TATGACACTG TCATCCAGGA 600
TGCAGTGGAT ATTCATTACC CTGAGAATGA AATGACCTGC AATTCGAAAA TCAGCTGTAT 660
CAGTTGGAGT AGTTACCATA AGAACCTGTT AGCTAGCAGT GATTATGAAG GCACTGTTAT 720
TTTATGGGAT GGATTCACAG GACAGAGGTC AAAGGTCTAT CAGGAGCATG AGAAGAGGTG 780
TTGGAGTGTT GACTTTAATT TGATGGATCC TAAACTCTTG GCTTCAGGTT CTGATGATGC 840
AAAAGTGAAG CTGTGGTCTA CCAATCTAGA CAACTCAGTG GCAAGCATTG AGGCAAAGGC 900
TAATGTGTGC TGTGTTAAAT TCAGCCCCTC TTCCAGATAC CATTTGGCTT TCGGCTGTGC 960
AGATCACTGT GTCCACTACT ATGATCTTCG TAACACTAAA CAGCCAATCA TGGTATTCAA 1020
AGGACACCGT AAAGCAGTCT CTTATGCAAA GTTTGTGAGT GGTGAGGAAA TTCTCTCTGC 1080
CTCAACAGAC AGTCAGCTAA AACTGTGGAA TGTAGGGAAA CCATACTGCC TACGTTCCTT 1140
CAAGGGTCAT ATCAATGAAA AAAACTTTGT AGGCCTGGCT TCCAATGGAG ATTATATAGC 1200
TTGTGGAAGT GAAAATAACT CTCTCTACCT GTACTATAAA GGACTTTCTA AGACTTTGCT 1260
AACTTTTAAG TTTGATACAG TCAAAAGTGT TCTCGACAAA GACCGAAAAG AAGATGATAC 1320
AAATGAATTT GTTAGTGCTG TGTGCTGGAG GGCACTACCA GATGGGGAGT CCAATGTGCT 1380
GATTGCTGCT AACAGTCAGG GTACAATTAA GGTGCTAGAA TTGGTATGAA GGGTTAACTC 1440
AAGTCAAATT GTACTTGATC CTGCTGAAAT ACATCTGCAG CTGACAATGA GAGAAGAAAC 1500
AGAAAATGTC ATGTGATGTC TCTCCCCAAA GTCATCATGG GTTTTGGATT TGTTTTGAAT 1560
ATTTTTTTCT TTTTTTCTTT TCCCTCCTTT ATGACCTTTG GGACATTGGG AATACCCAGC 1620
CAACTCTCCA CCATCAATGT AACTCCATGG ACATTGCTGC TCTTGGTGGT GTTATCTAAT 1680
TTTTGTGATA GGGAAACAAA TTCTTTTGAA TAAAAATAAA TAACAAAACA ATAAAAGTTT 1740
ATTGAGCCAC AGTTGAGCTT GGAAAGTTTT TGTCAAATGC NGCAAGAGAT AACTCTTTTT 1800
ANGAAGTAGC ATATGTGAAC TATAATGTAA CAGTGAATAA TTTGTAAAGT TCGTATTTCC 1860
CAACCTCTTT GGGAATTACA CATATCAATA TAAACAAAAT ATAAAGT 1907
(2) INFORMATION FOR SEQ ID NO: 79:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 1168 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 79:
GCTGGGGTGT CCCCKCSGCC ACCATCGTCA TCGCTTACTT GATGAAGCAC ACTCGGATGA 60 CCCATGACTG ATGCTTATAA ATTTGTCAAA GGCAAACGAC CAATTATCTC CCCAAACCTT 120 AACTTCATGG GGCAGTTGCT AGAGTTCGAG GAAGACCTAA ACAACGGTGT GACACCGAGA 180 ATCCTTACAC CAAAGCTGAT GGGCGTGGAG ACGGTTCTGT GACAATGGTC TGGATGGAAA 240 GGATTGCTGC TCTCCATTAG GAGACAATGA GGAAGGAGGA TGGATTCTGG TTTTTTTTCT 300 TTCTTTTTTT TTTTGTAGTT GGGAGTAAGT TTGTGAATGG AAACAAACTT GTTTAAACAC 360 TTTATTTTTA ACAAGTGTAA GAAGACTATA ACTTTTGATG CCATTGAGAT TCACCTCCCA 420 CAAACTGACA AATTAAGGAG GTTAAAGAAG TAATTTTTTT AAGCCAACAA TAAAAATATA 480 ATACAACTTG TTTCTCCCCC TTTTCCTTTT AAGCTATTTG TAGAGTTTAT GACTAAATAG 540 TCTGTGCAGG TTCATAGACC GAAGATACTA CACACTTTAA ACCAATTAAA AAGAACCAAA 600 AGTAAATAGA AAAGACATTG AATCACCAAG GCCTGGGATC AACCTGGGCT GTCCACACAG 660 AAAACAAAAA CCCAACCAAA CCAAGCCCTG TTGTGCTCAC TGGTGCAAAG AGAAGATCAG 720 GGCAGCTTAA GTGGTCTAAG RATCCTTCAG GCATTCTTTA AGGAGAAAAA GGATACCTTT 780 GATTTTGTGT GTTTCATGCT CTGGATTTTT TTTTTTTTTC CTTCTCTGGG TTTAAGAGAT 840 TTTTTTTGAA ATAGTGAGGA ACTGACCATT ATATGCCTTC ACTGGCTTCT TGTGCAATAA 900 TATGATGTTT TAAGTGTGCA AACAAGTTAG AGCTGGCAGC TGAATGATAG ACAAATAGTG 960 CAAATTTGCC AGCTTGGAGA TAGAAAGGAA TTCAACAATA TATCAAATAC TTTCCTTCCC 1020 ACCTTTTTCC TTTTTTTTTT TTTTTTCTGA TTTGATTCTG GTTACAGTGC CATAAACCTT 1080 GTTACATATG TATATCAGAA TGTAAGAAAA AAAAATTTAT TTAAAAATAT TTTTCGCAAA 1140 AAAAAAANNA AAAAACTCGA GGGGGGCC 1168
(2) INFORMATION FOR SEQ ID NO: 80:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1285 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 80: AGAAAATCAC ATCCTAACAA AGAAGTCTGT CTAAGACAGT ACATCTCCTG TTGAACTTGC 60 ATCTTTCCAC AGGACTTTCT GTTTTTAGGG ATGAGACTAT TCTCTGCTTC ATCAAGGAAA 120 GAGAAATGTT CAGGGTTGTA GGGATGGCAC ACTTATTAGT TCTGCCTGTC TGAAAGGTTC 180
CTGCAGGACA GTTTGGTCAG AGCTGCAATT CTTAGTCCAT GGTCTAATGC TTGAGTATCT 240
CTTCTTTCCC TTTCCTGTCT CAGGAATCAG CTGAGAATTC ATTCGATTGT CATGCCTCTA 300
GCCCCTTACT GTGATTTGTT GGTTGCACTT TCATTTGCTT TAGTTCTAGA ATCACCTGTT 360
GACTCCTCAG ACTTCACCTA ACTTTGGAAA CTCTCTTTTG GAGGCTTCTC ATTTCCCCCT 420
AATTCTCTGC TGCCTGAGCC CTAGAATTTT CCCACCAACG AATTATTCCA GGTAGATCCT 480
AAGTTGCTGG ATCTAGTTGA TATTTAAACA ATATCTAGTT GATATTTCTC ATTCAGTTGG 540
ATCCAGAAAC CAGTATCTCT NAAAAACAAC CTCTCATACC TTGTGGACCT AATTTTGTGT 600
GCGTGTGTGT GTGCGCGCAT ATGTATATAG ACAGGCACAT CTTTTTTACT TTTGTAAAAG 660
CTTATGCCTC TTTGGTATCT ATATCTGTGA AAGTTTTAAT GATCTGCCAT AATGTCTTGG 720
GGACCTTTGT CTTCTGTGTA AATGGTACTA GAGAAAACAC CTATATTATG AGTCAATCTA 780
GTTGGTTTTA TTCGACATGA AGGAAATTTC CAGATAACAA CACTAACAAA CTCTCCCTTG 840
ACTAGGGGGA CAAAGAAAAG CAAAACTGAC CATAAAAAAC AATTACCTGG TGAGAAGTTG 900
CATAAACAGA ATTAGGTAGT ATATTGAAGA CAGCATCATT AAACAGTTAT GTTGTTCTCC 960
TTGCAAAAAA CATGTACTGA CTTCCCGTTG AGTAATGCCA AGTTGTTTTT TTTATTATAA 1020
AACTTGCCCT TCATTACATG TTTCAAAGTG GTGTGGTGGG CCAAAATATT GAAATGATGG 1080
AACTGACTGA TAAAGCTGTA CAAATAAGCA GTGTGCCTAA CAAGCAACAC AGTAATGTTG 1140
ACATGCTTAA TTCACAAATG CTAATTTCAT TATAAATTGT TTTGCTAAAA TACACTTTGA 1200
AACTATTTTT CTGTATTCCA AGAGCTGAGA TCTTAGATTT TATGTAGTAT TAAGTGAAAA 1260
AATACGAAAA TAATAAACAT TGAAG 1285
(2) INFORMATION FOR SEQ ID NO: 81:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1290 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 81: TCTCCAGCCC CAATTTCTAC GCGCACCGGA AGACGGAGGT CCTCTTTCCT TGCCTAACGC 60 AGCCATGGCT CGTGGTCCCA AGAAGCATCT GAAGCGGGTG GCAGCTCCAA AGCATTGGAT 120 GCTGGATAAA TTGACCGGTG TGTTTGCTCC TCGTCCATCC ACCGGTCCCC ACAAGTTGAG 180 AGAGTGTCTC CCCCTCATCA TTTTCCTGAG GAACAGACTT AAGTATGCCC TGACAGGAGA 240 TGAAGTAAAG AAGATTTGCA TGCAGCGGTT CATTAAAATC GATGGCAAGG TCCGAACTGA 300
TATAACCTAC CCTGCTGGAT TCATGGATGT CATCAGCATT GACAAGACGG GAGAGAATTT 360
CCGTCTGATC TATGACACCA AGGGTCGCTT TGCTGTACAT CGTATTACAC CTGAGGAGGC 420
CAAGTACAAG TTGTGCAAAG TGAGAAAGAT CTTTGTGGGC ACAAAAGGAA TCCCTCATCT 480
GGTGACTCAT GATGCCCGCA CCATCCGCTA CCCCGATCCC CTCATCAAGG TGAATGATAC 540
CATTCAGATT GATTTAGAGA CTGGCAAGAT TACTGATTTC ATCAAGTTCC ATTCACCCAG 600
CCAGGTGGTC TCGTCACCTC AGAGGCTCCG CAGACTCCTG CCCAGGCCAG GACTGAGGCA 660
AGCCTCAAGG CACTTCTAGG ACCTGCCTCT TCTCACCAAG ATGAACTCAC TGGTTTCTTG 720
GCAGCTACTG CTTTTCCTCT GTGCCACCCA CTTTGGGGAG CCATTAGAAA AGGTGGCCTC 780
TGTGGGGAAT TCTAGACCCA CAGGCCAGCA GCTAGAATCC CTGGGCCTCC TGGCCCCSGG 840
GGAGCAGAGC CTGCCGTGCA CCGAGAGGAA GCCAGCTGCT ACTGCCAGGC TGAGCCGTCG 900
GGGGACCTCG CTGTCCCCGC CCCCCGAGAG CTCCGGGAGC CCCCAGCAGC CGGGCCTGTC 960
CGCCCCCCAC AGCCGCCAGA TCCCCGCACC CCAGGGCGCG GTGCTGGTGC AGCGGGAGAA 1020
GGACCTGCCG AACTACAACT GGAACTCCTT CGGCCTGCGC TTCGGCAAGC GGGAGGCGGC 1080
ACCAGGGAAC CACGGCAGAA GCGCTGGGCG GGGCTGAGGG CGCAGGTGCG GGGCAGTGAA 1140
CTTCAGACCC CAAAGGAGTC AGAGCATGCG GGGCGGGGGC GGGGGGCGGG GACGTAGGGC 1200
TAAGGGAGGG GGCGCTGGAG CTTCCAACCC GAGGCAATAA AAGAAATGTT GCGTAACTCA 1260
AAAAAAAAAA AAAAAAAANC TCGGGGGGGG 1290
(2) INFORMATION FOR SEQ ID NO: 82:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 684 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO : 82: TTTATTGTAT TCTGTAACTA TAGAACTTCT ATTTWATTCT TTTTTGGACT TGCTAAGTTG 60 TCTTTWATGG TTTTWAGTTC CATGCTGAAG TTTTCAGTAT TGACTTATCC CCTTGAACAT 120 GAGTTGTTTT ATAGACTCTR ATGATTCAAA AATCTTACAT CTTTTGGTAG TCTCTTTCAT 180 TTGTYCACTG TTTCTGTTGA TTCTWACTCA TGGTATTTTA ATTCTTCGTT WTTTTTTTTC 240 TGTTWAGAWA CATTCTTTGA AAAATAATTT GGAGGAATAT TTGATTCTTA TGAACAAGGC 300 ATTACTCACC AGAGAAGATT TTTTTGTTYT ACCARGTGCC TARGAATGCT AACAGTCTGG 360
GAMCACATAG AMCACCAGGT GATGAGACAA TCCTGGGART CCTGTTTTAC TTTGGSCCAT 420
CTTTTCTCCC AACCCTGTGG GAATARTCAT YCATATCCTA RCTGCAGGCT ARAAGGTGGT 480
TTATCAGAGC CCAACTTCGA GGGCTCTGGG CTTTAGCTAC TGTCACCCCA TCATAACTGA 540
GCTTCATGGA TTGATTCTCT TTTTATCTTT CAGATTTTCT TTTAAAAATC TTTGTTTTTT 600
TTTTTCTTCC GAAAGATTCC CCCAACATTA CCATTCCCCA CCTTCCGTTG AATTTTTTTG 660
GCTCTCATTT TGAATTTTTC AAGA 684
(2) INFORMATION FOR SEQ ID NO: 83:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2024 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 83:
CTGCAGGAAT TCGGCACAGC TGCGCTGGAG GCTTCATCTT TGCCGCCGCT GCCGTCGCCT 60
TCCTGGGATT GGAGTCTCGA GCTTTCTTCG TTCGTTCGYC GGCGGGTTCG CGCCCTTCTC 120 GCGCCTCGGG GCTGCGAGGC TGGGGAAGGG GTTGGAGGGG GCTGTTGATC GCCGCGTTTA 180
AGTTGCGCTC GGGGCGGCCA TGTCGGCCGG CGAGGTCGAG CGCCTAGTGT CGGAGCTGAG 240
CGGCGGGACC GGAGGGGATG AGGAGGAAGA GTGGCTCTAT GGCGATGAAA ATGAAGTTGA 300
AAGGCCAGAA GAAGAAAATG CCAGTGCTAA TCCTCCATCT GGAATTGAAG ATGAAACTCC 360
TGAAAATGGT GTACCAAAAC CGAAAGTGAC TGAGACCGAA GATGATAGTG ATAGTGACAG 420 CGATGATGAT GAAGATGATG TTCATGTCAC TATAGGAGAC ATTAAAACGG GAGCACCACA 480
GTATGGGAGT TATGGTACAG CACCTGTAAA TCTTAACATC AAGACAGGGG GAAGAGTTTA 540
TGGAACTACA GGGACAAAAG TCAAAGGAGT AGACCTTGAT GCACCTGGAA GCATTAATGG 600
AGTTCCACTC TTAGAGGTAG ATTTGGATTC TTTTGAAGAT AAACCATGGC GTAAACCTGG 660
TGCTGATCTT TCTGATTATT TTAATTATGG GTTTAATGAA GATACCTGGA AAGCTTACTG 720 TCAAAAACAA AAGAGGATAC GAATGGGACT TGAAGTTATA CCAGTAACCT CTACTACAAA 780
TAAAATTACG GTACAGCAGG GAAGAACTGG AAACTCAGAG AAAGAAACTG CCCTTCCATC 840
TACAAAAGCT GAGTTTACTT CTCCTCCTTC TTTGTTCAAG ACTGGGCTTC CACCGAGCAG 900
GAGATTACCT GGGGCAATTG ATGTTATCGG TCAGACTATA ACTATCAGCC GAGTAGAAGG 960
CAGGCGACGG GCAAATGAGA ACAGCAACAT ACAGGTCCTT TCTGAAAGAT CTGCTACTGA 1020 AGTAGACAAC AATTTTAGCA AACCACCTCC GTTTTTCCCT CCAGGAGCTC CTCCCACTCA 1080
CCTTCCACCT CCTCCATTTC TTCCACCTCC TCCGACTGTC AGCACTGCTC CACCTCTGAT 1140
TCCACCACCG GGTTTTCCTC CTCCACCAGG CGCTCCACCT CCATCTCTTA TACCAACAAT 1200
AGAAAGTGGA CATTCCTCTG GTTATGATAG TCGTTCTGCA CGTGCATTTC CATATGGCAA 1260
TGTTGCCTTT CCCCATCTTC CTGGTTCTGC TCCTTCGTGG CCTAGTCTTG TGGACACCAG 1320
CAAGCAGTGG GACTATTATG CCAGAAGAGA GAAAGACCGA GATAGAGAGA GAGACAGAGA 1380
CAGAGAGCGA GACCGTGATC GGGACAGAGA AAGAGAACGC ACCAGAGAGA GAGAGAGGGA 1440
GCGTGATCAC AGTCCTACAC CAAGTGTTTT CAACAGCGAT GAAGAACGAT ACAGATACAG 1500
GGAATATGCA GAAAGAGGTT ATGAGCGTCA CAGAGCAAGT CGAGAAAAAG AAGAACGACA 1560
TAGAGAAAGA CGACACAGGG AGAAAGAGGA AACCAGACAT AAGTCTTCTC GAAGTAATAG 1620
TAGACGTCGC CATGAAAGTG AAGAAGGAGA TAGTCACAGG AGACACAAAC ACAAAAAATC 1680
TAAAAGAAGC AAAGAAGGAA AAGAAGCGGG CAGTGAGCCT GCCCCTGAAC AGGAGAGCAC 1740
CGAAGCTACA CCTGCAGAAT AGGCATGGTT TTGGCCTTTT GTGTATATTA GTACCAGAAG 1800
TAGATACTAT AAATCTTGTT ATTTTTCTGG ATAATGTTTA AGAAATTTAC CTTAAATCTT 1860
GTTCTGTTTG TTAGTATGAA AAGTTAACTT TTTTTCCAAA ATAAAAGAGT GAATTTTTCA 1920
TGTTAAGTTA AAAATCTTTG TCTTGTACTA TTTCAAAAAT AAAAAGACAG CAATGACTTT 1980
ATATCCAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAGGGC GGCC 2024
(2) INFORMATION FOR SEQ ID NO: 84:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 931 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO : 84: CGCGCCMATA GCCGGACGGG GATCTGAGCT GGCAGGATGA ATGTGGGGGT GGCACACAGC 60 GAAGTAAACC CCAACACCCG AGTGATGAAT AGCCGAGGCA TCTGGCTGGC CTACATCATC 120 TTGGTAGGAT TGCTGCATAT GGTTCTACTC AGCATCCCCT TCTTCAGCAT TCCTGTTGTC 180 TGGACCCTGA CCAACGTCAT CCATAACCTG GCTACGTATG TCTTCCTTCA TACGGTGAAA 240 GGGACACCCT TTGAGACTCC TGACCAAGGA AAGGCTCGGC TACTGACACA CTGGGAGCAA 300 ATGGACTATG GGCTCCAGTT TACCTCTTCC CGCAAGTTCC TCAGCATCTC TCCTATTGTG 360 CTCTATCTCC TGGCCAGCTT CTATACCAAG TATGATGCTG CGCACTTCCT CATCAACACA 420
GCCTCATTGC TAAGTGTACT GCTGCCGAAG TTGCCCCAGT TCCATGGGGT TCGTGTCTTT 480
GGCATCAACA AATACTGAGG GATGGGTTTT GGGACAGCTC CATGGGCATG GGGAAGGCAC 540 TGAAACAGAG GACTATAAAA CATCCTTCTC TTATTCTCCA TACTGTCTTC TACACCTTTA 600
AAGCCTGAGA ACTATACAAC CTTTCCCAGA CTCCCAAGAA GAGAAGAGAT TGGCAAATGG 660
GGCTCCTGGG CCCAGTCCTG CTAGTGGCAA GTTTCTTTGA ATCAGGAAGG CAGGTGAGGT 720
AAGGGCCAAA TCACTCTCCT CCATAGCAGG AAGCCATTTG GGCAGCTCCT TTGGTGATTA 780
CATCTTTCCA TATCTTTTAC ACTTACCACC TTCCAGCTCT GTTTTGCTGT GTATTTTTCT 840 TACAATAATT TTTTTCAGCT ATAGCTGCAG TTTAATCAGG ATGGGTAGAG AGCTCTCCTC 900
ATAAGGCTGG GGGTGGGAAG ATGGAATACT G 931
(2) INFORMATION FOR SEQ ID NO: 85:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 825 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 85:
CGGGGCCGGC GGGGTCTTCA GGGTACCGGG CTGGTTACAG CAGCTCTACC CCTCACGACG 60 CAAACATGGC AGCGCAGAAG GACCAGCAGA AAGATGCCGA GGCGGAAGGG CTGAGCGGCA 120
CGACCCTGCT GCCGAAGCTG ATTCCCTCCG GTGCAGGCCG GGAGTGGCTG GAGCGGCGCC 180
GCGCGACCAT CCGGCCCTGG AGCACCTTCG TGGACCAGCA GCGCTTCTCA CGGCCCCGCA 240 ACCTGGGAGA GCTGTGCCAG CGCCTCGTAC GCAACGTGGA GTACTACCAG AGCAACTATG 300
TGTTCGTGTT CCTGGGCCTC ATCCTGTACT GTGTGGTGAC GTCCCCTATG TTGCTGGTGG 360
CTCTGGCTGT CTTTTTCGGC GCCTGTTACA TTCTCTATCT GCGCACCTTG GAGTCCAAGC 420
TTGTGCTCTT TGGCCGAGAG GTGAGCCCAG CGCATCAGTA TGCTCTGGCT GGAGGCATCT 480
CCTTCCCCTT CTTCTGGCTG GCTGGTGCGG GCTCGGCCGT CTTCTGGGTG CTGGGAGCCA 540 CCCTGGTGGT CATCGGCTCC CACGCTGCCT TCCACCAGAT TGAGGCTGTG GACGGGGAGG 600
AGCTGCAGAT GGAACCCGTG TGAGGTGTCT TCTGGGACCT GCCGGCCTCC CGGGCCAGCT 660
GCCCCACCCC TGCCCATGCC TGTCCTGCAC GGCTCTGCTG CTCGGGCCCA CAGCGCCGTC 720
CCATCACAAG CCCGGGGAGG GATCCCGCCT TTGAAAATAA AGCTGTTATG GGTGTCATTC 780 AGGAAAAAAA AAAAAAAAGG GGGGCCCCTC TAGGGGTCAA AGTTA 825
(2) INFORMATION FOR SEQ ID NO: 86:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1238 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 86:
CATGTAAAAG GATGAAATGT GACTTCTGGT GTTTTTTTAT TTCTATGGAG GGACTTTCTG 60
GGGACGGTTT CTGGCTCTCA GGCTCTGAGA AGCTGCAGTT TATGAGTGGC TCTGTGTGTG 120
CTGCCACCTA CTGGAGAAGC CATAAGCTGC AGCTTTAGGA AAAGGGAACC CGGGGCAGAG 180
TGTGGGGAAG TGGGATGGCA GCATGGCAGG GCTTTGGAAA ATGAGAGGTG AGAGTKTKTC 240
CAGGAAGGGT GTAAGGAGAG GATGGATCCT GATACATGGA TTCAGGATCA TTAGGGTCCT 300
GTCTGGGACA CTGGCCTTCC TGCTTACCTG CTCTTTCCTT CCTCCTTGGT CGGAGGAGGG 360
GCTGGCTCAC TGCTCTGGCT TCATTTTCCA GAGCTGCCTG CTGCAGTCAC ACTTAGGTCA 420
TCTTCTCTCA CTTTTCTCCT TTTGCCGATT AGTGGACGTG ACAGAGATGT GAATGGGGCA 480
GGGATGTCCT TTGATGGCAT CAAGACTTTA GCTTCTGGTG CGCTGTGTCC CAGCTCTGAT 540
TTCAGTTGCA GCCGTGATGG AMAGTTNGCA TGGAAGCTGA GACTCTCACT GACAGTGAAA 600
CCCTCAAATG AACACAATCC CTGCTTTCCT GCCAAGGATC CTTGTAGGGT NCCCCCAGCT 660
TCCCCACTTT TTTTCTGTGT CCTGACAAAG AAACACAGAG TAACTTGATT GCCCTGTGAC 720
CTGGCCAGTT GCATTTCCCC TGCAGGCTTG AGCCCAAGCC AGAGCCTTGA AAAGGTATTC 780
AGGTTGTTGC CCAAAACACT GAAAAAAACT GCCCTGGCCC TGAACCAAAT ACCTTGAACC 840
CTCGTAAACT CCATACCCTG ACCCCCTTGT TTTGGATATA CCCAGGTAGA ACAACTCTCT 900
CTCACTGTCT GTTGTGAGGA TACGCTGTAG CCCACTCATT AAGTACATTC TCCTAATAAA 960
TGCTTTGGAC TGATCACCCT GCCAGTCTTT TGTCTTGGGC AATCTATACT TTTNCTCAGA 1020
GGTTCCCAAG GCCTACTGAA GGGACTTAAC ATACTCTTAA TGGCTTTCCT CTCTCTTGTT 1080
TTACCTTATG CCCTCACTTC CTGAGTTAAC CTCCCAAATA CAGGATTCAC CTGTACCCAA 1140
GCCCTTAGCT TCAAGAATAC AGGATCACCT GTACCCAAGC CCTTAGCTCA AGCTCTGCTT 1200
TGGAAGAACC CAAACTAAGA CAGTGCTCCT GGTGCCCT 1238
(2) INFORMATION FOR SEQ ID NO: 87: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1460 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 87:
ATTGCCTTCT GGTCCCTGGT GACACTGGGG TCATCCTTCA TCCCCGGAGA GCATTTCTGG 60
CTGCTCCTCC TGACCCGGGG CCTGGTGGGG GTCGGGGAGG CCAGTTATTC CACCATCGCG 120
CCCACTCTCA TTGCCGACCT CTTTGTGGCC GACCAGCGCG ACCGGATGCT CAGCATCTTC 180
TACTTTGCCA TTCCGGTGGG CAGTGGTCTG GGCTACATTG CAGGCTCCAA AGTGAAGGAT 240
ATGGCTGGAG ACTGGCACTG GGCTCTGAGG GTGACACCGG GTCTAGGAGT GGTGGCCGTT 300
CTGCTGCTGT TCCTGGTAGT GCGGGAGCCG CCAAGGGGAG CCGTGGAGCG CCACTCAGAT 360
TTGCCACCCC TGAACCCCAC CTCGTGGTGG GCAGATCTGA GGGCTCTGGC AAGAAATCCT 420
AGTTTCGTCC TGTCTTCCCT GGGCTTCACT GCTGTGGCCT TTGTCACGGG CTCCCTGGCT 480
CTGTGGGCTC CGGCATTCCT GCTGCGTTCC CGCGTGGTCC TTGGGGAGAC CCCACCCTGC 540
CTTCCCGGAG ACTCCTGCTC TTCCTCTGAC AGTCTCATCT TTGGACTCAT CACCTGCCTG 600
ACCGGAGTCC TGGGTGTGGG CCTGGGTGTG GAGATCAGCC GCCGGCTCCG CCACTCCAAC 660
CCCCGGGCTG ATCCCCTGGT CTGTGCCACT GGCCTCCTGG GCTCTGCACC CTTCCTCTTC 720
CTCTCCCTTG CCTGCGCCCG TGGTAGCATC GTGGCCACTT ATATTTTCAT CTTCATTGGA 780
GAGACCCTCC TGTCCATGAA CTGGGCCATC GTGGCCGACA TTCTGCTGTA CGTGGTGATC 840
CCTACCCGAC GCTCCACCGC CGAGGCCTTC CAGATCGTGC TGTCCCACCT GCTGGGTGAT 900
GCTGGGAGCC CCTACCTCAT TGGCCTGATC TCTGACCGCC TGCGCCGGAA CTGGCCCCCC 960
TCCTTCTTGT CCGAGTTCCG GGCTCTGCAG TTCTCGCTCA TGCTCTGCGC GTTTGTTGGG 1020
GCACTGGGCG GCGCACTTCC TGGGCACCGC CATCTTCATT GAGGCCGACC GCCGGCGGGC 1080
ACAGCTGCAC GTGCAGGGCC TGCTGCACGA AGCAGGGTCC ACAGACGACC GGATTGTGGT 1140
GCCCCAGCGG GGCCGCTCCA CCCGCGTGCC CGTGGCCAGT GTGCTCATCT GGAGAGGCTG 1200
CCGCTCACCT ACCTGCACAT CTGCCACAGC TGGCCCTGGG CCCACCCCAC GAAGGGCCTG 1260
GGCCTAAACC CCTTGGCCTG GCCCAGCTTC CAGAGGGACC CTGGGCCGTG TGCCAGCTCC 1320
CAGACACTAC ATGGGTAGCT CAGGGGAGGA GGTGGGGGTC CAGGAGGGGG ATCCCTCTCC 1380
AACAGGGGCA GCCCCAAGGG CTCGGTGCTA TTTGTAACGG GATTAAAATT TGTAGCCAGA 1440
AAAAAAAAAA AAAAAAAAAA 1460
(2) INFORMATION FOR SEQ ID NO: 88:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1395 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 88:
CAGGTGCAAA GTGGGAAGTG TGAGTCCTCA GTCTTGGGCT ATTCGGCCAC GTGCCTGCCG 60
GACATGGGAC GCTGGAGGGT CAGCAGCGTG GAGTCCTGGC CTTTTGCGTC CACGGGTGGG 120 AAATTGGCCA TTGCCACGGC GGGAACTGGG ACTCAGGCTG CCCCCCGGCC GTTTCTCATC 180
CGTCCACCGG AYTCGTGGGC GCTCGCACTG GCGCTGATGT AGTTTCCTGA CCTCTGACCC 240
GTATTGTCTC CAGATTAAAG GTACGACATT TGGAGGCCCC AGCGAGAAAC GTCACCGGGA 300
GAAACGTCAC CGGGCGAGAG CGGKCCCGCT GTGTGCTCCC CCGGAAGGAC AGCCAGCTTG 360
TAGGGGGGAG TGCCACCTGA AAAAAAAATT TCCAGGTCCC CAAAGGGTGA CCGTCTTCCG 420 GAGACAGCGG ATCGACTACC ATGTGGGTGC CCACAAAAAT TYCACCTYTG AGTCCTCAAC 480
TGCTCACCCC GGGGTCAGTT CCAGAGAGAA GGACTCCCTC CTGCTTGGAA GAGACCTCAC 540
ACCGTCATCA CGATGCCAAC GGCTCTGAAG GTGGATGGCA TTCCTGCGTG GATTCATCAC 600
TCCCGCATCA AAAAGGCCAA CRGAGCCCAA CTAGAAACAT GGGTCCCCAG GGCTGGGTCA 660
GGCCCCTTAA AACTGCACCT AAGTTGGGTG AAGCCATTAG ATTAATTCTT TTTCTTAATT 720 TTGTAAAACA ATGCATAGCT TCTGTCAACT TATGTATCTT AAGACTCAAT ATAACCCCCT 780
TGTTATAACT GAGGGAATCA ATGATTTGAT TCCCCAAAAA CACAAGTGGG GAATGTAGTG 840
TCCAACCTGG TTTTTACTAA CCCTGTTTTT AGACTYTCCC TTTCCTTTAA TCACTCAGCC 900
TTGTTTCCAC CTGAATTGAC TCTCCCTTAG CTAAGAGCGC CAGATGGACT CCATCTTGGC 960
TCTTTCNACT GGCAGCCGCT TCCTYCAAGG ACTTAACTTG TGCAAGCTGA CTCCCAGCAC 1020 ATCCAAGAAT GCAATTAACT GATAAGATAC TGTGGCAAGC TATATCCGCA GTTCCCAGGA 1080
ATTCGTCCAA TTGATTACAC CCMAAAGCCC CGCGTCTATC ACCTTGTAAT AATCTTAAAG 1140
CCCCTGCACC TGGAACTATT AACGTTCCTG TAACCATTTA TCCTTTTAAC TTTTTTGCCT 1200
ACTTTATTTC TGTAAAATTG TTTTAACTAG ACCCCCCCTC TCCTTTCTAA ACCAAAGTAT 1260
AAAAGCAAAT CTAGCCCCTT CTTCAGGCCG AGAGAATTTC GAGCGTTAGC CGTCTCTTGG 1320 CCACCAGCTA AATAAACGGA TTCTTCATGT GTAAAAAAAA AAAAAAAAAA CTCGGAGGGG 1380
GGGCCCGGTA CCCAA 1395
(2) INFORMATION FOR SEQ ID NO: 89:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1186 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 89: GGCACGAGCC GGCAAGCCGA GCTAGGGTGA AAACTGGGGG CGCACCAGGA TGTNNGACAG 60 AAAAGCAGAA GATGAGACTC TGTTCATTCA CTTTTCCTAG GCCCATCCTG TGGTCATCTT 120 TCCCCCTCCC ATCATACCTC CTCCTTCCTG GAGCCTCTGC CGGCTTGGCT GTAATGGTGG 180 CACTTACCTG GATATTTCAG TGGGAGGATG AAAGGCGAGA CTCACCCTAC GCGGTGGGAC 240 AGATGGGGAG AGGAAAAAGG CAGAGATGGC CAGGAGAGGG GTGCAGGACA AACCAGAGAG 300 GTTGGGTCAG GGGAAAAGGG TGGGGAGAAA GAGGGGTGCA GGCCCTGCAG GCCGGTTAGC 360 CAGCAGCTGC GGCCTCCCCG GGCCCTTGGC ATCCAACTTC GCAGACAGGG TACCAGCCTC 420 CTGGTGTGTA TCATAGGATT TGTTCACATA GTGTTATGCA TGATCTTCGT AAGGTTAAGA 480 AGCCGTGGTG GTGCACCATG ACATCCAACC CGTATATATA AAGATAAATA TATATATATA 540 TGTATGTAAA TTATGGCACG AGAAATTATA GCACTGAGGG CCCTGCTGCC CTGCTGGACC 600 AAGCAAAACT AAGCCTTTTG GTTTGGGTAT TATGTTTCGT TTTGTTATTT GTTTGTTTTT 660 GTGGCTTGTC TTATGTCGTG ATAGCACAAG TGCCAGTCGG ATTGCTCTGT ATTACAGAAT 720 AGTGTTTTTA ATTCATCAAT GTTCTAGTTA ATGTCTACCT CAGCACCTCC TCTTAGCCTA 780 ATTTTAGGAG GTTGCCCAAT TTTGTTTCTT CAATTTTACT GGTTACTTTT TTGTACAAAT 840 CAATCTCTTT CTCTCTTTCT CTCCTCCCCA CCTCTCACCC TTGCCCTCTC CATCTCCCTC 900 TCCCGCCCTC CCCTCCTCCC TCTGGCTCCC CGTCTCATTT CTGTCCACTC CATTCTCTCT 960 CCCTCTCTCC TGCCTCCTGC TGCCCCCTCC CCAGCCCACT TCCCCGAGTT GTGCTTGCCG 1020 CTCCTTATCT GTTCTAGTTC CGAAGCAGTT TCACTCGAAG TTGTGCAGTC CTGGTTGCAG 1080 CTTTCCGCAT CTGCCTTCGT TTCGTGTAGA TTGACGCGTT TCTTTGTAAT TTCAGTGTTT 1140 CTGACAAGAT TTAAAAAAAA AAAAAGGAAA AAAAAAAAAA AAAAAA 1186
(2) INFORMATION FOR SEQ ID NO: 90:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1821 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
31 !
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 90:
AAAACATGCT TTCAGGGCGT CCCCTATGTA TTCGGGGGGC CCACGGACAC TCAGGCTGGA 60
KATCCGTCCT CACTGCGCTC AAGATCGCCT CAGCAGACAC CAGTTACCCA GCTGAAAGTC 120
ACAATCCCTC CCAGAAGTCT CCCAACACTA GTGCTGACCA GAGGTGGGGC TCTCAGGCTA 180
GGAGTTTCAC ACACAATGAC AGGCTGCTGG GGGACATTGC AGGACCCCTT TTCCTYTCCT 240
CTCCATGCTA GAAGCCAGCC CTAGGMAGCT GCAGTTACTC CCTGTGACTC AGCAGCAGGC 300
TGATTCAACA CAGCTGCCCA CACAAAGCCA GTGGTAATAC ATCTGTTTAC CTTTCCCTAT 360
CACCCAGACA CAAGCCCCTT TCCCAGGTCA AACCACAGGC CGATGCATCT CCAGTTTGAC 420
AGTCAAATCA CTACTTCCAT TGCTACTTTA GATCAGCCAA AGTGGTGACT GCTGCAGTGT 480
GTGGCTATCC CTACAAGGCC CACCCAAGGG ATGCCCAAAG CCCAACCTTC TCCAGGGCTG 540
CAGCAGNAGC AACCCCACCA GCCTAAGTCC AGCAGAGGAC CTCCCACCCA ATGTCTTGTT 600
CTAATTAGAA GGGGAAGTTA GCCACAGAAA ATCAACTTAT CTATAATTAC AAAATTCTCT 660
TGACTCACCT TAAAGTTCCT ATTGACATCT ACTGCTTTTA AACCTATTTG AAAACTCTGA 720
TACTAAAACA AATGACACTC TAAGAAAGTT TGGGAGCCCC ATGCTGAGAA CCATTTCTGT 780
GCAGTGAGGA TGTTTCCAGA AGCTACTTAC CTACATGTGA ATGTGCCATT TTCTTTCCTT 840
TTGTAGAGAA AATCCCCTTT ACTTTTTGGA ACAGTAATGG CAGCTTCTAG TACAGCCATT 900
ACAGTTTCAT ATGAGAAAAA TTAAGAATAA CTATAAAATT GTTAAAATAT CCAATAATGG 960
ATAATGATGG CCAGAAGATT TAACATACAA AGTAATTCTC AATGTAAAGC TATTCAGCTC 1020
TTCCAGGTTG AATGCCCTGT AACCCACCCT GACCTTCCAC ATCATCTTCA AAAAGCAGTT 1080
TCTCTGTTCC CCATGATTCT CCTATAAGGT AACTCTTTAG TCCTCCATTT AGCACATTTT 1140
AAATCCTCCA AAGAATAAGT ATCATGTGAT TATTTTAGCT TTACAAAAAA AAAGTTGAAT 1200
GGCGTTTTAT TTTCATGGCC TATAAGCAGG TACCTTAGTA GGGCAGATAT AGGAAAAACA 1260
AATTAGAGCA AAACAAATCC TCTACAAATC CAAGGCAGGA AAAGTGGTGG CAGAGTGACT 1320
CATTCTCCTG TCCCTCCCAT CAGGTCAAAT CAGGAGGCTG CAGTGAATGC CTGTTCTTTG 1380
AATGTGTAGC AGTTGTTCCT GTAACTCTTT AAAACTTGGC TATAGGCTGT TTAGCACAGT 1440
ACAGATTAAA GATACAGTTA CGTAAACAGC AAAGTAATTT TATAGTGCTT CATCCATTTA 1500
TCATGCTTTG GTTTGCTAAT TTTTTCACAT ACCTTTTTCT ATCACAGTCT GTTGCTTTTG 1560
TACACATTTC TCATATTGGG GTTCGACAGG TAAACACAAA CTGCTATTTC AGTAGAAAAA 1620
GTTATTGTTA TGGAATATTA AACCCAATAA ATTGTATAAA GGGTAAAAAA AAAAAAAAAA 1680
AAAAAAAAAA AAAAAAAAAA AAAAAAATTC CTGCGGGCCG CANGCTTTTT CCCTTTGGGT 1740
GAGGGGTTAT TTTNGGCTTG GGCACTGGGC CCTTCGTTTT TACAACGTCG TGANGGGGGG 1800 AACCCGGGGG GGGTTTCCCC C 1821
(2) INFORMATION FOR SEQ ID NO: 91:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 862 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 91: TGCCCTTTTT CCCACCGATT CGGGGCNTGG TGAAGGTGGG AGATGTGAAC TCCAATTAAG 60 GGACTGGAGA GAGGTGAAGA ATTTTGCAGG TGGGAGATTT GGATTTGAAT GTGGACTTGT 120 AAATGACTTG ACCTTGCCAT CTGTGTTCAA GGTCACGGTT TGCTGTGGGG TTCCTGGGAG 180 AGCTTACTCA CCCCGGAGTC TTTTCTTTCT CTTGCTCCAA GAAGAGCCCT GTTGGTGCTT 240 TACCACCGCT TGGAGTCTCC CGAGGACACA AACAGGCAGA GAGGGACGTG TAGGGAGAGT 300 TCTTTCCTGT TTTCTGTGCT TTCCTTTTTA CAGGACTCCC GGAAGGCCAC TCATGGCCAT 360 GCCAGGAGCT TTCTCAGAAA CAGTCATAAA CGATCTCTTG AGTCTCTTTC TTGTCCTCCC 420 AGCTGAGCTT TCTTATTCCA CCCTTTCTGG TGTCTATAGG AATGCATGAG AAGACCCTGG 480 GACGTTTTTC TGCTCTCTTC TGGCCCTCCA TGGAGCCATG GGCCTCGGCC TCGGCGGCTC 540 CTCACCCTCA CAATTTATTT CCTCCTCCCG TGCCAGCCCT TCTTTTGTGT CTGAAACCGG 600 TTTTAAAATG TGACTCTCCC AGAGAAGAAG CCGCTGGCTG TATGAAACTT GACGGCGCTT 660 TTGTAAGGTG CCACCCCCAA ACTTTAAGGT AGCTAAACCA ATTTTTAAAA GATTCAATGG 720 CTTGTTCATC CTCCAGATGT AGCTATTGAT GTACACTTCG CAACGGAGTG TCTGAAATTG 780 TGGTGGTCCT GATTTATAGG ATTTCATAAT TAAAATCTCT GCTGAATAAA AAAAAAAAAA 840 AAAAACTCGA GGGGGGCCCG GT 862
(2) INFORMATION FOR SEQ ID NO: 92:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 696 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 92:
CTGAGGCGAG TGAAGTGGAC TCTGAGGGCT ACCGCTACCG CCACTGCTGC GGCAGGGGCG 60 TGGAGGGCAG AGGGCCGCGG AGGCCGCAGT TGCAAACATG GCTCAGAGCA GAGACGGCGG 120
AAACCCGTTC GCCGAGCCCA GCGAGCTTGA CAACCCCTTT CAGGACCCAG CTGTGATCCA 180
GCACCGACCC AGCCGGCAGT ATGCCACGCT TGACGTCTAC AACCCTTTTG AGACCCGGGA 240
GCCACCACCA GCCTATGAGC CTCCAGCCCC TGCCCCATTG CCTCCACCCT CAGCTCCCTC 300
CTTGCAGCCC TCGAGAAAGC TCAGCCCCAC AGAACCTAAG AACTATGGCT CATACAGCAC 360 TCAGGCCTCA GCTGCAGCAG CCACAGCTGA GCTGCTGAAG AAACAGGAGG AGCTCAACCG 420
GAAGGCAGAG GAGTTGGACC GAAGGAGCGA GAGCTGCAGC ATGCTGCCCT GGGRGGCACA 480
GCTACTCGAC AGAACAATTG GCCCCCTCTA CCTTCTTTTT GTCCAGTTCA GCCCTGCTTT 540
TTCCAGGACA TCTCCATGGA GATCCCCCAA GAATTTCAGA AGACTGTATC CACCATGTAC 600
TACCTCTGGA TGTGCAGCAC GSTGGNTCTT CTCCTGAAYT TCMTCGSCTG CCTGGCCAGT 660 TCTGTGTGGA AACCAACAAT GGCGAGGCTT TGGGTT 696
(2) INFORMATION FOR SEQ ID NO: 93:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1886 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 93: CAGGCCACTG ACGCTTCTTT GCGAGGGATG CAGGAGGTCC TACAGAGAAA GGCGCTTCTT 60
GCATKTCAGA GGGCCCACAG CCTCTCACCC ACAGATCACC AAGCAGCTTT CTACCTGGCT 120
CTGCAGCTTG CCATCTCCAG ACAGATCCCA GAGGCTCTGG GGTATGTCCG CCAAGCTCTT 180
CAGCTTCAAG GTGACGATGC CAACTCCCTG CACCTCCTTG CCCTCCTGCT GTCAGCACAG 240
AAGCATTACC ATGACGCTCT GAACATCATC GACATGGCCC TGAGTGAATA CCCAGAAAAT 300 TTCATACTAC TGTTTTCCAA AGTGAAGTTG CAGTCACTCT GCCGAGGCCC GGACGARGCA 360
CTGCTGACTT GTAAGCACAT GCTGCAGATA TGGAAATCCT GCTACAACCT CACCAACCCC 420
AGTGATTCTG GACGTGGGAG CAGCCTCTTA GATAGAACCA TTGCTGACAG ACGACAGCTT 480
AATACAATTA CTTTGCCAGA CTTCAGCGAT CCCGAGACAG GCTCCGTCCA TGCCACATCG 540
GTAGCAGCCT CAAGAGTGGA GCAGGCACTG TCGGAAGTGG CTTCGTCTCT GCAGAGCATG 600 CCCCTAAGCA GGGCCCGCTG CACCCCTGGA TGACGCTGGC ACAGATCTGG CTCCATGCAG 660
CTGAAGTCTA TATCGGCATC GGGAAGCCTG CAGAAGCCAC AGCCTGTACC CAAGAAGCTG 720
CCAACCTCTT CCCAATGTCC CACAATGTCC TCTACATGCG CGGCCAGATT GCTGAGCTCC 780
GGGGAAGCAT GGACGAGGCG CGGCGGTGGT ATGAAGAGGC CTTAGCCANT CAGCCCCACC 840
CACGTGAAGA GCATGCAGCG ACTTGGCCCT GATCCTTCAC CAGYTAGGCC GYTACAGTYT 900
GGCGGAGAAG ATCCTCCGGG ACGCGGTGCA GGTGAACTCG ACAGCCCACG AGGTCTGGAA 960
CGGGCTCGGC GAGGTCCTCC AAGCTCAGGG CAACGATGCG GCGGCTACGG AGTGCTTCCT 1020
GACAGCCTTG GAGCTGGAGG CCAGCAGCCC CGCCGTGCCC TTCACCATCA TCCCCCGCGT 1080
GCTCTGAGCA GGCGCCTGCC AGCCTCACCT GCCGCTCAGC CTNCAGAGGC CCTGCCGGGC 1140
ACCAGGGCTT GTGCCATCGC CCCAAGGGGA TGAATCTGCC GCACTGAGGC CAGGGACGAG 1200
TGTTCAGTGG GCCACAGTGA ACCAACCAAA CCAACCCCGA ATCATCGCTC TCGCCATGTG 1260
CGTTTCTCTT GTTTTTTTTG CCAGCCCAAT GGTAGTTTCT GAACCTATTG ACATTGTTCA 1320
AAATGGATCA TGTGCCATAT TTTGTTAGTT GACATCTGAG TTTTCAGTAA AATGATTATG 1380
GAATTAATCA GCAAATGTAG AAGAATATAT TCAAAGTTAA AATTCAGTGG CAGCACAGAT 1440
TATTTTTATC AGAGCTGTAA AGAAAACAAC TGTCCTTTTC TCCCCACCAC CCCTCCTCCC 1500
CCACTTTGGC CCAGAAACCA AATGTGAACT TCCTGTCTCC CACCTCAGCA CTAGTCCATG 1560
CCAGGACACC AGCTGACAAT TTCTTGGTTT TACTGTCAAT AATTGTACCA TGTGATCAAT 1620
TACTGTCCTC ACTTAGAACA AAGCCTGAGT CCGAGAATAT TTATATTTTA CCAATATATG 1680
CCTCTTACAA GAGAAGGAAA TATGAGTTAT TTAAGTTTAA CTTTTTTATG TCAATTCAGA 1740
GTTTATTTAT CGAGGGAAAT ATGTACAAAG AAGCTTCAAA TGGAATATTT ACCGACATTC 1800
CTTATACATG ACAGACACTT GGCTACATGG GAAGATGATG TTAATAATAA AATGATTTTT 1860
AAATCGAAAA AAAAAAAAAA AAAAAN 1886
(2) INFORMATION FOR SEQ ID NO: 94:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1774 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 94: CTCAGCTACC GTATACAGTA GGACATAACC CCATTTCACA TGCACTACAC TGAGACTTCC 60 CTCCTCTCCC CCCACATTGA AGATGTTCTT TTTTCATAAC TATATACTAT TCCATTGCAT 120
GAATATTCTG TAATTTATTT AATCCCCTAT GGATTGATAA TTAGGTTCAT TATAGATAGA 180
AGTGTAATTA ACATTCCTGT ACATCTATTT TGCTACTTGT GTGGGTATTT CTGTAGGATG 240
AATAACTAGA AATTTATTGG ATCAGGTTTC ACATTTGCAG TTTTGAAAAC TACTACCAAA 300
AAGATTTCAC CAATTTACAA CTCCATCATT AGTAAGAATG CCTGTTTGCC TATAGTCTGC 360
CAACCCTGAA TCCTTAAAAA TTTTTGCCAA TCTGGTAGGC AAAATTTCTT TCTTTTCTTT 420
GAATATTAAT GAGGAGGAAC ATCTTTTCAT GTTTCTTGGC CATTTGCATT TCCTATTATG 480
AATTGCTTTT GCCCATTTTC CTTTTTTTAA TTATGAAAGT CTAATGACTA CCTTCTCATT 540
GTATAAAAAA CACAGTTCTT TGAATAGAGA GACCCTTTTC TCCAATGCTA CCAATCACAT 600
TCCACTTACC ACAGTTTAAC ATACATCCTC TAGTCACCTT TCCGTACGAA TATACATACA 660
CATAAAAACA CTTTTTACAT AAATAGGATC TCATATTCTG TAGCTTTTTA AAATTTTGGT 720
CTGAAAAAAA GATAACAGGT CTTTAAATTT CTTTAATGGT TGAATATGAT TAAATACTAT 780
GAAAATGCCA TTATTTATTC CCTTAATTTT TTTCCTCTCG CTATTACATT GCCAAAGTAA 840
ACATCCTATT CAGATGTCTT TGTGCATGTG TGTGAATATT TCTTTAGTCT GGAGTCCAGT 900
AAGGTGGATT TTTGGATCAA AGGGTTTGTT CTCTGTCCAC CTTCAGTCTT CCCAAAGGCC 960
TTCATAACTG TATTTTCACC AAGTGTATGG AGAATGTTCA TTTCCCCATA TAACCATACC 1020
TACACTTGAT AGTTTTTATC TGTTGGGCGA AAAAGAACCT TTTCTTATTT TGCATTTCCC 1080
TGATTATAAA AAAAAATGGT GAGATTGGGG TTATTTTCAT GTTTATTGGC CATTTATAGT 1140
TTACTGTGGA TTGTTTGTAT CCCTTACCTG CTTTCTATTG GGTTATGTGT GGATATATTG 1200
TTTTTATTTG TTCAGCATCT CCTTCCCCAT CTTCTGGTAA CACAACCTTT ATTTATTTCT 1260
GGGGAACCTA TTCCCTGTGG CTTAGGTGAG CATGTGACCA GGCCTGGCCT CCTGAGTCCC 1320
ACAGCTTCCT AGCCACAGTG ATAAAAGAAT GGGTATATAA CTTAAGCCAG GCTAAGGAAA 1380
GCCCTTAACA GAACTTCTGC TGGAACTACT GGAAAGAAGG CTTTATGGAG ATCCCAGGAA 1440
CCAAGGACCA TGTAAGCCTG AATTTGTGCC ATGTGGAGAG AGTCTGTCTG AGGAGAAACT 1500
CGGATGCTAG CAGAAATGGA AAGAGAACTA AGTTCTGATG TCATTTTTCT GGAGGCCCTA 1560
GATCCAGCTG TGCCTAAAGC CTGCCCTACT CCGGACTTTA AAGTTTTGTG AGCCAATAAA 1620
GTCCCTTTCT TGTTTAAGAT AATTGAATTG AGTTTCTGTT CTGATTAATA TAGGTTATTT 1680
GTATTTTCTT ATTGATTTGT AGAAAACCTT TGTAATTTTA AATTCTAGAC TTTATGCACT 1740
ATATAAGTTA ATAAAATTAG CATGGCCTTC CATG 1774
(2) INFORMATION FOR SEQ ID NO: 95:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2503 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 95:
GGCACGAGCG AAGGCAAGGG GGCACCAGCT CAGGACTGCA TCTGCCTGCC ATTTCCCTTC 60
CACTCCTCCT TTCTGGAGTC TGACATTAGA AAGCCAGCGA GAAGGAAGAT TCAAACAACC 120
AACCCTGATT TCCTGCTTCT CCTTTTCATG AGTGTTCCTG TGGTCTCTGC ACCTCCTTTC 180
TGTCCCCCGG CAGAGGGCAG TAGAGATGGC CGGCCCAAGG CCTCRGTGGC GCGACCAGCT 240
GCTGTTCATG AGCATCATAG TCCTCGTGAT TGTGGTCATC TGCCTGATGT TATACGCTCT 300
TCTCTGGGAG GCTGGCAACC TCACTGACCT GCCCAACCTG AGAATCGGCT TCTATAACTT 360
CTGCCTGTGG AATGAGGACA CCAGCACCCT ACAGTGTCAC CAGTTCCCTG AGCTGGAAGC 420
CCTGGGGGTG CCTCGGGTTG GCCTGGGCCT GGCCAGGCTT GGCGTGTACG GGTCCCTGGT 480
CCTCACCCTC TTTGCCCCCC AGCCTCTCCT CCTAGCCCAG TGCAACAKTG ATGAGAGAGC 540
GTGGCGSCTG GCAGTGGGCT TCCTGGCTGT KTCCTCTGTG CTGCTGGCAG GCGGCCTGGG 600
CCTCTTCCTC TCCTATGTGT GGAATGGGTC ARGCTCTCCC TCCCGGGGCC TGGGTTTCTA 660
GCTCTGGGCA GCGCCCAGSC CTTACTCATC CTCTTGCTTA TAGCCATGGC TGTGTTCCCT 720
CTGAGGGCTG AGAGGGCTGA GAGCAAGCTT GAGAGCTGCT AAAGGCTTAC GTGATTGCAA 780
GGGTTCAGTT CCAACCATGG TCAGAGGTGG CACATCTGCT CAGCCATCTC ATTTTACAGC 840
TAACGCTGAT CTCCAGCTCC AGCGATGGAA CCCACTACAG AGGAGGTGGG GCCCCTGTGT 900
CAAAGAGGCC GAGGGGCAGC AAGGGCAGMC AGGGCACCTG TGACTTCTTA GTACAAGATT 960
GTCTGTCCTT CAGGACTTCC AAGGCTCCCA AAGACTCCCT AAACCATGCA GCTCATTGTC 1020
ACACCAATTC CTGCTTTAAT TAATGGATCT GAGCAAATCT TCCTCTAGCT TCAGGAGGGT 1080
GGGGAGGGAG TGATTGCTGT CATGGGGCCA GACTTCCAGG CTGATTTGCC AAATGCCAAA 1140
ATGAAACCTA GCAAAGAACT TACGGCAACA AACGAGGACA TTAAAAGAGC GAGCACCTCA 1200
GTGTCTCTGG GGACATGGTT AAGGAGCTTC CACTCAGCCC ACCATAGTGA GTGGGCCGCC 1260
ATAAGCCATC ACTGGAACTC CAACCCCAGA GGTCCAGGAG TGATCTCTGA GTGACTCAAC 1320
AAAGACAGGA CACATGGGGT ACAAAGACAA GGCTTGACTG CTTCAAAGCT TCCCTGGACC 1380
TGAAGCCAGA CAGGGCAGAG GCGTCCGCTG ACAAATCACT CCCATGATGA GACCCTGGAG 1440
GACTCCAAAT CCTCGCTGTG AACAGGACTG GACGGTTGCG CACAAACAAA CGCTGCCACC 1500
CTCCACTTCC CAACCCAGAA CTTGGAAAGA CATTAGCACA ACTTACGCAT TGGGGAATTG 1560
TGTGTATTTT CTAGCACTTG TGTATTGGAA AACCTGTATG GCAGTGATTT ATTCATATAT 1620 TCCTGTCCAA AGCCACACTG AAAACAGAGG CAGAGACATG TACTCTGGTG TGATCTCTTG 1680 TCCTCAGTCT CTCTTCTGGG CTCCTCTCCC TCTTGCTTTA TAGCTAGCTG CCCGGGGACC 1740 AAGGTACAGG TGAAAGCAAG GTAGCAGCTT GCGGGAGGAG GCCTGTCTGG CTTACCAGTC 1800 TATACACTGT GGCCTCAACC TCCCAGACAG GGCAGAGAAC TGTGGGCAGC TCGTTTGCTT 1860 TCTAGGCTGG CTGGAGAGGT GGGAGCTCAT TGATAGACTC ATGATGGAAA CTATTTTTGA 1920 AACAGGCTTC CTCCTTCAGG AGAGATCATG CGGACTAAAC TGTAGCAATT CCAGTGCACC 1980 TGGCAGTGAT CCTTTTCTTT GCAAAGTACT GTCTCTTTGG TTCCAGTAAG TTGGACCACC 2040 ACATGACATY ATTTTCCCTG GAACCTGGTC ACTGACTAAC ACAGACAATT GGGACTCCAG 2100 AGCCTCAAGA GCCAGGAGAG GGCACAGTAC ATACAGAGGG AGTCAAATGG GATCTCATTT 2160 TGAGTCCTGC CTTCCGCACA CTCAGAACGG CANCCCCAAG GCCCGGAGTG TCCAGGGCTT 2220 CTGGCCTGAG GTGAATCTGC CAGGCCCAAG AAGGCACAAA GGTAGGAGCA CAGAGAGCCC 2280 CATTCCCACA GGCGGKCGGC CCAGCAGCAC CAGTGGAAGC TCAGCTGTCC TCCAGCTGCT 2340 CTCGGCAGAC AGTTCAGTGC ACAGTTTATG CCCTAGCTGA AAAAGATCTC CCGGACGTAT 2400 TTCAGCACAT CCTCTTCCTC CTCCTCCTCA GGGCTCCTGC TACAGGCAGA GCTGGAACCC 2460 CCCGGCCTCT GGGAAGGGCT GAGGCCTGGA GYCAGTGCCT GTC 2503
(2) INFORMATION FOR SEQ ID NO: 96:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2801 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO; 96: CTGGAAAGCC GAGGGTAGCC GAGCGGGGCG GGCGCTCTGG AGCGGCGGGT GCTCGGGCTG 60 CCGTCCGCTC CGCCAGAAGC ACCGAGCAGC CGAGCCGGGG CCCGCCGCCC TCCTCCTCCA 120 TGAGGCCCGA GTGAGGCGCG GCGGCTATAG CCGACCCGCG GCGCCTTCCC CCCGCGTCCT 180 ATCGCGAGCG CACGACMAGC GGCCCCTGGA GGAGGAGGCG GAGGAGGAGG AGCATGTCGG 240 ACGGTTTCGA TCGGGCCCCA GGTGCTGGTC GGGGCCGGAR CCGGGGCCTG GGCCGCGGAG 300 GGGGCGGGCC TRAGGGCGGC GGTTTYCCGA AMGGARCGGR GCCTGCTGAG CGGRCGCGGC 360 ACCAGCCGCC GCAACCCAAA GCCCCGGGCT TYCTGCARCC AMCGCCGCTG CGCCARCCCA 420
GGACGACCCC GCCGCCAGGG GCCCAGTGCG AGGTCCCCGC CAGCCCCCAG CGGCCTTCCC 480
GGCCCGGGGC GCTCCCAGAG CAAACGAGGC CCCTGAGAGC TCCACCTAGT TCACAGGATA 540
AAATCCCACA GCAGAACTCG GAGTCAGCAA TGGCTAAGCC CCAGGTGGTT GTAGCTCCTG 600
TATTAATGTC TAAGCTGTCT GTGAATGCCC CTGAATTTTA CCCTTCAGGT TATTCTTCCA 660
GTTACACAGA ATCCTATGAG GATGGTTGTG AGGATTATCC TACTCTATCA GAATATGTTC 720
AGGATTTTTT GAATCATCTT ACAGAGCAGC CTGGCAGTTT TGAAACTGAA ATTGAACAGT 780
TTGCAGAGAC CCTGAATGGT TGTGTTACAA CAGATGATGC TTTGCAAGAA CTTGTGGAAC 840
TCATCTATCA ACAGGCCACA TCTATCCCAA ATTTCTCTTA TATGGGAGCT CGCCTGTGTA 900
ATTACCTGTC CCATCATCTG ACAATTAGCC CACAGAGTGG CAACTTCCGC CAATTGCTAC 960
TTCAAAGATG TCGGACTGAA TATGAAGTTA AAGATCAAGC TGCAAAAGGG GATGAAGTTA 1020
CTCGAAAACG ATTTCATGCA TTTGTACTCT TTCTGGGAGA ACTTTATCTT AACCTGGAGA 1080
TCAAGGGAAC AAATGGACAG GTTACAAGAG CAGATATTCT TCAGGTTGGT CTTCGAGAAT 1140
TGCTGAATGC CCTGTTTTCT AATCCTATGG ATGACAATTT AATTTGTGCA GTAAAATTGT 1200
TAAAGTTGAC AGGATCAGTT TTGGAAGATG CTTGGAAGGA AAAAGGAAAG ATGGATATGG 1260
AAGAAATTAT TCAGAGAATT GAAAACGTTG TCCTAGATGC AAACTGCAGT AGAGATGTAA 1320
AACAGATGCT CTTGAAGCTT GTAGAACTCC GGTCAAGTAA CTGGGGCAGA GTCCATGCAA 1380
CTTCAACATA TAGAGAAGCA ACACCAGAAA ATGATCCTAA CTACTTTATG AATGAACCAA 1440
CATTTTATAC ATCTGATGGT GTTCCTTTCA CTGCAGCTGA TCCAGATTAC CAAGAGAAAT 1500
ACCAAGAATT ACTTGAAAGA GAGGACTTTT TTCCAGATTA TGAAGAAAAT GGAACAGATT 1560
TATCCGGGGC TGGTGATCCA TACTTGGATG ATATTGATGA TGAGATGGAC CCAGAGATAG 1620
AAGAAGCTTA TGAAAAGTTT TGTTTGGAAT CAGAGCGTAA GCGAAAACAG TAAAGTTAAA 1680
TTTCAGCATA TCAGTTTTAT AAAGCAGTTT AGGTATGGTG ATTTAGCAGA ACACAAGAGA 1740
GCAAGAAAAT GTGTCACATC TATACCAAAT TRAGGATGTT GAGTTATGTT ACTAATGTAT 1800
GCAACTTTAA TTTTGTTTAA CACTATCTGC CAAAATAAAC TTTATTCCCT ATAACTTAAA 1860
ATGTGTATAT ATATATAATA GTTTATTATG TACAGTTAAT TCTACTGTTT TGGCTGCAAT 1920
AAAATCGATT TTGAAATAAA TGAAATGTTG AAAATTTTGC TAGTTGGTTA GATGCTTATC 1980
CTTTAAATTC TACTTTTCTT GAGGGGAAAA AGTCTTCGTC TGGAAATACA TATTACTGCA 2040
AAAATGTAGC ATCCTTTTTT AGGTAGGAGT ATTATAGCTT YCATTTTAGT TKGACATTTA 2100
GTGTCCCAAT GAATTCAATT TCAAATATGA ATCATAATCT TGAAAATCTT TAGCACTAAA 2160
GTCTTGGGAA TATATCAACA ACTGATTTAC ATATGCAGAT GCTATTTGNA TACCAAGGGC 2220
TTTTTAAATG TCATGGGGGG GAAAAACCCA ACTTGGTGGA ACTCCCAGCT AAACAACCAA 2280 GACTTCACTG GAAGATTTAT TCCAATTCTA GGAATTGTTC TTTTTTATTT TTATTTTTTC 2340 AACTGRCTAA CTTCATTACC TTAAAGCCTA GAACATTATT CTGCTTTATT TATATGGCTT 2400 TCTCACTTTT ATTTTGTAGC AKGGGTTGCA TCGACTTTTT TACTAGAGAA TTTTACTAGA 2460 TATTTGTCAT TCAAGTTTTC ATCTGCTTTA TAATTGATAC ACCTTGAGGG TCACTTTTCT 2520 AATACTTTTA CTATAATGTG GTACCACCTC AGCCCTAATA AATAATATTT TTACCTAATG 2580 TCAAATCTTT TTCCAGCTAA CTAAAAACTG TGTACAAAAG GATTGCTTGT AAATATGCAT 2640 GTAAATAGTT CTGTTAATAA CCCACTGTTT TACATTTGGT ACATCTGTGT CTGCTAATAC 2700 AGTTAGCTTT CTCACTTTTC TGCTTGTTTG TTCAGTCTGA ATTAAAATTA GACTTTGAAA 2760 ATAAAGCTTA AAAAAAAAAA AAAAAAAAAA AAAAACTCGA G 2801
(2) INFORMATION FOR SEQ ID NO: 97:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1631 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 97:
ATGGAGCCAA AGACAATCAC TGATCCTTTG GCTTCTAGTA TAATTAAGAG TCTGCTCCCT 60
AATTTTCTTC CATACAATGT CATGCTCTAC AGTGATGCTC CAGTGAGTGA ACTCTCCCTC 120
GAGCTGCTTC TGCTTCAGGT TGTCTTGCCA GCATTACTCG AACAGGGACA CACGAGGCAG 180
TGGCTGAAGG GGCTGGTGCG AGCGTGGACT GTGACCGCCG GATACTTGCT GGATCTTCAT 240
TCTTATTTAT TGGGAGACCA GGAAGAAAAT GAAAACAGTG CAAATCAACA AGTTAACAAT 300
AATCAGCATG CTCGAAATAA CAACGCTATT CCTGTGGTGG GAGAAGGCCT TCATGCAGCC 360
CACCAAGCCA TACTCCAGCA GGGAGGGCCT GTTGGYTTTC AGCYTTACCG CCGACCTTTA 420
AATTTTCCAC TCAGGATATT TCTGTTGATT GTCTTCATGT GTATAACATT ACTGATTGCC 480
AGCCTCATCT GCCTTACTTT ACCAGTATTT GCTGGCCGTT GGTTAATGTC GTTTTGGACG 540
GGGACTGCCA AAATCCATGA GCTCTACACA GCTGCTTGTG GTCTCTATGT TTGCTGGCTA 600
ACCATAAGGG CTGTGACGGT GATGGTGGCA TGGATGCCTC AGGGACGCAG AGTGATCTTC 660
CAGAAGGTTA AAGAGTGGTC TCTCATGATC ATGAAGACTT TGATAGTTGC GGTGCTGTTG 720
GCTGGAGTTG TCCCTCTCCT TCTGGGGCTC CTGTTTGAGC TGGTCATTGT GGCTCCCCTG 780
AGGGTTCCCT TGGATCAGAC TCCTCTTTTT TATCCATGGC AGGACTGGGC ACTTGGAGTC 840
CTGCATGCCA AAATCATTGC AGCTATAACA TTGATGGGTC CTCAGTGGTG GTTGAAAACT 900 GTAATTGAAC AGGTTTACGC AAATGGCATC CGGAACATTG ACCTTCACTA TATTGTTCGT 960 AAACTGGCAG CTCCCGTGAT CTCTGTGCTG TTGCTTTCCC TGTGTGTACC TTATGTCATA 1020 GCTTCTGGTG TTGTTCCTTT ACTAGGTGTT ACTGCGGAAA TGCAAAACTT AGTCCATCGG 1080 CGGATTTATC CATTTTTACT GATGGTCGTG GTATTGATGG CAATTTTGTC CTTCCAAGTC 1140 CGCCAGTTTA AGCGCCTTTA TGAACATATT AAAAATGACA AGTACCTTGT GGGTCAACGA 1200 CTCGTGAACT ACGAACGGAA ATCTGGCAAA CAAGGCTCAT CTCCACCACC TCCACAGTCA 1260 TCCCAAGAAT AAAGTAGTTG TCTCAACAAC TTGACCTTCC CCTTTACATG TCCTTTTTTG 1320 TGGACTTCTC TCTTTCGAGA TTTTTCCCAG TGATCTCTCA GCGTTGTTTT TAAGTTAAAT 1380 GTATTTGACT TGTGTTCTCA GCATTCAGAG AGCAGCGGTG TAAGATTCTG CTGTTCTCCC 1440 TGGATCTTCT GACATTACTG CTGTCTGAGA TTTGTATATG TGTAAATACA AGTTCCTTGA 1500 TACCCTAAAA CCTTGGATTA AACAGAATGT GCATTGTACA TCTTTAAACA AAATGTATAT 1560 TAATTTATTA AATCTAGTTG TCACTTTAAA AAAAAAAAAA AAAAAACTCG AGGGGGGCCC 1620 GGTACCCAAA T 1631
(2) INFORMATION FOR SEQ ID NO: 98:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 504 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 98: CCGAGCTGGG CGAGAAGTAG GGGAGGGCAC GAGCCGCCGC GGTGGCGGTT GCTATCGCTT 60 CGCAGAACCT ACTCAGGCAG CCAGCTGAGA AGAGTTGAGG GAAAGTGCTG CTGCTGGGTC 120 TGCAGACGCG ATGGATAACG TGCAGCCGAA AATAAAACAT CGCCCCTTCT GCTTCAGTGT 180 GAAAGGCCAC GTGAAGATGC TGCGGCTGGA TATTATCAAC TCACTGGTAA CAACAGTATT 240 CATGCTCATC GTATCTGTGT TGGCACTGAT ACCAGAAACC ACAACATTGA CAGTTGGTGG 300 AGGGGTGTTT GCACTTGTGA CAGCAGTATG CTGTCTTGCC GACGGGGCCC TTATTTACCG 360 GAAGCTTCTG TTCAATCCCA GCGGTCCTTA CCAGAAAAAG CCTGTGCATG AAAAAAAAGA 420 AGTTTTGTAA TTTTATATTA CTTTTTAGTT TGATACTAAG TATTAAACAT ATTTCTGTAT 480 TCTTCCAAAA AAAAAAAAAA AAAA 504
(2) INFORMATION FOR SEQ ID NO: 99:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1416 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 99:
GGCACGAGGG AGGGAGCCCT CTCCGTTGGG TGACTCTTGT GTGCCCTTTA GACAGGCTGG 60
CCTGCCGGTT CCACAGGGTA CAGTTAGGAC TTCAGTCTTT CTTTTTCTGT TTTGAGTTGG 120
TCAGTGAGTG ATAGGGTAAC ATGGGCCTTC AGGATGACCC CTTGGAACTG TGCCGAGTTC 180
CTTAAATCTC AGCTGGGATC CTGGACCTGG GAGGCCCCTG TGAGGGCCAG CTCTGGAAAA 240
ACCTGGGAGT TGATGCCGGA GCTGTGGAAG AACTCTGCTC GAGGGCAGGG TGCCCTGGAA 300
CACTGGTAGT TCTGGGGCTG GGAGGGAGAG GGGCTCCGGC TTTCTCTGAA ATGAACACTG 360
CTCTTCAGCA GTTCAAGTAC TTGTTCTCAA AACATTTTCT AATTGATTGG TAGGTTTTCA 420
TAAGCATTGT TTCTTTAAGG CATGGAAAGG GAAGAATGCT CAAGCAAGTC ATCTTTGTTT 480
TCAGTGGGAT GGGCCCGCGT TCTCACTGCT GGGGGCTTCC CCTTCATGTG GCACCTTTGT 540
GCAGGGGCCA CCAGGCAGAC TCTTCCCACC TTCTCCCACT GAAGCACCAA GGGGCTTGGA 600
ACCGTAATTT GGCTAATCAG AGGCATTTTT TTTGTCCTAG TATCTTTCAC ACTTGTCCAA 660
CCGTCTTATT TTTTTAAAAG TTCTGTTGCT TGTATTAACA CGAAACTAGA GAGAAATAGT 720
TTCTGAAGCC AGTTTATTGT GAAGATCCCC AAGGGGAGGT TCGGTAGAGA AAAATAGTAA 780
GCTGGTTTAG AAACTGACGA GGGCAAACAG CCAGGACGCA TTGGAGAGGA ATTTGCCAAA 840
GATCTACCCT GAGATAACGC CTGTCCAGTG TCTTCACCAC GTGAATAACC AGCGCTCCAA 900
AGTCTTTTTC TGCTTTGAAA AAAAAAATTC CACAAGCTTT TAAAGGTGCA TTTAAGAATC 960
CATGTGACTT TAGAATGGAA CTGCCGGCCC TGGCAACTGT CACGTGTGCT AGAAGGTTCG 1020
ATGCCTCTCG AATGCATGTG ATACTCATCT CCATTTTGTT TCCTTGATTG CATTTTTGTT 1080
CTTTTAGCAG ATCTGTCCCT GTGGGTGGTG TCTAAGAAGT CGGACACCTT GGTTTTTGTG 1140
TTAGATTGAG CTGGGCAGCT GCAATCAGCT TCTTTATATG CAAATTAGGC ACGACCCATC 1200
TGTGGTTCCT GGTTGGTGGC TAATGAAGTG AGGGGAGGGA GGGATGTCAC CCCAAAAGTA 1260
GGCCCTCCCA TTGGCTTTGG CCAGGCCAGA CACTTCACAT CGTTTACATG GTTCTGTGTA 1320
ATTTTAAAGT TTATGTGTAT AAAGCGAAGC TGTTTCTGTG AAACTGTATA TTTTGTAAAT 1380
AAATATATTG CTACTTGAAA AAAAAAAAAA AAAAAA 1416
(2) INFORMATION FOR SEQ ID NO: 100:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2847 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 100:
GGCTAGGACA ATTTTGGTGC TTTACCTATC TCTGCAAAGA CTGGAGAATT TGGCATACCA 60
TTAATTACAA CCACCAATCA TATCCAACAA AAGTACCCTA AAAGAAGGAC CAGTGGCCAC 120
TCTCGAAAAA ATTTAAGTAT CAGAAGATTA AAAAGATTTT AGGATTTGGA AGCTTGTATT 180
GTCTTTCCCC AATAATCATT GTTTGATCTC CAAATAGTAG CCTTATATTA GCAATRGACA 240
GATCATTGGT TCTCCATATC TGATCATATG TTACTACTTT GGAATCAGTA TTTGGGCAAA 300
TTCAAGCATT TATGCAGTGG ATATAAATGG AAATATAAAA ATATTTGCCA ACCTGTCTCA 360
GTAACTTATC ATATCTCTGT GNATCCTCAA GGAAAGCACT TTTGCTTTTA CTTAGAAAGC 420
GTTTCAGATT TGCTTTATAG ACTCCTGCTG TCTTCAGTAC CTGATAAAAC TTTAACCAGG 480
GAAGCATTAA ACACAGTGCA GCAGCTTTTG CCCAGGCTTC TAAGTTCCTG CCGGCAGCAT 540
TTATCAATGT AAGAACTAGG ATGCTTCCTG CAGTGGCACT ACCTTCCCCT AGAGCTGGAG 600
CATGCTGCTT GGCCTTAAGC CCCAGCATGA TGAGGCTTCC CTCCTGCCAG GTCAGTAAAA 660
GTTAGAGAGC TCAGAATTGG GTCTTGCCTG GGTGCAGGTG GCAGGGTTTG CTGAAACCCC 720
TAAAGAGAAG TCACCAAGGG AGGCAGGTAA TGAATGTTTC CAGAATCAGT CKGATACTCA 780
TAGCAATTTC TGGCTATCTT TCAAATGTTG AATTTCTGGA TGCTGAGAGG GACTTTGATT 840
TGATATCATT AAATCCAGGA CAGTCCCAAG AAGTGCTTGG AGTCTCGGCT CTGACAGCCC 900
AAGAAGGGAA ATAACTTGTA TTAAGGAACA ACTATGAGCC AGGCCCTGAG CTGTCTCTTA 960
GATAATAAAA CAGATGGGGA GTGGAAGAGT CATTTGCTTC AAGTTATACA GCTAGGAAAT 1020
ACTCAAGCCA AATCTTGAAC GCAGCTCCCC CTAATTCTGT GGACAGGCAC TTTGTACCAC 1080
ACACCATGGT CCACCTAAAA ACAGAAGGAT AAAAAGACTT CAGGTTTTCC CACTGTGTGC 1140
TGACCATCCC AATTTATGAA TCTTCTTCAA AATGACATTT CACAGTTATA GTTAGGGCTC 1200
AGAAATGGCA TTGAGGTAGC CTTATTTCTC CCCTTTAGCA GATGCTTTAA GTACACATTG 1260
CTGACTTGAG CCCACCCCCA GGAGTTAGGA GAACATTTCC TTTTTCATGC CATCTTCCAT 1320
AAATAAGGTG TTTCTTGGCC TTCAAAGATA TAGAACTTTG CAGCAGTAGT AAAAGTGAAG 1380
GGTGTTCTGC TCTCTACTCA ACTTTATTTG AAAATGTCTG CAGCTTCACT CCTGTAGAAA 1440
AGGAAATCTT CATATTTTAG TAAACTTAGC CGCCAGTGTA CTCTGTGAGG ATGTGGCAAT 1500
TCAAAGTCCA GTGAATCTCG CTCTCTTACT GATTCCTGGT TTTAGTGTCT GTGTCGGGGG 1560
AGTGTGTACC TATATATAAA GGACAAGTGT GATATGTCTG TATATGTATA TACATACATA 1620
CATCTCCACA CACACACACA CAATATTTGA GAGCTAAGGA AAACTCAAAG CAGCCCCTTC 1680
ATTATCTTGC GTACTACTTC AAAGATTTCT GTCAGCCCTA ATTACAAGTG TCACCATATA 1740
GTTGGGGCTT AGGTACTTGC TTACAGGAAG AGCAATTCCC TAGCAAAGGT CATTAGCTCC 1800
TAAGGCACTG AGTCAAAGTG ACAGCCCTGA AGGAAATTGC ACTCCAGCCC TCCTCCAGGA 1860
TGTCTAATAA GATGGGAAAC TTGGATGCCC AGCCΛTTTTG GTGACCTCAG AGTCTAACTA 1920
CTCCAGTTAG ACCTAAGGGC ACAAATGCAG AATTCATGAC CTTCTAGTTC TGGCAGGGTC 1980
TAGGAAGTCC TCTCTCCCCA AGTAGAAAAT ATTCTCTTGC CATTCCTGAA ATTCCACATT 2040
CATATAATGG CTGTGCAATA CATGCTTCTC AATAAGAAAA TTAACTGCAT GTTTACTGTG 2100
TGCTGATCAC ATCAGATTTT TATGTTTAAA AAAATCTCAT TATGGNTTCA GTCCAGCCCA 2160
GCTCTAAGAG AAAAAGAAGG CCCATATGGG AGACTTCAGT CTCATTATTA TTGCCTTTAT 2220
CCAGCAGTGC TTATRAAGCC CCCTACCCTG TCCCATTCCA GAAACCATAA GACTCAGGCA 2280
GTTCTTGATT CTGGAGGCCT GCCTGGTAAG ATAAGATAGT ATAATTTGGA ACTGAGAACA 2340
TACCAGAAAC AGCAGAACGA GGGCCAGAGC AGAAAAATGA AAATAAGTGG AGACACTTAT 2400
GGATACATTG GTGCAAAAAA AGCCACGGGS CCCATACTGG GCTTGATATG ACTTTGAGGG 2460
GACAGCAGAT TAATACTTAA TGAGGGTTAA ACCTGACCAG TCTTTCTACA GTGACAGGCC 2520
ACACTGCATC AATGGGGAGA ACCAATGAAT CCATTGTCCT CTGCCTATTT TCCTGTGCAC 2580
AGTCACATTC CCTCCTTAGG AATCTTCCCC TTCCACCCTT TACATTAAAC AAGGGAACAC 2640
TGAATCTTTC AAGGGAATTA CACGTTTGGG TTAATGTTTC AGTATATCAT TTTCATACTG 2700
TAAATTATTT TGTAAGAGAG ATTTACTGCT ATCCCAGGAT GTTCGGACTT GGTGCCCCTG 2760
TGCATTTGGA AATCAATAAA CTATTACTGG AAATGCCAAA AAAAAAAAAA AAAAAAAAAN 2820
NAAAAAACTC GAGGGGGGCC CGTACCC 2847
(2) INFORMATION FOR SEQ ID NO: 101:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1394 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 101:
GAGATTGGTC GAGGAGAGTA AATAATCTAG AGGCAAGAGT TCAGTGAGGG CCAAGGGGGA 60
CCCCCAGAAA AAGGTATGGA GCTAACTCAT CrCTTTTACA AGGGGTGGCC ATGACTTACT 120
GTTGCAAAGT ACTCAGTGTA TATTTAATGT TGATTGTTGA ATTTTAGTTA CGAGAGGGAA 180
GAACAATTTT ACTTCTGTCC TTATTTCACT TGCTGAAAAG CTGTGGGACA AAATGTATGG 240
AATAGACAAG GCCACTTTCT TTCTCATTTC TGCTTTTCAT GCATATTATT TTATTTACCC 300
ATAATTTCCA AGAGGTTTGG CGTTCCGCTC TCCTGCTTTT TTCTTTCATC CACCCCTTTC 360
CTTTTTTTGG AAGGGGGTTA TATATGAGAG TTCATTGAAG AAGTCCAGTG AGGCTGAAGT 420
AAAGGGGCAA GATAGGGCAG TTAACTAAAG AGCACTTTAT TTCTTTGAAG CCTTTCTAAG 480
AAAGAAATGG GGGTGCGAGT GGCTTGAATC TCCCATGATG TTGGAGGGCA CTTAGTGGGG 540
TTGAAGTATG ACATAATATT TCCCATTGGG GAAAGGAGAA TTTCTCTTAG AGGGTGGCAA 600
AATGCCTTTC CCCAGTGTCC CTATTTTAGG CATCTTTTCC TTCCTTATTC CTTCCAGTCA 660
GGGTGTGTCC TATACAAAAC TTCCCATCAG TTCTCCTCAA TATTCCCCAT TTGTAAATGA 720
TCACTTCTCT TTTCTAAACC CTTTTCCTGT TCAGATCCAT ACAGGATTTG CAAGGGTAGG 780
ATCATACATG CAAATGCCCC TTGTTCATCT GTGTCTTCTG CAAACTAGTC TCATGAAGAA 840
TTCTGGCGTG CAGCAGGGTA GCTGAAGTTT GGGTCTGGGA CTGGAGATTG GCCATTAGGC 900
NTCNCTGAGA TTCCAGCTCC CTTCCACCAA GCCCAGTCTT GCTACGTGGC ACAGGGCAAA 960
CCTGACTCCC TTTGGGCCTC AGTTTCCCCT CCCCTTCATG AAATGAAAAG AATACTACTT 1020
TTTCTTGTTG GTCTAGCATT GCTGGACACA AAGTGTAGTC ATTATTGTTG TATTGGGTGA 1080
TGTGTGCAAA ACTGCAGAAG CTCACTGCCT ATAAGAGGAA ATAAGAGAGA AAGTGGAGGA 1140
GAGGGACAAA AGGAGTAATT ATTTGGTATA GATCCACCCA TCCCAACCTT TCTCTCCTCA 1200
GTCCCTGCTC CTCATGTTTC TGGTTTGGTG AGTCCTTTGT GCCACCACCC ATAATCCTTT 1260
GCATTGCTGC ATCCTGGGAA GGGGGTATAT GGTCTCACAA GTTCTTGTCA TTGTTTTTTT 1320
GCATGCTTTC TTAATAAAAA AAAAAAAAAA ATGTTTANAG TTTTATCTTA AAAAAAAAAA 1380
AAAAAAAAAA ACCC 1394
(2) INFORMATION FOR SEQ ID NO: 102:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 794 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 102: GGMRCGAGGC GGAGTAAAGG GACTTGAGCG AGCCAGTTGC CGGATTATTC TATTTCCCCT 60
CCCTCTCTCC CGCCCCGTAT CTCTTTTCAC CCTTCTCCCA CCCTCGCTCG CGTACCATCG 120
CGGAGCGTCG GCGGCCACTC AGTCCCATTC CATCTCCTCG TCGTCCTTCG GAGCCGAGCC 180
GTCCGCGCCC GGCGGCGGCG GGAGCCCAGG AGCCTGCCCC GCCCTGGGGA CGAAGAGCTG 240
CAGCTCCTCC TGTGCGGTGC ACGATCTGAT TTTCTGGAGA GATGTGAAGA AGACTGGGTT 300 TGTCTTTGGA CACGCTGATC ATGCTGCTTT CCCTGGCAGC TTTCAGTGTC ATCARTGTGG 360
GTTTCTTAMC TCATCCTGGC TCTTCTCTCT GTCACCATCA RCTTCAGGAT CTACAAGTCC 420
GTCATCCAAG CTGTWCAGAA RTCAGAARAA GGCCATCCAW TCCAAAGCCT ACCTGGACGT 480
AGACATTACT CTCTCCTCAG AAGCTTTCCA TAATTACATG AATGCTGCCA TGGTGCACAT 540
CAACAGGGCC CTGAAACTCA TTATTCGTCT CTTTCTGGTA GAAGATCTGG TTGACTCCTT 600 GAAGCTGGCT GTCTTCATGT GGCTGATGAC CTATGTTGGT GCTGTTTTTA ACGGAATCAC 660
CCTTCTAATT CTTGCTGAAC TGCTCATTTT CAGTGTCCCG ATTGTCTATG AGAAGTACAA 720
GACCCAGATT GATCACTATG TTGGCATCGC CCGAGATCAG ACCAAGTCAA TTGTTGAAAA 780
GATCCCAAGC AAAA 794
(2) INFORMATION FOR SEQ ID NO: 103:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1544 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 103:
TTTGCTTGCT AGTCTGAACC AAAGAGTTGT TTGGGCATTT GCTGTGTTCG CCATTTCTGG 60
AGCAAGAGGG TCTTCTTCCT CCTTCCCCCA GCCAGCCAGC TGTCCTGGGG CCAGGCTTTC 120 CTGGGTGGAA AGAAGTATAC CTTTCCCTGG GGCCCTAGGA TAGCAAAGTC AGCCATAGTG 180
GGCCAGGCTG CCCTCCATGC TGGGCCCCAG CCCAGGTCTG CACTCGCCTG GATCACCTTC 240
TTTGAGCCTT AGCCATCTCC TGTCAGGTAG GAATGAACTT GCCAGCCTTC AGGYTCGTTC 300
AGCTATGACC ATCTGTGCGG TCAGGGTACA CTCAGCTCTC CTCCCCAACT CCAGCAGCCT 360
TTAAGAAGTG TCCCTTTGGC GCCCCCTGGA GGCAGAGCAC TGAGCTGGAC CCTGGGTAGA 420 CTCCCACAGG GAGGACGGAG CTGGCCTCAG GAGTGGGACA CCCAGACTTG GCAGGGCCTT 480
CAAGAGGCCT GTGTGGGGGC CCCAGGAATC CTTAGCTGAA GCGGGGAGAC TCACTCTCCA 540 TCTCAGGAAA TTCTAGCCCT TGCCCTCAGG GAGCCACGGT TGAGGGTGAG GCCCAACACC 600 TGCCTTAGGG CCCTGGGTGG GCAAGTCTGG GCCCTGGGGT AGGGAGGGAG ACTCAGGCCC 660 ACACTTGGGT ATTTTCTAAT TTCAGACAAA CACACACTCA GCGCGCACTC ACTGATTCCT 720 ACACATTGCC AAGATTTCAC ACATGTGACC AGGGGCCACC AAAGTCCCTG TGACCTTTGT 780 GACTAGGATC CTAATTTCTC TATTTTCTCC TGGGTGCCTG GGTCTGTGTC ACCTGGGGCA 840 GTCTGGATAA TGTTTAGTTC TGTGACACTG TTTTTTGGGG GTGGCACCTG GTTCTCCGAT 900 GCCTGGGCTG GTGTCAGGCC CAGGACTGTA GTGCTGGGAG CAGTAAAGCT CAGCTCTGTG 960 TAATGAGTGA TGCTATGGCT TGCTCGTGTC TTATGATCCA ATCCTTTTCT ACATCAGCCC 1020 TTGTTTTGTT TTATGGCTAG TCTTATCTGG CCTGGTTATT TCCTTGCGGG GAGGAGAGGG 1080 TTTGCTAATC TGCTCCCAGC CCAACCTATT ACCACCCCAC CTCGCTGGGA CCTACTGCTC 1140 GGGAGGCAGC AGACAGGGAG CCACCAGCAG TGGCTTCCTG GCCCTGTGCT GGGGGTGGGG 1200 GGAAGCTGGG GGCACATGTG GCCCTTGCCT TCTGAGCAGC TCCCAGTGCC AGGC-CTTTGA 1260 GACTTTCCCA CATGATAAAA GAAAAGGGAG GTACAGAAGT TCCAATTCCC TTTTTATTTT 1320 GCTGGTTGGT ATCTGTAAAT GTTTAATAAA TATCTGAGCA TGTATCTATC AACGCCAAGA 1380 ATTTCAAAGT CTCCTTCAAC AATATGAGGC TTTTAGGATG TTTATATTCC TTCATCCCTC 1440 TTGTTTCCCA GGTTTTGCAG GGAAAAAAAG TCTGGAATTA TAGATACAGC TTATTATTAA 1500 ATTTGTTCTT GCATAAAAAA AAAAAAAAAA AACNCNNGGG GGGG 1544
(2) INFORMATION FOR SEQ ID NO: 104:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 871 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 104: ACCCACGCGT CCGNCTTGTC CACCCGGGGG CGTGGGAGTG AGGTACCAGA TTCAGCCCAT 60 TTGGCCCCGA CGCCTCTGTT CTCGGAATCC GGGTGCTGCG GATTGAGGTC CCGGTTCCTA 120 AGGTGGGTCG CTGTCCACCC GGGGGCGTGG GAGTGAGGTA CCAGATTCAG CCCATTTCGC 180 CCCGACGCCT CTGTTCTCGG AATCCGGGTG CTGCGGATTG AGGTCCCGGT TCCTAACGGA 240 CTGCAAGATG GAGGAAGGCG GGAACCTAGG AGGCCTGATT AAGATGGTCC ATCTACTGGT 300
CTTGTCAGGT GCCTGGGGCA TGCAAATGTG GGTGACCTTC GTCTCAGGCT TTCCTGCTTT 360
TCCGAAGCCT TCCCCGACAT ACCTTCGGAC TAGTGCAGAG CAAACTCTTC CCCTTCTACT 420 TCCACATCTC CATGGGCTGT GCCTTCATCA ACCTCTGCAT CTTGGCTTCA CAGCATGCTT 480
GGGCTCAGCT CACATTCTGG GAGGCCAGCC AGCTTTACCT GCTGTTCCTG AGCCTTACGC 540
TGGCCACTGT CAACGCCCGC TGGCTGGAAC CCCGCACCAC AGCTGCCATG TGGGCCCTGC 600
AAACCGTGGG AGAAGGAGCG AGGCCTGGGT GGGGAGGTAC CAGGCAGCCA ACAGGTTCCC 660
GATCCTTAAC GCCAGNTGCG AGAGAAGGAC CCCAAGTACA GTGCTCTCCG CCAGAATTTC 720 TTCCGCTACC ATGGGCTGTC CTCTCTTTGC AATCTGGGCT GCGTCCTGAG CAATGGGCTC 780
TGTCTCGCTG GCCTTGCCCT GGAAATAAGG AGCCTCTAGC ATGGGCCCTG CATGCTAATA 840
AATGCTTCTT CAGAAAAAAA AAAAAAAAAA A 871
(2) INFORMATION FOR SEQ ID NO: 105:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 404 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 105:
GGCACGAGTT ATAGCATGGC ATTCATACTT TTGTTTTATT GCCTCATGAC TTTTTTGAGT 60
TTAGAACAAA ACAGTGCAAC CGTAGAGCCT TCTTCCCATG AAATTTTGCA TCTGCTCCAA 120
AACTGCTTTG AGTTACTCAG AACTTCAACC TCCCAATGCA CTGAAGGCAT TCCTTGTCAA 180 AGATACCAGA ATGGGTTACA CATTTAACCT GGCAAACATT GAAGAACTCT TAATGTTTTC 240
TTTTTAATAA GAATGACGCC CCACTTTGGG GACTAAAATT GTGCTATTGC CGAGAAGCAG 300
TCTAAAATTT ATTTTTTTAA AAAGAGAAAC TGCCCCATTA TTTTGGTGGG GTTGGTTTTT 360
AATTTNTAAT NTGAAAAATT TTTTTGGGGT TTTTGGGGCC ATGG 404
(2) INFORMATION FOR SEQ ID NO: 106:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1542 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 106:
GTCAGACAGG TGGAGCCGCC GGGGCAGGAG TCTCAAAGAG CCAGGCTCCA GGAGAGGAAG 60
GGCTCTRCGA GAGGAGAGAG GAGAGCGCTG GAGAGGAGAG GCTGGAGAGT CCTTAGCCAG 120
GATGGAGGCT GTTGTGAACT TGTACCAAGA GGTGATGAAG CACGCAGATC CCCGGATCCA 180
GGGCTACCCT CTGATGGGGT CCCCCTTGCT AATGACCTCC ATTCTCCTGA CCTACGTGTA 240
CTTCGTTCTC TCACTTGGGC CTCGCATCAT GGCTAATCGG AAGCCCTTCC AGCTCCGTGG 300
CTTCATGATT GTCTACAACT TCTCACTGGT GGCACTCTCC CTCTACATTG TCTATGAGTT 360
CCTGATGTCG GGCTGGCTGA GCACCTATAC CTGGCGCTGT GACCCTGTGG ACTATTCCAA 420
CAGCCCTGAG GCACTTAGGA TGGTTCGGGT GGCCTGGCTC TTCCTCTTCT CCAAGTTCAT 480
TGAGCTGATG GACACAGTGA TCTTTATTCT CCGAAAGAAA GACGGGCAGG TGACCTTCCT 540
ACATCTCTTC CATCACTCTG TGCTTCCCTG GAGCTGGTGG TGGGGGGTAA AGATTGCCCC 600
GGGAGGAATG GGCTCTTTCC ATGCCATGAT AAACTCTTCC GTGCATGTCA TAATGTACCT 660
GTACTACGGA TTATCTGCCT TTGGCCCTGT GGCACAACCC TACCTTTGGT GGAAAAAGCA 720
CATGACAGCC ATTCAGCTGA TCCAGTTTGT CCTGGTCTCA CTGCACATCT CCCAGTACTA 780
CTTTATGTCC AGCTGTAACT ACCAGTACCC AGTCATTATT CACCTCATCT GGATGTATGG 840
CACCATCTTC TTCATGCTGT TCTCCAACTT CTGGTATCAC TCTTATACCA AGGGCAAGCG 900
GCTGCCCCGT GCACTTCAGC AAAATGGAGC TCCAGGTATT GCCAAGGTCA AGGCCAACTG 960
AGAAGCATGG CCTAGATAGG CGCCCACCTA AGTGCCTCAG GACTGCACCT TAGGGCAGTG 1020
TCCGTCAGTG CCCTCTCCAC CTACACCTGT GACCAAGGCT TATGTGGTCA GGACTGAGCA 1080
GGGGACTGGC CCTCCCCTCC CCACAGCTGC TCTACAGGGA CCACGGCTTT GGTTCCTCAC 1140
CCACTTCCCC CGGGCAGCTC CAGGGATGTG GCCTCATTGC TGTCTGCCAC TCCAGAGCTG 1200
GGGGCTAAAA GGGCTGTACA GTTATTTCCC CCTCCCTGCC TTAAAACTTG GGAGAGGAGC 1260
ACTCAGGGCT GGCCCCACAA AGGGTCTCGT GGCCTTTTTC CTCACACAGA AGAGGTCAGC 1320
AATAATGTCA CTGTGGACCC AGTCTCACTC CTCCACCCCA CACACTGAAG CAGTAGCTTC 1380
TGGGCCAAAG GTCAGGGTGG GCGGGGGCCT GGGAATACAG CCTGTGGAGG CTGCTTACTC 1440
AACTTGTGTC TTAATTAAAA GTGACAGAGG AAACCANAAA AAAAAAAAAA AAAAACTCGA 1500
GGGGGGCCCG TACCCAAATC GCCGGTATGA TCGTAAACAA TC 1542
(2) INFORMATION FOR SEQ ID NO: 107:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2327 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 107:
GGTAGCTCAN TGCAGTGAAA TAGTCTTACT GGAAACAAAG CCCTTTATCA AGAATAATTA 60
ACTCTTCCCT TTTCTTTTTG GAGAGGTGCT TTGTTTCTGA TCGGACCATT TCACTGCAGC 120
AAGCAACACA GTATTCTRAG CAGAAGATCG GGACTTGAGG CCATGTTGCG GAGGGCCAGT 180
RACATTATCT GGACTCTGGA GTGTGAGGAA TATGGACTCC ACTCTTCACT ATATTCACAR 240
CGATTCAGAC TTGAGCAACA ATAGCAGTTT TAGCCCTGAT GAGGAAAGGA GAACTAAAGT 300
ACAAGATGTT GTACCTCAGG CGTTGTTAGA TCAGTATTTA TCTATGACTG ACCCTTCTCG 360
TGCACAGACG GTTCACACTG AAATTGCTAA GCACTGTGCA TATAGCCTCC CTGGTGTGGC 420
CTTGACACTC GGAAGACAGA ATTGGCACTG CCTGAGAGAG ACGTATGRGA CTYTGGCCTC 480
AGACATGCAG TGGAAAGTTC GACGGAACTC TAGCATTCTC CATCCACGRG CTTGCAGTTA 540
TTCTTGGAGA TCAATTGACA GCTGCAGATC TGGTTCCAAT TTTTAATGGA TTTTTAAAAG 600
ACCTCGATGA AGTCAGGATA GGTGTTCTTA AACACTTCCA TGATTTTCTG AAGCTTCTTC 660
ATATTGACAA AAGAAGAGAA TATCTTTATC AACTTCAGGA GTTTTTGGTG ACAGATAATA 720
GTAGAAATTG GCGGTTTCGA GCTGAACTGG CTGAACAGCT GATTTTACTT CTAGAGTTAT 780
ATAGTCCCAG AGATGTTTAT GACTATTTAC GTCCCATTGC TCTGAATCTG TGTGCAGACA 840
AAGTTTCTTC TGTTCGTTGG ATTTCCTACA AGTTGGTCAG CGAGATGGTG AAGAAGCTGC 900
ACGCGGCAAC ACCACCAACG TTCGGAGTGG ACCTCATCAA TGAGCTTGTG GAGAACTTTG 960
GCAGATGTCC CAAGTGGTCT GGTCGGCAAG CCTTTGTCTT TGTCTGCCAG ACTGTCATTG 1020
AGGATGACTG CCTTCCCATG GACCAGTTTG CTGTGCATCT CATGCCGCAT CTGCTAACCT 1080
TAGCAAATGA CAGGGTTCCT AACGTGCGAG TGCTGCTTGC AAAGACATTA AGACAAACTC 1140
TACTAGAAAA AGACTATTTC TTGGCCTCTG CCAGCTGCCA CCAGGAGGCT GTGGAGCAGA 1200
CCATCATGGC TCTTCAGATG GACCGTGACA GCGATGTCAA GTATTTTGCA AGCATCCACC 1260
CTGCCAGTAC CAAAATCTCC GAAGATGCCA TGAGCACAGC GTCCTCAACC TACTAGAAGG 1320
CTTCAATCTC GGTGTCTTTC CTGCTTCCAT GAGAGCCGAG GTTCAGTGGG CATTCGCCAC 1380
GCATGTGACC TGGGATAGCT TTCGGGGGAG GAGAGACCTT CCTCTCCTGC GGACTTCATT 1440
GCAGGTGCAA GTTGCCTACA CCCAATACCA GGGATTTCAA GAGTCAAGAG AAAGTACAGT 1500
AAACACTATT ATCTTATCTT GACTTTAAKG KKWAWKMMWW KCTCAGMSRA TTATAMTTSW 1560
CWMMRARGSM WYMAAWSCTK SWGCTCYWCC KSRSTGRMKG MMRCTCTAGA AYTRGYRGAK 1620
CMYYYKSGCT KMWGGAAKKS GGCASGAGCC AGAGACCTGC ATTGCTTTCT CCTGGTTTTA 1680
TTTAACAATC GACAAATGAA ATTCTTACAG CCTGAAGGCA GACGTGTGCC CAGATGTGAA 1740
AGAGACCTTC AGTATCAGCC CTAACTCTTC TCTCCCAGGA AGGACTTGCT GGGCTCTGTG 1800
GCCAGCTGTC CAGCCCAGCC CTGTGTGTGA ATCGTTTGTG ACGTGTGCAA ATGGGAAAGG 1860
AGGGGTTTTT ACATCTCCTA AAGGACCTGA TGCCAACACA AGTAGGATTG ACTTAAACTC 1920
TTAAGCGCAG CATATTGCTG TACACATTTA CAGAATGGTT GCTGAGTGTC TGTGTCTGAT 1980
TTTTTCATGC TGGTCATGAC CTGAAGGAAA TTTATTAGAC GTATAATGTA TGTCTGGTGT 2040
TTTTAACTTG ATCATGATCA GCTCTGAGGT GCAACTTCTT CACATACTGT ACATACCTGT 2100
GACCACTCTT GGGAGTGCTG CAGTCTTTAA TCATGCTGTT TAAACTGTTG TGGCACAAGT 2160
TCTCTTGTCC AAATAAAATT TATTAATAAG ATCTATAGAG AGAGATATAT ACACTTTTGA 2220
TTGTTTTCTA GATGTCTACC AATAAATGCA ATTTGTGACC TGTAAAAAAA AAAWAAAAAA 2280
ACTCGAGGGG GGCCCGGTAC CCAAATCGCC GATATGATCT AANCATC 2327
(2) INFORMATION FOR SEQ ID NO: 108:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1062 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO 108: GGCCGCCGAG GCGCAACAGC CGTTCTGTCA GCTCTGGGTC CAACCGGACT AGCGAANATC 60 TTCCTCATCC TCATCATCGT CTTCCTCATC CCGATCTCGG TCCAGGTCCC TCTCCCCCCC 120 ACACAAGAGG TGGCGAAGGT CCAGCTGTAG TTCCTCTGGA CGTTCTCGAA GATGCTCTTC 180 CTCTTCTTCG TCATCATCTT CCTCTTCGTC TTCCTCATCC TCATCATCCA GTTCTCGAAG 240 CCGCTCACGA ATCCCCATCC CCCCGCCGGA GRAAGTGACA GGAGGCGGCG GTACAGCTCT 300 TATCGTTCAC ATGACCATTA CCAAAGGCAA AGAGTGCTAC AAAAGGAGCG TGCAATAGAA 360 GAAAGAAGGG TGGTCTTCAT TGGAAAGATA CCTGGCCGCA TGACTCGATC AGAGCTGAAA 420 CAGAGGTTCT CCGTTTTTGG AGAGATTGAG GAGTGCACCA TCCACTTCCG TGTCCAAGGG 480 GACAACTACG GCTTCGTCAC TTATCGCTAT GCTGAGGAGG CATTTGCAGC CATTGAGAGT 540 GGCCACAAGC TGCGGCAGGC AGATGAGCAG CCCTTTGATC TCTGCTTTGG GGGCCGAAGG 600 SWGTNCTGCA AGAGGAGCTA TTCTGATCTT GACTCCAACC GGGAAGACTT TGACCCAGCA 660 CCTGTAAAGA GCAAATTTGA TTCTCTTGAC TTTGACACAT TGTTGAAACA GGCCCAGAAG 720
AACCTCAGGA GGTAACCTTG GGCCCTTCCC TGCTATCCTT TTTCTCCTTT GGAGGTGCCC 780
AACCTCCTCC ACCCCCTTCC CCTACTCTAG GGGAGAGAGC TGCTAGTGAG ATGACTGTTT 840
TATAAAGAAA TGGAAAAAAG TGAAATAAAA AATATGTTGA ATCAGATTTT TTAAAAGGGG 900
TATTTCTTTT TTTATAACAG GTATTGAAAC AAGTTAACTT GCATTCCTAT GTAAGATAGG 960
AGGGGCTGAG GGGATCCCCA GTGTTTGGAA CATAAGTCAC TATGCAGACT AATAAACATC 1020
AACTAGAGAG NAAAAAAAAA AAAAAAAAAA ATTTAAAAAA CT 1062
(2) INFORMATION FOR SEQ ID NO: 109:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2539 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 109:
GAGAGACTCA CACTTCTTTT CCATTATCAC TGACGATGTA GTGGACATAG CAGGGGAAGA 60
GCACCTACCT GTGTTGGTGA GGTTTGTTGA TGAATCTCAT AACCTAAGAG AGGAATTTAT 120
AGGCTTCCTG CCTTATGAAG CCGATGCAGA AATTTTGGCT GTGAAATTTC ACACTATGAT 180
AACTGAGAAG TGGGGATTAA ATATGGAGTA TTGTCGTGGC CAGGCTTACA TTGWCTCTAG 240
TGGATTTTCT TCCAAAATGA AAGTTGTTGC TTCTAGACTT TYAAGMKMRA TWKCCCCMAK 300
Y AWCKGAAC AMAMKCTGSW CYTCCWSYGC SKTRRMKRYC GYKSTATRRC WARWKSAKYM 360
CCYGKKMTGS RRGTAWYTSK TGCAYKAGGG AACAATTGAG GAAGTTTGTT CTTTTTTCCA 420
TCGATCACCA CAACTGCTTT TAGAACTTGA CAACGTAATT TCTGTTCTTT TTCAGAACAG 480
TAAAGAAAGG GGTAAAGAAC TGAAGGAAAT CTGCCATTCT CAGTGGACAG GCAGGCATGA 540
TGCTTTTGAA ATTTTAGTGG AACTCCTGCA AGCACTTGTT TTATGTTTAG ATGGTATAAA 600
TAGTGACACA AATATTAGAT GGAATAACTA TATAGCTGGC CGAGCATTTG TACTCTGAGT 660
GCAGTGTCAG ATTTTGATTT CATTGTTACT ATTGTTGTTC TTAAAAATGT CCTATCTTTT 720
ACAAGAGCCT TTGGGAAAAA CYYCMAGGGG CAAACCTCTG ATGTCTTCTT TGCKKMMSRT 780
ARMTTTTGAY ATRMARYACT RMMTKSAYTY AAYGRWGTGA CWSGAWAATA TTRAASTYTA 840
TACAATKAAT YWTRRYTTSM KRMAGMYAAT CCGAAAYTGT GGMAAMYAAA CTTGATATTC 900
AAATGAAACT CCCTGGGAAA TTCCGCAGAG CTCACCAGGG TAACTTGGAA TCTCAGCTAA 960
CCTCTCAGAG TTACTATAAA GAAACCCTAA GTGTCCCAAC AGTGGAGCAC ATTATTCAGG 1020
AACTTAAAGA TATATTCTCA GAACAGCACC TCAAAGCTCT TAAATGCTTA TCTCTGGTAC 1080
CCTCAGTCAT GGGACAACTC AAATTCAATA CGTCGGAGGA ACACCATGCT GACATGTATA 1140
GAAGTGACTT ACCCAATCCT GACACGCTGT CAGCTGAGCT TCATTGTTGG AGAATCAAAT 1200
GGAAACACAG GGGGAAAGAT ATAGAGCTTC CGTCCACCAT CTATGAAGCC CTCCACCTGC 1260
CTGACATCAA GTTTTTTCCT AATGTGTATG CATTGCTGAA GGTCCTGTGT ATTCTTCCTG 1320
TGATGAAGGT TGAGAATGAG CGGTATGAAA ATGGACGAAA GCGTCTTAAA GCATATTTGA 1380
GGAACACTTT GACAGACCAA AGGTCAAGTA ACTTGGCTTT GCTTAACATA AATTTTGATA 1440
TAAAACACGA CCTGGATTTA ATGGTGGACA CATATATTAA ACTCTATACR AKTAMGTCAG 1500
MGCTYYCTAC AKAYRAYTCM SWAWMTGTGG AAARYWSSTA MGMSWGCWKK TAMMRRTMCG 1560
GMWWTYYYMK RKTYGAYMYW YGCGWMCGAG AAAAAGCCGT AAGGTGTATG TAGACCACTT 1620
AATCACTAAA TATCTTTGCC TATAGGACTC CATTGAATAC ATTAGCCATT GATAATCTAC 1680
CTGTTTAAAT GGCCCCTGTT TGAACTCTCA AGCTTTGAAG ACCTACCTGT TCTTCCAGAA 1740
GAGAACGTTG AAAGTGCCAT GTTTCCTTTT GCGTGATCTC TGTTGATGGC ACTCTGGAAT 1800
TGTTTCCAGT TTAAKTCATT TTAGACATAG CATTTATTAT CACTGTGGAT CTCTACTTGT 1860
TGGGTGTTAT GAATTCTTTG AAGAATATAT TTTGAAGAGG TGTGGGAGGA AGGAATACAT 1920
TTTATAAAAT GTTGTAGTGA AGCCCACAAT TGACCTTKGA CTAATAGGAG TTTTAAGTAT 1980
GTTAAAAATC TATACTGGAC AGTTACAAGA AATTACCGGA GAAAAGCTTG TGAGCTCACC 2040
AAACAAGGAT TTCAGTGTAG ATTTTGTCTT TCTTGAACTT AAAGAAACAA ATGACAAAGT 2100
TTGAATGGAA AAGCCTGCTG TTGTTCCACA TCTCGTTGCT GTTTACATTC CTTTGTGGAG 2160
CCTACATCTT CCTAAGCTTT TTAGCAGGTA TATGTTGAAC ACTTCTGTTT CATGGTTGAG 2220
ACAGAATCAG AGGCCATGGA TACTGACAAC TGATTTGTCT GTTTTTTTTC TCTGTCTTTT 2280
TCCATGACTC TTATATACTG CCTCATCTTC ATTTATAAGC AAAACCTGGA AAACCTACAA 2340
AATAAGTGTT GTGGTTTATC TAGAAAAATA TGGAAAATAT TGCTGTTATT TTTGGTGAAG 2400
AAAATCAATT TTGTATAGTT TATTTCAATC TAAATAAAAT GTGAATTTTG TTWWATTAAA 2460
AATTWGGSAC AAABTBGHGG GGGDTCCAAA CHTWVTCGHG KAAMTTCTCT WAARMATYTK 2520
ATAAACMSCT TCACAATTC 2539
(2) INFORMATION FOR SEQ ID NO: 110:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1751 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 110:
AGCATGAAGC CGATGGCCGT GGTGGCCAGT ACCGTCCTGG GCCTGGTGCA AAACATGCGT 60
GCGTTTGGCG GGATCCTGGT GGTGGTCTAC TACGTATTTG CCATCATTGG GATCAACTTG 120
TTTAGAGGCG TCATTGTGGC TCTTCCTGGA AACAGCAGCC TGGCCCCTGC CAATGGCTCG 180
GCGCCCTGTG GGAGCTTCGA GCAGCTGGAG TACTGGGCCA ACAACTTCGA TGACTTTGCG 240
GCTGCCCTGG TCACTCTGTG GAACTTGATG GTGGTGAACA ACTGGCAGGT GTTTCTGGAT 300
GCATATCGGC GCTACTCAGG CCCGTGGTCC AAGATCTATT TTGTATTGTG GTGGCTGGTG 360
TCGTCTGTCA TCTGGGTCAA CCTGTTTCTG GCCCTGATTC TGGAGAACTT CCTTCACAAG 420
TGGGACCCCC GCAGCCACCT GCAGCCCCTT GCTGGGACCC CAGAGGCCAC CTACCAGATG 480
ACTGTGGAGC TCCTGTTCAG GGATATTCTG GAGGAGCCCG GGGAGGATGA GCTCACAGAG 540
AGGCTGAGCC AGCACCCGCA CCTGTGGCTG TGCAGGTGAC GTCCGGGCTG CCATCCCAGC 600
AGGGGCGGCA GGAGAGAGAG GCTCGCCTAA CACAGGTGCC CATCATGGAA GAGGCGGCCA 660
TGCTGTGGCC AGCCAGGCAG GAAGAGACCT TTCCTCTGAC GGACCACTAA GCTGGGGACA 720
GGAACCAAGT CCTTTGCGTG TGGCCCAACA ACCATCTACA GAACAGCTGC TGGTGCTTCA 780
GGGAGGCGCC GTGCCCTCCG CTTTCTTTTA TAGCTGCTTC AGTGAGAATT CCCTCGTCGA 840
CTCCACAGGG ACCTTTCAGA CAAAAATGCA AGAAGCAGCG GCCTCCCCTG TCCCCTGCAG 900
CTTCGGTGGT GCCTTTGCTG CCGGCAGCCC TTGGGGACCA CAGGCCTGAC CAGGGCCTGC 960
ACAGGTTAAC CGTGAGTCTG TCTCATCTAT TCACAGCTGG GAATGATACT AATACCTCCG 1020
ATTTTAGCCC AGCACCACAG GGTACGTTCC AGTTTTTCTC TCTTTCCATA GCTGTAAGGC 1080
CCTTTCTGGG AATGGTTCTC ATTCTCCTTA ATCTATTATT GGGTCAGTTT TCCTGCATGT 1140
CCCCAGCCTC CCATCACTGC CACCCACTCC CCACAGAGAT GCCCTGCTCA TCCGACTGGG 1200
GCTTTGACTC CCACACTGTC TACCCCTCTT GTGTGGACGC CCTGCTGCCA AAACCTTCAG 1260
CAAACAGCTT TCCAAATGGA AGTTGTCACT GTCAGGCCTT TACAATCAGC AACAGCAAAA 1320
TCTACATGCT GCTGAGGGTC CTGCCTCATT AAGATGCAAT AAATATGTAA GTACATAAAA 1380
ACAGCAATAG AAGAAACGTA ATGCTTTATT CTCAAATATG ATGTCTACAT AGAAAAGCCA 1440
AAATTATTAA GAATAGTAAG AATTCACCCA GCACTTTGGG AGGCCGAGGC GGGTGGATCA 1500
TGAGGTCAGG AGATCGAGAC CATCCTGGCT AACAGGGTGA AACCCCGTCT CTACTAAAAA 1560
TACAAAAAAT TGGCCGGGCG CAGTGGCGGG CGCCTGTGGT CCCAGCTACT GGGGAGGCTG 1620
AGGCAGGAGA ATGGCGTGAA CCCGGGAAGC GGAGCTTGCA GTGAGCCGAG ATTGCGCCAC 1680
TGCAGTCCGC AGTCCAGCCT GGGCGACAGA GCGAGACTCC GTCTCAAAAA AAAAAAAAAA 1740 AAAAAAAAAA A 1751
(2) INFORMATION FOR SEQ ID NO: 111:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1117 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 111:
AATGTTGTGG TCGTAGCATT TGGGTTAATT CTRATTATAG AGTCTCTTGG AGAGCAATGT 60
CCATAAACTA ATCCCAAACA ACATTGTCTT TTTRATGTTG TAGTGAACAG CAGAGAATTT 120
CAAAGGACCT TGCTAATATC TGTAAGACGG CAGCTACAGC AGGCATCATT GGCTGGGTGT 180
ATGGGGGAAT ACCAGCTTTT ATTCATGCTA AACAACAATA CATTGAGCAG AGCCAGGCAG 240
AAATTTATCA TAACCGGTTT GATGCTGTGC AATCTGCACA TCGTGCTGCC ACACGAGGCT 300
TCATTCGTTA TGGCTGGCGC TGGGGTTGGA GAACTGCAGT GTTTGTGACT ATATTCAACA 360
CAGTGAACAC TAGTCTGAAT GTATACCGAA ATAAAGATGC CTTAAGCCAT TTTGTAATTG 420
CAGGAGCTGT CACGGGAAGT CTTTTTAGGA TAAACGTAGG CCTGCGTGGC CTGGTGGCTG 480
GTGGCATAAT TGGAGCCTTG CTGGGCACTC CTGTAGGAGG CCTGCTGATG GCATTTCAGA 540
AGTACTCTGG TGAGACTGTT CAGGAAAGAA AACAGAAGGA TCGAAAGGCA CTCCATGAGC 600
TAAAACTGGA AGAGTGGAAA GGCAGACTAC AAGTTACTGA GCACCTCCCT GAGAAAATTG 660
AAAGTAGTTT ACAGGAAGAT GAACCTGAGA ATGATGCTAA GAAAATTGAA GCACTGCTAA 720
ACCTTCCTAG AAACCCTTCA GTAATAGATA AACAAGACAA GGACTGAAAG TGCTCTCAAC 780
TTGAAACTCA CTGGAGAGCT GAAGGGAGCT GCCATGTCCG ATGAATGCCA ACAGACAGGC 840
CACTCTTTGG TCAGCCTGCT GACAAATTTA AGTGCTGGTA CCTGTGGTGG CAGTGGCTTG 900
CTCTTCTCTT TTTCTTTTCT TTTTAACTAA GAATGGGGCT GTTCTACTCT CACTTTACTT 960
ATCCTTAAAT TTAAATACAT ACTTATGTTT GTATTAATCT ATCAATATAT GCATACATGA 1020
ATATATCCAC CCACCTAGAT TTTAAGCAGT AAATAAAACA TTTCGCAAAA GATTAAAGTT 1080
GAATTTTACA GTTAAAAAAA AAAAAAAAAA AAAAAAA 1117
(2) INFORMATION FOR SEQ ID NO: 112:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1313 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 112:
GGCAGAGGTT TTCTTATATT TTAAGTAAAT TTAAAGTGGC TATCAGAATA TTTATTCTTG 60
TTTGAGACTA CCAACATAAC TACGTGTTGA AGGTGCTTCA CAGAGAATAT ATTGCCTTTA 120
ATGTGAAATA ATTTTCACCA ATCTTGCTAA CTTTAATAAA GTATAAAATT TCTAGAATAT 180
TCAGTTAAGT AGTTGGTAAC CCTTTTCTAT TTTAGTAAAA CTTAATGCAT GTTTACTTTT 240
TTTTCAAAGA TGCAGACAAT CTCTTTGAAC ATGAATTGGG GGCTCTCAAT ATGGCTGCAT 300
TACTACGAAA AGAAGAAAGA GCAAGTCTTC TTAGTAATCT TGGCCCATGT TGTAAGGCGT 360
TGTGCTTCAG ACGGGATTCT GCAATTCGAA AGCAGCTTGT TAAAAATGAG AAGGGCACCA 420
TAAAACAAGC TTACACGAGT GCTCCAATGG TAGACAATGA ATTACTTCGA TTGAGTCTTC 480
GGTTATTTAA GCGGAAGACT ACTTGCCATG CTCCAGGACA TGAAAAGACT GAAGATAATA 540
AACTTTCACA GTCCAGTATC CAACAGGAAC TGTGTGTGTC TTAAGACCGA AGTTACAATA 600
TGGTATTTTT GGTACTGTCT TCCTTCAGCA GTGCATATTC TTTTGCAAAG TTCTTTGGTT 660
TGACAAGCAT TAGTGACAAA GGCAGAAAAG ATTTATCAGC CATGCTAAAA GAGTGAAGAA 720
TTTTGATCTT TAGAGACACT AGTTTTGGCC AACTTAAGAT TTTACGTTAA TTTTTACATA 780
GTATTTGACA CTCATGCAAA ATAATGTGAA AACATCTAGA TTTAGTAGTT TATTCTGCGC 840
CTTTTGTTAA AACTGAAGAT TTTGGAAAAT GGTTGTCACT GCTCTTCCAG CCTATGAATA 900
TTTTTGTGAA ATGGAACCAT GGATTTATGT CTGGATCATC CATACAGAAC CAACAATTTT 960
ATTCAAAAAC AATGTGTTCA TCAAAGTAAT TGCTCACATT GTGCAGTACT ATGTTGTACA 1020
GACCACGTGA AAGGGAATGC TGGTCTAGCT GGCGTGGTAT GTTTATAGGC GAATTTCAGC 1080
AGAAGGAAGC CAAAATAGTT TTTTCCTTTT GΛAAAGTTTTT TAAAAATTAT TTCATGGGTC 1140
TTTTTTTTAA TTAATATGTG TGCATTGTTA CAATGTATGT TCGGATGTCT TTTGACCCTA 1200
AATGCTTTTT TTGTTATCAG AGATTGTGTA CTATTTTTAT TTTTAATAAA TGTATCTTCC 1260
CTTTTMAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAA 1313
(2) INFORMATION FOR SEQ ID NO: 113:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1654 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 113:
ACAGGGACAG AATACTTTCT TTCCTTCCTT CAAGTACAAG AAGGCTTTCT CTACCATTTG 60
CGTCTACACT TTATTTTAAA AGCTATCCTT TTCTAGTAGT ATTTTATCAT GGCAATGGCA 120
TGATGACAAC AACAGTCTTT CATTACAGAC TGAAGGGAAG CATGTCCTTA CTTAAAATAG 180
TTCTGCTACT TTCCCTCCTA TTATAAGGAA ATTTTACAGA TTCTAAAAAT ACCTTAATTT 240
TTCTTTGATT TTTATTTTAC CAAGTCACAA ATGTCTTTTT GATGTTTTGA GAATTGTTCT 300
CATAGAATCA CAAATACTGA CATTTCATTA GATGATTATT TTCCTAGAAT CCCCAAAGAG 360
CAGTGGCAGT CCATGGCTTG GTTGAAGCTA GAAATTTTCC TGCCCCTGGT GACCTGGTAA 420
GCCTCCTGCT CGGAACCGTG TGAGTGGGTG AGGAAGATGA GAGATGGTCA GATGGAAGAG 480
AGRAATACAT GAACTGCTCT GGCCTCTCTG GTTCTGTTCT TGGCCCAGAG TTTTTGAAAA 540
GCAGCGGANA TNGACTGACT TCACATGCTC AGCTTTCTCA GCCTTTTGTT TATTTTGTTG 600
TCCTTAGATT TCCCTGTTGT AAAAGGGGCA AGAAAAGTAA CTCATCATCT CTAACACACC 660
ATGGCAGCTT AGCCAGGTAG TCTTAGTGGT GGTGTTTAGG CATAAGATAT GCTGATCATC 720
AGTCTCAGGC CACAGTTTCC TTCACTAATC GTCCAGCTTG AGTGTTCTGT TCTCTTCCTG 780
CCCATTTCCT TGAACCTCCT GCTCTAGCCT TGGCGGAGGG AGAGTGCTAT TTCCTTTTCT 840
TCTCCCTCTG TCTTAGGAAA AGCCATCTTT AATATAGTTC TTCACCACTG TTGGGGTTGT 900
TTTGTGATTT TTTTTTTCTT CCGAAGAACT CCTGGTTGTT ATTGGATTTT GTATTTTAAT 960
ACAAATTATT GAATTTTATA AGCTTGTACA CAATATTTAA TTAGTGTGAA AGGAAACAAA 1020
GAATGCAGGA AAAATAATTT AATATCAACC TCAGTTGACA AGGTGCTCAG ATTATTCAAT 1080
TCGGGATCCT CCTTTTGTTA GGTTTTTGAG ACAACCCTAG ACCTAAACTG TGTCACAGAC 1140
TTCTGAATGT TTAGGCAGTG CTAGTAATTT CCTCGTAATG ATTCTGTTAT TACTTTCCTA 1200
TTCTTTATTC CTCTTTCTTC TGAAGATTAA TGAAGTTCAA AATTGAGGTG GATAAATACA 1260
AAAAGGTAGT GTGATAGTAT AAGTATCTAA GTGCAGATGA AAGTGTGTTA TATACATCCA 1320
TTCAAAATTA TGCAAGTTAG TAATTACTCA GGGTTAACTA AATTACTTTA ATATGCTGTT 1380
GAAYCTACTC TGTTCCTTGG CTAGAAAAAA TTATAAACAG GACTTTGTAG TTTGGGAAGC 1440
CAAATTGATA ATATTCTATG TTCTAAAAGT TGGGCTATAC ATAAATTATT AAGAAATATG 1500
GATTTTTATT CCCAGGATAT GGTGTTCATT TTATGATATT ACGCAGGATG ATGTATTGAG 1560
TAAAATCAGT TTTGTAAATA TGTAAATATG TCATAAATAA ACAATGCTTT GACTTATTTC 1620
CAAAAAAAAA AAAAAATAAA NTTCGAGGGG GGGC 1654
(2) INFORMATION FOR SEQ ID NO: 114:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1171 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 114:
GGCAAACTTT CCCCCAANGC TTCGAAACTT GCAAGCCGAA ACCTTGAATC GTTAAAAGTT 60
GGGTTGCGNC GGCGCCCTGG CCCGAAGAAG CGCAATTGGC GTTCCGCGAA CGTTGGCCCT 120
CAACGGCTCG GCAGCCAGCC ATGTCCTGCA CCCAGGACAG CGGCCCTGGG CTACAAGGAC 180
CTGGMCCTCA TCTTCCTGCG CCGACCTGCG CGGGGTAAGG GGWAGTTTCA GACTGTGAAG 240
GACGTCGTGC TGGACTGCCT GTTGGACTTC TTACCCGAGG GGGTGAACAA AGAGAAGATC 300
ACACCACTCA CGCTCAAGGA AGCTTATGTG CAGAAAATGG TTAAAGTGTG CAATGACTCT 360
GACCGATGGA GTCTTATATC CCTGTCAAAC AACAGTGGCA AAAATGTGGA ACTGAAATTT 420
GTGGATTCCC TCCGGAGGCA GTTTGAATTC AGTGTAGATT CTTTTCAAAT CAAATTAGAC 480
TCTCTTCTGC TCTTTTATGA ATGTTCAGAG AACCCAATGA CTGAGACATT TCACCCCACA 540
ATAATCGGGG AGAGCGTCTA TGGCGATTTC CAGGAAGCCT TTGATCACCT TTGTAACAAG 600
ATCATTGCCA CCAGGAACCC AGAGGAAATC CGAGGGGGAG GCCTGCTTAA GTACTGCAAC 660
CTCTTGGTGA GGGGCTTTAG GCCCGCCTCT GATGAAATCA AGACCCTTCA AAGGTATATG 720
TGTTCCAGGT TTTTCATCGA CTTCTCAGAC ATTGGAGAGC AGCAGAGAAA ACTGGAGTCC 780
TATTTGCAGA ACCACTTTGT GGGAATTGGA AGACCGCAAG TATGAGTATC TCATGACCCT 840
TCATGGAGTG GTAAATGAGA GCACAGTGTG CCTGATGGGA CATGAAAGAA GACAGACTTT 900
AAACCTTATC ACCATGCTGG CTATCCGGGT GTTAGCTGAC CAAAATGTCA TTCCTAATGT 960
GGCTAATGTC ACTTGCTATT ACCAGCCAGC CCCCTATGTA GCAGATGCCA ACTTTAGCAA 1020
TTACTACATT GCACAGGTTC AGCCAGTATT CACGTGCCAG CAACAGACCT ACTCCACTTG 1080
GCTACCCTGC AATTAAGAAT CATTTAAAAA TGTCCTGTGG GGAAGCCATT TCAGACAAGA 1140
CAGGAGAGAA AAAAAAAAAA AAAAAAAAAA A 1171
(2) INFORMATION FOR SEQ ID NO: 115: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 842 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 115:
GGTCTGCGCC GGAAGTGCAT GAGCTGCCGA TGTGGTGCTT AGTGATTGCG GTTTCGGTCG 60 CTCTCCCGTG TTTCCCGGGC TGGGTATTTG CCTCGCACCA TGGCGCCCAA GGGCAAAGTG 120
GGCACGAGAG GGAAGAAGCA GATATTTGAA GAGAACAGAG AGACTCTGAA GTTCTACCTG 180
CGGATCATAC TGGGGGCCAA TGCCATTTAC TGCCTTGTGA CGTTGGTCTT CTTTTACTCA 240
TCTGCCTCAT TTTGGGCCTG GTTGGCCCTG GGCTTTAGTC TGGCAGTGTA TGGGGCCAGC 300
TACCACTCTA TGAGCTCGAT GGCACGAGCA GCGTTCTCTG AGGATGGGGC CCTGATGGAT 360 GGTGGCATGG ACCTCAACAT GGAGCAGGGC ATGGCAGAGC ACCTTAAGGA TGTGATCCTA 420
CTGACAGCCA TCGTGCAGGT GCTCAGCTGC TTCTCTCTCT ATGTCTGGTC CTTCTGGCTT 480
CTGGCTCCAG GCCGGGCCCT TTACCTCCTG TGGGTGAATG TGCTGGGCCC CTGGTTCACT 540
GCAGACAGTG GCACCCCAGC ACCAGAGCAC AATGAGAAAC GGCAGCGCCG ACAGGAGCGG 600
CGGCAGATGA AGCGGTTATA GCCATTGACA TTGTGGCCAC AGGCCACTGG CCCTGGGTGG 660 CTCTGTGAGG GTGCACAGCC CCTCATGCCT GGAGCAATGA GGGTCTAGTC CAGGGGCCAA 720
AAGCAGTCTG AGGTATTGGG TATACTTATA CTCTATAGGG TCGTTGAATA AATGGCTTAG 780
AATGTGAAAA AAAAAAAAAA AAAAAACTCG AGGGGGGCCC GGTACCCAAT TTCNCCTANA 840
AT 842
(2) INFORMATION FOR SEQ ID NO: 116:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1640 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 116:
GGCACGAGGC GGCGGCAGCG GTGGCGGCGG CGCCCCCCGG CGGGAGCCGT TCCCTTTCCC 60
GTCGGGGAGC GCGGGGYCGG GGCCCAGGGG ACCCCGGGCC ACGGAGAGCG GGAAGAGGAT 120 GGATTGCCCG GCCCTCCCCC CCGGATGGAA GAAGGAGGAA GTGATCCGAA AATCTGGGCT 180
AAGTGCTGGC AAGAGCGATG TCTACTACTT CAGTCCAAGT GGTAAGAAGT TCAGAAGCAA 240
GCCTCAGTTC GCAAGGTACC TGGGAAATAC TGTTGATCTC AGCAGTTTTG ACTTCAGAAC 300
TGGAAAGATG ATGCCTAGTA AATTACAGAA GAACAAACAG AGACTGCGAA ACGATCCTCT 360
CAATCAAAAT AAGGGTAAAC CAGACTTGAA ATACAACATT GCCAATTAGA CAAACAGCAT 420
CAATTTTCAA ACAACCGGTA ACCCAAAGTC ACAAATCATC CTAGTAATAA AGTGAAATCA 480
GACCCACAAC GAATGAATGA ACAGCCACGT CAGCTTTTCT GGGAGAAGAG GCTACAAGGA 540
CTTTAGTGCA TCAGATGTAA CAGAACAAAT TATAAAAACC ATGGAACTAC CCAAAGGTCT 600
TCAAGGAGTT GGTCCAGTAG CAATGATGAG ACCCTTTTAT CTGCTGTTGC CAGTGCTTTG 660
CACACAAGCT CTGCGCCAAT CACAGGGCAA GTCTCCGCTG CTGTGGAAAA GAACCTGCTG 720
TTTGGCTTAA CACATCTCAA CCCCTCTGCA AAGCTTTTAT TGTCACAGAT GAAGACTCAG 780
GAAACAGAAG AGCGAGTACA GCAAGTACGC AAGAAATTGG AAGAAGCACT GATGGCAGAC 840
ATCTTGTCGC GAGCTGCTCA TACAGAAGAG ATGGATATTG AAATGGACAG TGGAGATGAA 900
GCCTAAGAAT ATGATCAGGT AACTTTCGAC CGACTTTCCC CAAGAGAAAA TTCCTAGGAA 960
ATTGAACAAA AATGTTTCCA CTGGCTTTTG CCTGTAAGAA AAAAAATGTA CCCGAGCACA 1020
TAGAGCTTTT TAATAGCACT AACCAATGCC TTTTTAGATG TATTTTTGAT GTATATATCT 1080
ATΓATTCAAA AAATCATGTT TATTTTGAGT CCTAGGACTT AAAATTAGTC TTTTGTAATA 1140
TCAAGCAGGA CCCTAAGATG AAGCTGAGCT TTTGATGCCA GGTGCAATCT ACTGGAAATG 1200
TAGCACTTAC GTAAAACATT TGTTTCCCCC ACAGTTTTAA TAAGAACAGA TCAGGAATTC 1260
TAAATAAATT TCCCAGTTAA AGATTATTGT GACTTCACTG TATATAAACA TATTTTTATA 1320
CTTTATTGAA AGGGGACACC TGTACATTCT TCCATCGTCA CTGTAAAGAC AAATAAATGA 1380
TTATATTCCA CAGAAAAAAA AAAAAAAAAW MWSTYGARRR GSRGCMCRSW AYMMARWWCC 1440
CCWMRTWRGS MKTCSTMTKA YTTACATTCA ACTCTGATCC CGGGGCCTTA GGTTTGACAT 1500
GGGAGGTGGG AGGAAGATAG CGCATATATT TGCAGTATGA ACTATTGCCT CTGGGACGTT 1560
GTGAGGAATT GTGCTTTCAC CAGAATTTCT AAGGATTTCT GGCTTAAATA TCACCTAGCC 1620
TGTGGTAATT TTTTTTCCCT 1640
(2) INFORMATION FOR SEQ ID NO: 117:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 952 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 117: TGAATTTAGN AAACACTTTG GAAAACTCAT AACCTCATCA GAAACTGCCT TTAGCCACAC 60
TCCTGACCTT CTAGATGAGT AACAAAAAAA TGAAATAAGT TCTTGGAAAT TAAGCCATTT 120
ATTTTAATTT GCTATTTTTT TCAATGTTCT AGGTATCTTT AAATTTGTTA TTGTGGAATC 180
ATTTTCCTGC CAGATACCTT TATCAAAATT ATTGGCCTCA TGAGAGCTGA AGTAAGTCAG 240
CTTTTTGGTG AACTTTAGTG GACTTCTGTG AGATTGTAGT TGTACTTTGT ATCTCTAAAT 300 CTAAAGATAG TTTTTTAAAA CTCCCAAAGA AAATCTGCTC TCCTTTCTGA TCTAAAAACT 360
CATCTTTGGG GTAAAGAGTT AAGTGTCCAA AGGTTGTCAC AGTTCATGAG GTCAGAGGGA 420
GCTAGCCTGG CACCTGGACT CTGCCCATCC ACAGCTGACA GATTCCAACA GAAGTGTATT 480
TAAATTCTCC AGTAGACAAT GCTGGGTAAG GGAGGGGGTA GGGCTGGGTT ATTAAGATAC 540
AGGCTGCTGT ATTTTACATT GGTTGTGGGG GAAGGGGAGC CTGGAGAAAA CAAAGTCACT 600 ATTCCCTTTT TTGAAACAGG AAAAAAAATT ATTTTTTGTT CAGTAAAAAT GGTAGAGAAT 660
TCCAATGTCC CTAGCCACAA GGGACCAGTT CCACTGAGAA GTGAACAGTG GGAACTCAAA 720
ATTTCAGAAA CATTGGGGGA AGGGAAAATT GGCTTTCTCT TAATTGGCAG ATGTTCCAGT 780
GGGGSGGGGG GGCTCTGTTT TTGTTGGGAT GTGTTATGTT GTATGTACGC ATATATGGAC 840
CGGAGTCTGC TGAGTTTATA AGGTTCCAAA AATATGGTAA AATCTTGGTT TTTGTTAATT 900 TATCTCAATA AAAGCCCACT GGRACTCCAA AAAAAAAAAA AAAAAAAAGA NN 952
(2) INFORMATION FOR SEQ ID NO: 118:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1256 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 118: GACGTCATAG GTAAACAGGC TCTGTATCCG TGGCAGCGGC CGTGGCAGGC TGGCTGGGTA 60
CCGGCTGTCG CTGACCCAGG AGAAGCTGCC TGTCTACATC AGCCTGGGCT GCAGCGCGCT 120
GCCGCCGCGG GGCCGGCAGC TGAACTATGT GCTCTTCAGG GCGGGCACCG TGTTGCATTC 180
ATCTTTGTAC CCCCAGCATC TAGCAGTGTT GGCATGTAGT AGGCACTCAA GAAATGTGTG 240
TTGAATGAAC GATGCCTGTG ACAAGCAAGC GGACTTTATT CTTTCCTGAC CCTTGCTCCT 300 ATGACACACC TCCTCCTGAC TGCCACTGTC ACTCCTTCAG AGCAGAACTC CTCTAGGGAA 360
CCTGGATGGG AAACAGCCAT GGCCAAGGAC ATCCTGGGTG AAGCAGGGCT ACACTTTGAT 420
GAACTGAACA AGCTGAGGGT GTTGGACCCA GAGGTTACCC AGCAGACCAT AGAGCTGAAG 480
GAAGAGTGCA AAGACTTTGT GGACAAAATT GGCCAGTTTC AGAAAATAGT TGGTGGTTTA 540
ATTGAGCTTG TTGATCAACT TGCAAAAGAA GCAGAAAATG AAAAGATGAA GGCCATCGGT 600
GCTCGGAACT TGCTCAAATC TATAGCAAAG CAGAGAGAAG CTCAACAGCA GCAACTTCAA 660
GCCCTAATAG CAGAAAAGAA AATGCAGCTA GAAAGGTATC GGGTTGAATA TGAAGCTTTG 720
TCTAAAGTAG AAGCAGAACA AAATGAATTT ATTGACCAAT TTATTTTTCA GAAATGAACT 780
GAAAATTTCG CTTTTATAGT AGGAAGGCAA AACAAAAAAA AGCCTCTCAA AACCAAAAAA 840
ACCTCTGTAG CATTCCAGCG GCTTGACCAA TGACCTATGT CACAAGAGGT GGCGTGTAAG 900
GAATGCAGCC CCCTGAAGAC AGCACTACAA GTCTGGGGGA GCCAGTTTTA ACATCAGTGC 960
ACAGCTGCTG CTGGTGGCCC TGCAGTGTAC GTTCTCACCT CTTATGCTTA GTTGGAACTA 1020
AGCAGTTTGT AAACTTTCAT CCTTTTTTTT GTAAATTCAC AAAGCTTTGG AAGGAGAAGC 1080
AATAAATTTT TGTTTTCAAA TGGCTTGATG TACCTTTTTT CCTGTTGCTC TTGAAATATG 1140
TTTAACTCCT CATGAGAGAA CCCTGGATTC TCTATCCCCT AGTCCACAAA ACAAACCAGG 1200
CAGTGGTCAG CAGCTACCTT TNATTTGGAT CACACACGTG AGTCAGACAG TACCAC 1256
(2) INFORMATION FOR SEQ ID NO: 119:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1143 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 119:
GGCCGTAGCA GCCGGGCTGG TCCTGCTGCG AGCCGGCGGC CCGGAGTGGG GCGGCGGCAT 60
GTACCTTCCA CATTGAGTAT TCAGAAAGAA GTGATCTGAA CTCTGACCAT TCTTTATGGA 120
TACATTAAGT CAAATATAAG AGTCTGACTA CTTGACACAC TGGCTCGAGC AAACATGAAC 180
GTTGGAGTTG CCCACAGTGA AGTGAATCCA AATACCCGTG TCATGAACAG CCGGGGTATG 240
TGGCTGACAT ATGCATTGGG AGTTGGCTTG CTTCATATTG TCTTACTCAG CATTCCCTTC 300
TTCAGTGTTC CTGTTGCTTG GACTTTAACA AATATTATAC ATAATCTGGG GATGTACGTA 360
TTTTTGCATG CAGTGAAAGG AACACCTTTC GAAACTCCTG ACCAGGGTAA AGCAAGGCTC 420
CTAACTCATT GGGAACAACT GGACTATGGA GTACAGTTTA CATCTTCACG GAAGTTTTTC 480
ACAATTTCTC CAATAATTCT ATATTTTCTG GCAAGTTTCT ATACGAAGTA TGATCCAACT 540
CACTTCATCC TAAACACAGC TTCTCTCCTG AGTGTACTAA TTCCCAAAAT GCCACAACTA 600
CATGGTGTTC GGATCTTTGG AATTAATAAG TATTGAAATG TTTTGAAACT GAAAAAAAAT 660
TTTACAGCTA CTGAATTTCT TATAAGGAAG GAGTGGTTAG TAAACTGCAC TGTTTCTSTG 720 ATAATGTGAA ATGAGAAGTA TTTACATTGG AGGGCCAATG GCTGGTCCTT CAAGTGCTGT 780 TTTGAAGTGC AGATTTCCAT TAAATGATGC CTCTGTTTAA TACACCTGGT ACATTTCTGA 840 AGAGGGGCTT TATAAGCAGG CTGGGCAGGC CCAGCTTATA AGTTAAAGGG CATCACAGTG 900 AGGGTGTAGT AGATAAATTC AAGGAAATAA GAGATTTGTA AGAAACTAGG ACCAGCTTAA 960 CTTATAATGA ATGGGCATTG TGTTAAGAAA AGAACATTTC CAGTCATTCA GCTGTGGTTA 1020 TTTAAAGCAG ACTTACATGT AAACCGGAAT CCTCTCTATA CAAGTTTATT AAAGATTATT 1080 TTTATTACCG TAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAANA 1140 GAN 1143
(2) INFORMATION FOR SEQ ID NO: 120:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1782 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 120:
CAGGCCCCGG CCCCCCACCC ACGTCTGCGT TGCTGCCCCG CCTGGGCCRG GCCCCAAAGG 60
CAAGGACAAA GCAGCTGTCA GGGAACCTCC GCCGGAGTCG AATTTACGTG CAGCTGCCGG 120
CAACCACAGG TTCCAAGATG GTTTCCGGGG GCTTCGCGTG TTCCAAGAAC TGCCTGTGCG 180
CCCTCAACCT GCTTTACACC TTGGTTAGTC TGCTGCTAAT TGGAATTGCT GCGTGGGGCA 240
TTGGCTTCGG GCTGATTTCC AGTCTCCGAG TGGTCGGCGT GGTCATTGCA GTGGGCATCT 300
TCTTGTTCCT GATTGCTTTA GTGGGTCTGA TTGGAGCTGT AAAACATCAT CAGGTGTTGC 360
TATTYTTTTA TATGATTATT CTGTTACTTG TATTTATTGT TCAGTTTTCT GTATCTTGCG 420
CTTGTTTAGC CCTGAACCAG GAGCAACAGG GTCAGCTTCT GGAGGTTGGT TGGAACAATA 480
CGGCAAGTGC TCGAAATGAC ATCCAGAGAA ATCTAAACTG CTGTGGGTTC CGAAGTGTTA 540
ACCCAAATGA CACCTGTCTG GCTAGCTGTG TTAAAAGTGA CCACTCGTGC TCGCCATGTG 600
CTCCAATCAT AGGAGAATAT GCTCGAGAGG TTTTGAGATT TCTTGGTGGC ATTGGCCTGT 660
TCTTCAGTTT TACAGAGATC CTGGGTGTTT GGCTGACCTA CAGATACAGG AACCAGAAAG 720
ACCCCCGCGC RAATCCTAGT GCATTCCTTT GATGAGAAAA CAAGGAAGAT TTCCTTTCGT 780
ATTATGATCT TGTTCACTTT CTGTAATTTT CTGTTAAGCT CCATTTGCCA GTTTAAGGAA 840
GGAAACACTA TCTGGAAAAG TACCTTATTG ATAGTGGAAT TATATATTTT TACTCTATGT 900 TTCTCTACAT GTTTTTTTCT TTCCGTTGCT GAAAAATATT TGAAACTTGT GGTCTCTGAA 960 GCTCGGTGGC ACCTGGGAAT TTACTGTATT CATTGTCGGG CACTGTCCAC TGTGGCCTTT 1020 CTTAGCATTT TTACCTGCAG AAAAACTTTG TATGGTACCA CTGTGTTGGT TATATGGTGA 1080 ATCTGAACGT ACATCTCACT GGTATAATTA TATGTAGCAC TGTGCTGTGT AGATAGTTCC 1140 TACTGGAAAA AGAGTGGRAA TTTATTAAAA TCAGAAAGTA TGAGATCCTG TTATGTTAAG 1200 GGAAATCCAA ATTCCCAATT TTTTTTGGTC TTTTTAGGAA AGATGTGTTG TGGTAAAAAG 1260 TGTTAGTATA AAAATGATAA TTWACTKGTA GTCTTTTATG ATWACACCAA TGTATTCTAG 1320 AAATAGTTAT GYCYTAGGAA ATTGTGGTTT AATTTTTGAC TTTTACAGGT AAGTGCAAAG 1380 GAGAAGTGGT TTCATGAAAT GTTCTAATGT ATAATAACAT TTACCTTCAG CCTCCATCAG 1440 AATGGAACGA GTTTTCAGTA ATCAGGAAGT ATATCTATAT GATCTTGATA TTGTTTTATA 1500 ATAATTTGAA GTCTAAAAGA CTGCATTTTT AAACAAGTTA GTATTAATGC GTTGGCCCAC 1560 GTAGCAAAAA GATATTTGAT TATCTTAAAA ATTGTTAAAT ACCGTTTTCA TGAAAGTTCT 1620 CAGTATTGTA ACAGCAACTT GTYAAACCTA AGCATATTTG AATATGATCT CCCATAATTT 1680 GAAATTGAAA TCGTATTGTG TGGCTCTGTA TATTCTGTTA AAAAATTAAA GGACAGAAAC 1740 CTTTCTTTGT GTATGCATGT TTGAATTAAA AGAAAGTAAT GG 1782
(2) INFORMATION FOR SEQ ID NO: 121:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 610 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO; 121: GTTGGCTCCA GATTTGTGGT GCGTTCTGAG CCGTCTGTCC TGCGCCAAGA TGCTTCAAAG 60 TATTATTAAA AACATATGGA TCCCCATGAA GCCCTACTAC ACCAAAGTTT ACCAGGAGAT 120 TTGGATAGGA ATGGGGCTGA TGGGCTTCAT CGTTTATAAA ATCCGGGCTG CTGATAAAAG 180 AAGTAAGGCT TTGAAAGCTT CAGCGCCTGC TCCTGGTCAT CACAACCAGA TTTACTTGGA 240 GTACATGTGA AAGAAAACGT CAGTCTGCCT GTAAATTTCA GCAAGCCGTG TTAGATGGGG 300 AGCGTGGAAC GTCACTGTAC ACTTGTATAA GTACCGTTTA CTTCATGGCA TGAATAAATG 360 GATCTGTGAG ATGCACTGCT ACCTGGTACT GCTTTCAGTG TGTTCCCCCT CAGCCCTCCG 420 GCGTGTCAGG CATACTCTGA GTAGATAATT TGTCATGCAG CGCATGCAAT CAGAATCTCA 480
CTGAGCCACC CATCATTGTG AAATAATTAC CTCAGTTGTA CAGGACTTGG TGATCAGGAT 540 CCAGGCACTC ACTTGTATTC TACTGCTCAA TAAACGTTTA TTAAACTTGA AAAAAAAAAA 600 AAAAAAAAAA 610
(2) INFORMATION FOR SEQ ID NO: 122:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 526 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 122:
GGTACGCCTG CAGGTACCGG TCCGGAATTC CGGGTCGCCC ACGCGTCNGG CCACGCGTCC 60
ACCCACGCGT CCGSCCACGC GTCGGAGCCG AGCCGGACTG GTCAGGATGA TCACGGACGT 120 GCAGCTCGCC ATCTTCGCCA ACATGCTGGG CGTGTCGCTC TTCTTGCTTG TCGTTCTCTA 180
TCACTACGTG GCCGTCAACA ATCCCAAGAA GCAGGAATGA AAGTGGCGCT TTCTCCGCCC 240
CAGGGTTCCA GGACATAGTC TCAGGCAAGA TGGAGGGTAT GAGGGGCCTT CACACTTCAC 300
TTCATCCCTT CTACCCATCA CAACATACAA AGCAACTACA CCTGGATTTT TCCAAACAAC 360
TTTTATTTCC TCAGAGTCTT CCTTAATCCT ATGGAACAAG AAGCTGCCAC TGAATAGGGC 420 CCAGTATAGG GGCTTGCTTT TCTACTCCCT CCCCCCAATA TAAAAATATA GACTTTTTAA 480
AAAAAAAAAA AAAAANTTCG NGGGGGGSCC GGTACCCATC CCCCTA 526
(2) INFORMATION FOR SEQ ID NO: 123:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 2081 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 123:
TGTACCGGTC CGGAAATTCC CGGGTCGACC CACGTCGTCS GGGGAACATG GCGGCTKCGG 60 AGCCGGCGGT CCTTGCGCTC CCCAACAGCG GCGCCGGGGG CGCGGGGGCG CCGTCGGGCA 120
CAGTCCCGGT GCTCTTCTGT TTCTCAGTCT TCGCGCGACC CTCGTCGGTG CCACACGGGG 180
CGGGCTACGA GCTGCTCATC CAGAAGTTCC TCAGCCTGTA CGGCGACCAG ATCGACATGC 240 ACCGCAAATT CGTGGTGCAG CTGTTCGCCG AGGAGTGGGG CCAGTACGTG GACTTGCCCA 300
AGGGCTTCGC GGTRAGCGAG CGCTGCAAGG TGCGCCTCGT GCCGYTGCAG ATCCAGCTCA 360
CTACCCTGGG AAATCTTACA CCTTCAAGCA CTCTCTTTTT CTGCTGTGAT ATGCAGGAAA 420
GGTTCAGACC AGCCATCAAG TATTTTGGGG ATATTATTAG CGTGGGACAG AGATTGTTGC 480
AAGGGGCCCG GATTTTAGGA ATTCCTGTTA TTGTAACAGA ACAATACCCT AAAGGTCTTG 540
GGAGCACGGT TCAAGAAATT GATTTAACAG GTGTAAAACT GGTACTTCCA AAGACCAAGT 600
TTTCAATGGT ATTACCAGAA GTAGAAGCGG CATTAGCAGA GATTCCCGGA GTCAGGAGTC 660
TTGTATTATT TGGAGTAGAA ACTCATGTGT GCATCCAACA AACTGCCCTG GAGCTAGTTG 720
GCCGAGGAGT CGAGGTTCAC ATTGTTGCTG ATGCCACCTC ATCAAGAAGC ATGATGGACA 780
GGATGTTTGC CCTCGAGCGT CTCGCTCRAR CCGGGATCAT AGTGACCACG AGTGAGGCTG 840
TTCTGCTTCA GCTGGTAGCT GATAAGGACC ATCCAAAATT CAAGGAAATT CAGAATCTAA 900
TTAAGGCGAG TGCTCCAGAG TCGGGTCTGC TTTCCAAAGT ATAGGACATT TGAAGAACTG 960
GTATGCTACT CACTGGTGAA GGACAGTCAG GTGAAGGACT GTAAGCCCAC ACAAGCTCTT 1020
CTTATCTCTA CTAGAATTAA AATGTTAAGT CAAAAACGGC TCCTTTTTTG CGCCTCCTAG 1080
TGAAACTTAA CCAGCTAGAC CATTTGAGTA CCAGCATTTA GTTACAAACG TCAAAGGCTT 1140
CCGGTGCTGC TTACCTTCCT TTTTTGTTAA TGTGCTTTTA TTTATTAAAA AAAATTACAA 1200
TGAAGATGCC TGTTTTGTCT CTACTGTGTA CTCTGATCGT ATCTTTCCAA AGTCCAGACT 1260
CTTGTGAAGT TTTCTTAAAT TGTTCACTTT AAAGAAAATG ACGTACCAAC AATGATTTGG 1320
CTTTTATATT ACTGTAAGAT GTTATAATGT TAATGTGGAT GTAGTGCTTT TACTTTACAG 1380
ATTGATTGGA ATAAGATTAT TGCATATGAA TTTACCCACA GGACTCTGAA TCATGTTACC 1440
CACTCCCCTC ACAATGTTGT CCACTTAGTG AGTTGCATTG ATCTATCCGT ACCAAATGAT 1500
GTTCAATAAT TACATATCTT TCTTGACTAT ACTGATTTCT TATTTTGGTC ACTATTACTA 1560
AATCTCTGTT AATATTCTCT CTTTTAACTG AAAAGGGATG GGATAGAAGG GTTTGCAATG 1620
CCATATTATT GGTGGAGGGC TGTTTTAACA TCTTTGAAGT ATGGCTTGCT GAATATCTTT 1680
ACCAACATCT TGAATATATA TTCTAGTGTC CACAAGATTT AGCAAAAAGA TAAAGCTTGG 1740
GTGGAATATC ATTTTAAAAT GTTCATGTTC TGTTCTATAT TTTCTTCACC TACTCTCCAA 1800
ATATTGTAAT GCAAAAAGTC TCAGTAATGA TTTGGTAGTA TTAATTTTGT GGTCATTCTT 1860
TCTCTTCGAT AAATTTATTT TCATTAAATA CTTRTTAGAG GGTTTTGAAA TGTTTTTCAA 1920
ATATGTGAAA TGTGAAACTG CTGTCTTTTA TATTAAAGTA ATTAAAGAAA ATGTATTGTG 1980
ATTGAAATTA TTTTGNCCTC CACAAGATGG CTCTATGAGT ATTCTTCCAG GGATTCTAAT 2040
ATTTATTTAA GGTNATAAAA TCTTGACATT TATAATCTTT C 2081
(2) INFORMATION FOR SEQ ID NO: 124:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1717 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 124:
CCCCGGCGGA GCTCGACCCG CGGTGGGCTA GGGGCAGGGC CGGAGCCGCG GCGGCGGAGC 60
TGTGGATCCT TCATGATGAG AGATTTGGGG ACACTTCTCT CTCCTGTGTG TAGTTGATAG 120
TTTGGTGGTG AAGAGATGGC TGACAGTGTC AAAACCTTTC TCCAGGACCT TGCCAGAGGA 180
ATCAAAGACT CCATCTGGGG TATTTGTACC ATCTCAAAGC TAGATGCTCG AATCCAGCAA 240
AAGAGAGAGG AGCAGCGTCG AAGAAGGGCA AGTAGTCTCT TGGCACAGAG AAGAGCCCAG 300
AGTATAGAGC GGAAGCAAGA GAGTGAGCCA CGTATTGTTA GTAGAATTTT CCAGTGTTGT 360
GCTTGGAATG GTGGAGTGTT CTGGTTCAGT CTCCTCTTGT TTTATCGAGT ATTTATTCCT 420
GTGCTTCAGT CGGTAACAGC CCGAATTATC GGTGACCCAT CACTACATGG AGATGTTTGG 480
TCGTGGCTGG AATTCTTCCT CACGTCAATT TTCAGTGCTC TTTGGGTGCT CCCCTTGTTT 540
GTGCTTAGCA AAGTGGTGAA TGCCATTTGG TTTCAGGATA TAGCTGACCT GGCATTTGAG 600
GTATCAGGGA GGAAGCCTCA CCCATTCCCT AGTGTCAGCA AAATAATTGC TGACATGCTC 660
TTCAACCTTT TGCTGCAGGC TCTTTTCCTC ATTCAGGGAA TGTTTGTGAG TCTCTTTCCC 720
ATCCATCTTG TCGGTCAGCT GGTTAGTCTC CTGCATATGT CCCTTCTCTA CTCACTGTAC 780
TGCTTTGAAT ATCGTTGGTT CAATAAAGGA ATTCAAATGC ACCAGCGGTT GTCTAACATA 840
GAAAGGAATT GGCCTTACTA CTTTGGGTTT GGTTTGCCCT TGGCTTTTCT CACAGCAATG 900
CAGTCCTCAT ATATTATCAG TGGCTGCCTT TTCTCTATCC TCTTTCCTTT ATTCATTATC 960
AGCGCCAATG AAGCAAAGAC CCCTGGCAAA GCRTATCTCT TCCAGTTGCG CCTCTTCTCC 1020
TTGGTCGTCT TCTTAAGCAA CAGACTCTTC CACAAGACAG TCTACCTGCA GTCGGCCCTG 1080
AGCAGCTCTA CTTCTGCAGA GAAGTTCCCT TCACCGCATC CGTCGCCTGC CAAACTGAAG 1140
GCTACTGCAG GTCACTCAGT TGCCTGCCAT CCAAAGGGGA TGGGCGGGAT TGGAAGAAGC 1200
TGTGGCAGCT CTTTTCCCTG TTCACCTCCC GCCTGCCAGG GAAGGCAGGA CCCGCTCTGC 1260
CAAGGGCCCT CTGCGTATTC CCTTCTCTCT GAGGAATTGA AATTTTTGTC TCTGGTGCAC 1320
GTAAGGCAGA ATGTTCCCTG ACACCAGTGT GTGGATTTTT AACATCACCG TGAGTCTGAA 1380
AGGACCACAG GTTTTTCTGC AGCTATTTTC TAGCATTTGC CAGTCCCTGT GCCTGGACTG 1440
ATTGGAACAC TTTGTTTTTC TCCCTGTGCC ATTTACCCTT CCACCTTTCC ATCCTGCCTT 1500
CTACCACCCT TGGATGAATC GATTTTGTAA TTCTAGCTGT TGTATTTTGT GAATTTGTTA 1560
ATTTTGTTGT TTTTCTGTGA AACACATACA TTGGATATGG GAGGTAAAGG AGTGTCCCAG 1620
TTGCTCCTGG TCACTCCCTT TATAGCCATT ACTGTCTTGT TTCTTGTAAC TCAGGTTAGG 1680
TTTTGGTCTC TCTTGCTCCA CTGCAAAAAA AAAAAAA 1717
(2) INFORMATION FOR SEQ ID NO: 125:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 804 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO : 125: CCACGCGTCC GGTCACTATG TAGTGGAGGG GCAGACACCC TCCCGCAAAT TCTGGAAGGT 60 TCTTAGTCTC GACTAGGGCA GTAGCCCCAG GACTCCTAGT CGCCGGCTTC AGGTCACTGC 120 CGGCTGAACG GAGCTGCCGT CGCCATGTTT GGCTGCTTGG TGGCGGGGAG GCTGGTGCAA 180 ACAGCTGCAC AGCAAGTGGC AGAGGATAAA TTTGTTTTTG ACTTACCTGA TTATGAAAGT 240 ATCAACCATG TTGTGGTTTT TATGCTGGGA ACAATCCCAT TTCCTGAGGG AATGGGAGGA 300 TCTGTCTACT TTTCTTATCC TGATTCAAAT GGAATGCCAG TATGGCAACT CCTAGGATTT 360 GTCACGAATG GGAAGCCAAG TGCCATCTTC AAAATTTCAG GTCTTAAATC TGGAGAAGGA 420 AGCCAACATC CTTTTGGAGC CATGAATATT GTCCGAACTC CATCTGTTGC TCAGATTGGA 480 ATTTCAGTGG AATTATTAGA CAGTATGGCT CAGCAGACTC CTGTAGGTAA TGCTGCTGTA 540 TCCTCAGTTG ACTCATTCAC TCAGTTCACA CAAAAGATGT TGGACAATTT CTACAATTTT 600 GCTTCATCAT TTGCTGTCTC TCAGCCCCAG ATGACACCAA GCCCATCTGA AATGTTCATT 660 CCGGCAAATG TGGTTCTGAA ATGGTATGAA AACTTTCAAA GACGACTAGC ACAGAACCCT 720 NTNTTTTGGN AAACATAATT TGAATAAAAT AATTTTTAAT GGATTNTGNA AAAAAAAAAA 780 AAAAAAAAAA AAAAAAAAAA AAAA 804
(2) INFORMATION FOR SEQ ID NO: 126:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 431 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 126:
GGCACAGCCC AGGGCCTTGA AGCCAGCTGG CCCTGGAGAG GGGCTGCTGT GCCAGCTTGG 60
GGAGGGTCTG GGATGGGGCT GCCCCTGATG GCCCTGATGT GGAGTACCTT GCCAGCATCT 120
GCTGGGGTGA ACTTTATTTT AGCCCTTCCC TTGTTGCTCT TATGGAAGAA CAGAGGAGGG 180
GTGGGCAGGT CAGTGATGTC AGCAGTGGAG TGATTCCCAG CACAGCGGCT TCTGGGAAGA 240 GGGCATGGAG GCATTTCTTT CAGGGAAATG GTCCATNATT TCAGCCAGAA GGCATTGCAT 300
TAAGTTAAGT CCNGGACTTT TGTGGCCCAG CTCTGTGTTA TTAAGGGCCC TTGGCGAAGA 360
CTTCAAGGAG GGGGCAAAAN GACCTTTAAG TTTTTAGGTT TAACACAGGG AACCCNCAAA 420
GGGTTATTTT G 431
(2) INFORMATION FOR SEQ ID NO: 127:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 3752 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 127:
NGGCACGAGG AGAGTCACCT GGACTCAGAA CTAGAGATAT CCAATGACCC AGACAAAATT 60
AAACTTCAGC TTTCTAAGCA TAAGGAGTTT CAGAAGACTC TTGGTGGCAA GCAGCCTGTG 120 TATGATACCA CAATTAGAAC TGGCAGAGCA CTGAAAGAAA AGACTTTGCT TCCCGAAGAT 180
ASTCAGAAAC TTGACAATTT CCTAGGAGAA GTCAGAGACA AATGGGATAC TGTTTGTGGC 240
AAGTCTGTGG AGCGGCAGCA CAAGTTGGAG GAAGCCCTGC TCTTTTCGGG TCAGTTCATG 300
GATGCTTTGC AGGCATTGGT TCACTGGTTA TACAAGGTGG AGCCACAGCT GGCTGAGGAC 360
CAGCCCGTGC ACGGGGGACC TTGACCTCGT CATGAACCTC ATGGATGCAC ACAAGGTTTT 420 CCAGAAGGAA CTGGNGAAAG CGAACAGGAA CCGTTCAGGT CCTGAAGCGG TCAGGCCGAG 480
AGCTGATTGA GAATAGTCGA GATGACACCA CTTGGGTAAA AGGACAGCTC CAGGAACTGA 540
GCACTCGCTG GGACACTGTC TGTAAACTCT CTGTTTCCAA ACAAAGCCGG CTTGAGCAGG 600
CCTTAAAACA AGCGGAAGTG TTTCGAGACA CAGTCCACAT GCTGTTGGAG TGGCTTTCTG 660
AAGCAGAGCA AACGCTTCGC TTTCGGGGAG CACTTCCTCG ATGACACAGA GGCCCTGCAG 720 TCTCTCATTG ACACCCATAA GGAATTCATG AAGAAAGTAG AAGAAAAGCG AGTGGACGTT 780
AACTCAGCAG TAGCCATGGG AGAAGTCATC CTGGCTGTCT GCCACCCCGA TTGCATCACA 840
ACCATCAAAC ACTGGATCAC CATCATCCGA GCTCGCTTCG AGGAGGTCCT GACATGGGCT 900
AAGCAGCACC AGCAGCGTCT TGAAACGGCC TTGTCAGAAC TGGTGGCTAA TGCTGAGCTC 960
CTGGAAGAAC TTCTGGCATG GATCCAGTGG GCTGAGACCA CCCTCATTCA GCGGGATCAG 1020
GAGCCAATCC CGCAGAACAT TGACCGAGTT AAAGCCCTTA TCGCTGAGCA TCAGACATTT 1080
ATGGAGGAGA TGACTCGCAA ACAGCCTGAC GTGGACCGGG TCACCAAGAC ATACAAAAGG 1140
AAAAACATAG AGCCTACTCA CGCGCCTTTC ATAGAGAAAT CCCGCAGCGG AGGCAGGAAA 1200
TCCCTAAGTC AGCCAACCCC TCCTCCCATG CCAATCCTTT CACAGTCTGA AGCAAAAAAC 1260
CCACGGATCA ACCAGCTTTC TGCCCGCTGG CAGCAGGTGT GGCTGTTAGC ACTGGAGCGG 1320
CAAAGGAAAC TGAATGATGC CTTGGATCGG CTGGAGGAGT TGAAAGAATT TGCCAACTTT 1380
GACTTTGATG TCTGGAGGAA AAAGTATATG CGTTGGATGA ATCACAAAAA GTCTCGAGTG 1440
ATGGATTTCT TCCGGCGCAT TGATAAGGAC CAGGATGGGA AGATAACACG TCAGGAGTTT 1500
ATCGATGGCA TTTTAGCATC CAAGTTCCCC ACCACCAAGT TAGAGATGAC TGCTGTGGCT 1560
GACATTTTCG ACCGAGATGG GGATGGTTAC ATTGATTATT ATCAATTTGT GGCTGCTCTT 1620
CATCCCAACA AGGATGCGTA TCGACCAACA ACCGATGCAG ATAAAATCGA AGATGAGGTT 1680
ACAAGACAAG TGGCTCAGTG CAAATGTGCA AAAAGGTTTC AGGTGGAGCA GATCGGAGAG 1740
AATAAATACC GGTTCTTCCT CGGCAATCAG TTTGGGGATT CTCAGCAGTT GCGGCTGGTC 1800
CGTATTCTGC GCAACCGTGA TGGTTCGCGT TGGTGGAGGA TGGATGGCCT TGGATGAATT 1860
TTTAGTGAAA AATGATCCCT GCCGAGCACG AGGTAGAACT AACATTGAAC TTAGAGAGAA 1920
ATTCATCCTA CCAGAGGGAG CATCCCAGGG AATGACCCCC TTCCGCTCAC GGGGTCGAAG 1980
GTCCAAACCA TCTTCCCGGG CAGCTTCCCC TACTCGTTCC AGCTCCAGTG CTAGTCAGAG 2040
TAACCACAGC TGTACATCCA TGCCATCTTC TCCAGCCACC CCAGCCAGTG GAACCAAGGT 2100
TATCCCATCA TCAGGTAGCA AGTTGAAACG ACCAACACCA ACTTTTCATT CTAGTCGGAC 2160
ATCCCTTGCT GGTGATACCA GCAATNAGTT CTTCCCCGGC CTCCACAGGT GCCAAAACTA 2220
ATCGGGCAGA CCCTAAAAAG TCTGCCAGTC GCCCTGGGAG TCGGGCTGGG AGTCGAGCCG 2280
GGAGTCGAGC CAGCAGCCGG CGAGGAAGTG ACGCTTCTGA CTTTGACCTC TTAGAGACGC 2340
ATTGCTTGTT CCGACACTTC AGAAAGCAGC GCTGCAGGGG GCCAAGGCAA CTCCAGGAGA 2400
GGGCTAAACA AACCTTCCAA AATCCCAACC ATGTCTAAGA AGACCACCAC TGCCTCCCCC 2460
AGGACTCCAG GTCCCAAGCG ATAACACTGT CTAAGCACCC CCAAGCCACT ATCCACTTTG 2520
AATCCTGCTC CATACATTGG GTGTATATTT ATTCTGAACG GGAGAAGTTA TATTGTTAAA 2580
AGTGTAAAAG AATAATTGTG TTATGAAGCT GCCTTATTTT TTTTCTTTTT GTAAGTTACT 2640
ATTTTCATGT GAATATTTAT GTAGATAAAA TTTGCCTCCT GGTAACCCTG TAATGGATGG 2700
GGCCCAGAAA TGAAATATTT GAGAAAAACA AGTCAAAAGG TCAAGATACA AATGTCTATT 2760
AAAAAAAAAA AAGCCTATTA ATAGGGTTTC TGCGCGGTGC AGGGTTGTAA ACCTGCTTTA 2820
TCTTTTAGGA TTATTCCTAA ATGCATCTTC TTTATAAACT TGACTTGCTA TCTCAGCAAG 2880
ATAAATTATA TTAAAAAAAT AAGAATCCTG CAGTGTTTAA GGAACTCTTT TTTTGTAAAT 2940
CACGGACACC TCAATTAGCA AGAACTGAGG GGAGGGCTTT TTCCATTGTT TAATGTTTTC 3000
TGATTTTTAG CTAAAGAGAG GGAACCTCAT CTAAGTAACA TTTGCACATG ATACAGCAAA 3060
AGGAGTTCAT TGCAATACTG TCTTTGGATA TTGTTTCAGT ACTGGGTGTT TAAAGGACAA 3120
ATAGCTGCTA GAATTCAGGG GTAAATGTAA GTGTTCAGAA AACGTCAGAA CATTTGGGGT 3180
TTTAAACTGA TTTGTTGCTC CCTATCCAGC CTAGACACCA GTAACTCTTG TGTTCACCAG 3240
GACCCAGACC CTTGGCAAGG GATAGGCTCG TTGGTGACAT TGTGAATTTC AGATTTGTTT 3300
TATCCACTTT TTTTGCTATT TATTTAAATG GTCGATCAAC TTCCCACAAA CTGAGGAATG 3360
AATTCCACGA GCCTGTTCTG AAAATGTGGA CGTAAGACAA ACACGTGCTC GTCCTTTAAT 3420
GGAGTTCACC AGCACACTTG TTAACCAGTC CTGTTTGCTT TCGTCTTTTT TTGTGCGTAA 3480
TAAAGTCAAC TGACCAAGTG ACCATGAAAA GGGGCTGTCT GGGGCTCCTG TTTTTTAGCT 3540
GCTGTTCTTC AGCTCCGACC ATGTTGCTGT GTGATTATCT CAATTGGTTT TAATTGAGGC 3600
AGAAACTGAA GCTCTACCAA TGAACTGTTT AGAAACAAGA CΛC^CTTTTG TATTAAAATT 3660
GCTTGCAGTA ACAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAACTCGAGG GGGGCCCGGT 3720
ACCCAATTCG CCGTATATGA TCGTAAACAA TC 3752
(2) INFORMATION FOR SEQ ID NO: 128:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1144 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 128: TGACCCTCTG CCTGCCGGGC TCAGTGCTGG ACGCTTTCTG TTTTGTCGCA GTCGGTCCTC 60 GGTAACACCA GCGGCCTGTG GTCCACCACT CCATTCAGCA GCTCCATTTG GTCCAGCAAC 120 CTTAGCAGCG CCTTCCCTTC ACCACTCCAG CAAACACGCT GGCAAGCATC GGCCTCATGG 180
GCACAGAAAA CTCCCCTGCT CCTCACGCTC CCTCCACCTC CAGTCCAGCT GACGACTTGG 240
GACAGACCTA CAACCCGTGG CGGATATGGA GCCCCACGAT TGGAAGAAGA AGCTCGGACC 300
CTTGGTCTAA TTCGCACTTT CCTCACGAGA ATTAAATTAA GCAAAAAACA AACAAACATA 360
GTGGGCCCTC GTCTAGATCA TGATGTGCCA GTTTCTGAGA CATCTTTTTA AGGCTCTTAC 420
TGCAGCTCCC CTCCCCACCC TCCTCTTCTT TGCAAAACAG ACCCAAGCAG GGCAGGCTCA 480
GACCACTCGC TTCTTTCAGA TCTTTCTTGC AATTATGATA ACATGAGATT TGCTCTTCTC 540
CTTTTAGAGA AAAGTCTGGA CTCAGCCACA AACTCTAATA AGACCTGTAC ATCTGAGAAC 600
CTTTCCCGTT ACTGCGTTTT CACCACCTGT CTTCCCCATG CTTTATTTAT CTGTATGAAC 660
ACAGATTTGA CATTACAGCT AAGGAAATAA TTTGAGTTGA TTCAGAAATC CTGGCATGTG 720
ACAATTTTGT TAAATTACCA AGTTTGGTTT TTAATAATTT CTCAATATTA TGCGCCAAGA 780
TCTAATTTTA AAACTGTATG AGGACTTTGT GCTGAAAATA GAGTATTTTT TTAAAGTAAG 840
GCTGTCTTGG TTTAAAAGCA GATTACAGAA ATGTAAGTCA ACTTAAGAAC RGTGAATGAA 900
TGTAAAAACA TTCAGTYGAG ACCATATGCA TTTTCTGTGC TGTTTGTACT TGAGGTATGT 960
AACATTTGTA TACCTGAACT TATTTTAAAG ATGAACTGAA ATGCACATAG CCAAGTCTTG 1020
AGATACAAGA TTGAATGTGT ATTTCTTAAA AATACAACTT TGTGTTGTAC TTTGAAATAA 1080
ATGATGCTTT TTTCAAAAAA AAAAAAAAAA AAAAAAAAAC TCGAGGGGGG GCCCGGTACC 1140
CAAT 1144
(2) INFORMATION FOR SEQ ID NO: 129:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1830 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO : 129: GCATGCAGAG GAGCACCCTG AGCGTGTYCC TGGAGCAGGC GGCCATSTTG GCACGGAGCC 60 ACGGGTTGCT GCCCAAGTGC ATCATGCAGG CCACGGACAT CATGCGGAAC AGGGCCCAAG 120 GGTGGAGATT CTGGCCAAAA ACCTGCGAGT CAAGGACCAG ATGCCCCAGG GTGCTCCGCG 180 CCTCTACCGC CTCTGCCAGC CGCCGGTGGA TGGGGACCTC TGAACACCCA AATGCCCCAC 240 GCTCGGCCGC GGCCTCTGGA GCTGGGATTT GGGAGGACAC AGCAGGCAGC GCTGGCCTTC 300 TCCAGGGATG GCCCAANGCT TCCGCARCCG CCCGTTCCGG GACCTGCCCA GCGTCCTCCC 360 TGCCTCCTTC CGGGACAAGC CTGGCCACCC TCGCTGTGAT GACGAGCTGG CTGATTGGCC 420
CTGGGCCGGC CCATTCTTCA CACGCCTGCC AGAAGCTGGA GGGGTGCTGG AGACCCATAG 480 AGCTGATGGG AGCAGCTGGT GCCTGGCCTT CGGCTCCTGC GTCCCCAGAA CCCAAGGGAA 540 CGTCATGGAG GCCACATGGG GCCACCCGGC TCCCTCGGGA TGGCTCCGCT GCACTTTTGA 600 AACCCCGGTT TCCTTCAACG TCCACATTCC AGGTGACCAC ACGTGTCTCC TCCTCCTCAT 660 CTTAGCTTCC AGGTTCACCC TAACCCTGTA CTAACCTGCT TGGTGGACTT GGAAAAGACT 720 TGGCTCTGTC GGGAAAGGAG AGACGGGGCC TCCATCACGC CTGTTACCAG AGGATCCCCG 780 AGAGCCACAC CAGCTCTGGA CATCACCGCC CCTGGAACTG GGGCCACCAG CCCTGGGCAC 840 GAGATTTGCT CTGACTTTAT TTATATGGCA TGAAATCTCT GGTTTATTTT GGGATTTTTT 900 GTTGTTGGTG TTGTCAAAGT TTCTTTTTTC TAAAGTTGTG TGATTATATA TTTGACATTT 960 TACATTTCAA AGAAAGGTAT GTTGTCTAAC AGGGGACCAA CAGAAGGTAG TATTGACAAC 1020 TGTTCCTGCT TCTACTAAAA AAAAAAGAGC ACAAAAGAAA AACTAAATTA TTGAAAAATT 1080 AAAAAATGTC ATTGTTTCCT GTTTGTTAAT ATTAGGGTTG TAAGGTGTCG TTTTGAGGTA 1140 TCGACTGTGA TTCCTTCCCC CACCCTCCAT TCTCCAGCGG TTGGCCGGTG TTAGAACTCG 1200 CTCTCTTTGA GTGACTGGCT ACAAGGGCCT GAGAGGTGGC CAGCCAGGGT TGGAGCTGGA 1260 GGGGATGGAG CCCCACCTGA GGTGCCGTGT CACACGGGTT AGAGGGTCAC TGGGAAACAC 1320 CGGGCGGTGG CTTCTGTGAT TTATTTTCTT GATGGTAACT TCTCAGAGCA GGGCRATTGG 1380 GACATCACCA GCCAGAGCAC AGGAAGCCAC CCTGCCTGCT GGGGAGGAGG GACCCACACA 1440 AGCCCCCTCG GCAGTTTGTC CCCCCAGCTT CGGTATGCCT TCAGGGAAAG GTCACAGCTG 1500 GGGAGGAAGC GGGGGGACGC CTGTCACCCC TGGCAGGTGG TGAGTTCAGG TGGGGGCTCC 1560 CTGCTKCCCC CAGGCCTGGG AGCTTGAAGC CCTCCCGGCA TCTGGCATCC GAGCCTCCCG 1620 CCCTCCAGGG TGCGCTTCCC TCTCTTGCCG CAGCATACAC GAGGGCAGGC AGTGGCCTTG 1680 TCACTGTATC TTGCATCAGA GACAAAGGAG GACCCGCTTT AGCCCTGCTG CGGGAAATGG 1740 GGGATGGCCC AGGGCCAGCG CATTGTGCAC TGGTTTACTT TAAAATGTAC AGATTCTTCT 1800 CGTTAAATTC TTGATAGATT TTTTATTATT 1830
(2) INFORMATION FOR SEQ ID NO: 130:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1864 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 130:
GGCCGCCCGG ATGGCGACCC CAGCCTCGGC CCCAGACACA CGGGCTCTGG TGGCAGACTT 60
TGTAGGTTAT AAGCTGAGGC AGAAGGGTTA TCTCTCTGGA GCTGGCCCCG GGGAGGGCCC 120
AGCAGCTGAC CCGCTGCACC AAGCCATGCG GGCAGCKGGA GATGAGTTCG AGACCCGCTT 180
CCGGCGCACC TTCTCTGATC TGGCGGCTCA GCTGCATGTG ACCCCAGGCT CAGCCCAACA 240
ACGCTTCACC CAGGTCTCCG ATGAACTTTT TCAAGGGGGC CCCAACTGGG GCCGCCTTGT 300
AGCCTTCTTT GTCTTTGGGG CTGCACTGTG TGCTGAGAGT GTCAACAAGG AGATGGAACC 360
ACTGGTGGGA CAAGTGCAGG AGTGGATGGT GGCCTACCTG GAGACGCGGC TGGCTGACTG 420
GATCCACAGC AGTGGGGGCT GGTTATCCCA GATCACTGAA GCTGAGATGG CTGATGAAGT 480
AATTTGCAGT GAAATTTTAA GCGACTGTGA CTCTGCTGCA AGTTCCCCAG ATCTTGAGGA 540
GCTGGAAGCT ATCAAAGCTC GAGTCAGGGA GATGGAGGAA GAAGCTGAGA AGCTAAAGGA 600
GCTACAGAAC GAGGTAGAGA AGCAGATGAA TATGAGTCCA CCTCCAGGCA ATGCTGGCCC 660
GGTGATCATG TCCATTGAGG AGAAGATGGA GGCTGATGCC CGTTCCATCT ATGTTGGCAA 720
TGTGGACTAT GGTGCAACAG CAGAAGAGCT GGAAGCTCAC TTTCATGGCT GTGGTTCAGT 780
CAACCGTGTT ACCATACTGT GTGACAAATT TAGTGGCCAT CCCAAAGGGT TTGCGTATAT 840
AGAGTTCTCA GACAAAGAGT CAGTGAGGAC TTCCTTGGCC TTAGATGAGT CCCTATTTAG 900
AGGAAGGCAA ATCAAGGTGA TCCCAAAACG AACCAACAGA CCAGGCATCA GCACAACAGA 960
CCGGGGTTTT CCACGAGCCC GCTACCGCGC CCGGACCACC AACTACAACA GCTCCCGCTC 1020
TCGATTCTAC AGTGGTTTTA ACAGCAGGCC CCGGGGTCGC GTCTACAGGG GCCGGGCTAG 1080
AGCGACATCA TGGTATTCCC CTTACTAAAA AAAGTGTGTA TTAGGAGGAG AGAGAGGAAA 1140
AAAAGAGGAA AGAAGGAAAA AAAAAAGAAT TAAAAAAAAA AAAAAAAAAA ACAGAAGWTG 1200
MCCTTCATGG AAAAAAAATA TTTTTTAAAA AAAAGATATA CTGTGGAAGG GGGGAGAATC 1260
CCATAACTAA CTCCTGAGGA GGGACCTGCT TTGGGGAGTA GGGGAAGGCC CAGGGARTGG 1320
GGCAGGGGGC TGCTTATTCA CTCTGGGGAT TCGCCATGGA CACGTCTCAA CTGCGCAACT 1380
GCTTGCCCAT GTTTCCCTGC CCCACCCCAC CCCTCTTCTC CGGCTCCCTG CCCCTCCAGA 1440
TTGCCTGGTG ATCTATTTTG TTTCCTTTTG TGTTTCTTTT TCTGTTTTGA GTCTCTTTCT 1500
TTGCAGGTTT CTGTAGCCGG AAGATCTCCG TTCCGCTCCC AGCGGCTCCA GTGTAAATTC 1560
CCCTTCCCCC TGGGGAAATG CACTACCTTG TTTTGGGGGG TTTAGGGGTG TTTTTGTTTT 1620
TCAGTTGTTT TGTTTTTTTG TTTTTTTNTT TTTCCTTTGC CTTTTTTCCC TTTTATTTGG 1680
AGGGAATGGG AGGAAGTGGG AACAGGGAGG TGGGAGGTGG ATTTTGTTTA TTTTTTTAGC 1740
TCATTTCCAG GGGTCGGAAT TTTTTTTTAA TATGTGTCAT GAATAAAGTT GTTTTTGAAA 1800
AKAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA 1860 AAAA 1864
(2) INFORMATION FOR SEQ ID NO: 131:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2041 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 131:
GGCACGAGCG CGCGGCAGGG CCCTCGACCC GCGCGGCTCC CGGGGATGGT GAGCAAGGCG 60
CTCCTGCGCC TTCTCTCTGC CGTCAACCGC AGGAGGATGA AGCTGCTGCT GGGCATCGCC 120
TTGCTGGCCT ACGTCGCCTC TGTTTGGGGC AACTTCGTTA ATATGAGGTC TATCCAGGAA 180
AATGGTGAAC TAAAAATTGA AAGCAAGATT GAAGAGATGG TTGAACCACT AAGAGAGAAA 240
ATCAGAGATT TAGAAAAAAG CTTTACCCAG AAATACCCAC CAGTAAAGTT TTTATCAGAA 300
AAGGATCGGA AAAGAATTTT GATAACAGGA GGCGCAGGGT TCGTGGGCTC CCATCTAACT 360
GACAAACTCA TGATGGACGG CCACGAGGTG ACCGTGGTGG ACAATTTCTT CACGGGCAGG 420
AAGAGAAACG TGGAGCACTG GATCGGACAT GAGAACTTCG AGTTGATTAA CCACGACGTG 480
TGGAGCCCCT CTACATCGAG GTTGACCAGA TATACCATCT GGCATCTCCA GCCTCCCCTC 540
CAAACTACAT GTATAATCCT ATCAAGACAT TAAAGACCAA TACGATTGGG ACATTAAACA 600
TGTTGGGGCT GGCAAAACGA GTCGGTGCCC GTCTGCTCCT GGCCTCCACA TCGGAGGTGT 660
ATGGAGATCC TCAAGTCCAC CCTCAAAGTG AGGATTACTG GGGCCACGTG AATCCAATAG 720
GACCTCGGGC CTGCTACGAT GAAGGCAAAC GTGTTGCAGA GACCATGTGC TATGCCTACA 780
TGAAGCAGGA AGGCGTGGAA GTGCGAGTGG CCAGAATCTT CAACACCTTT GGGCCACGCA 840
TGCACATGAA CGATGGGCGA GTAGTCAGCA ACTTCATCCT GCAGGCGCTC CAGGGGGAGC 900
CACTCACGGT ATACGGATCC GGGTCTCAGA CAAGGGCGTT CCAGTACGTG AGCGATCTAG 960
TGAATGGCCT CGTGGCTCTC ATGAACAGCA ACGTCAGCAG CCCGGTCAAC CTGGGGAACC 1020
CAGAAGAACA CACAATCCTA GAATTTGCTC AGTTAATTAA AAACCTTGTT GGTAGCGGAA 1080
GTGAAATTCA GTTTCTCTCC GAAGCCCAGG ATGACCCACA GAAAAGAAAA CCAGACATCA 1140
AAAAAGCAAA GCTGATGCTG GGGTGGGAGC CCGTGGTCCC GCTGGAGGAA GGTTTAAACA 1200
AAGCAATTCA CTACTTCCGT AAAGAACTCG AGTACCAGGC AAATAATCAG TACATCCCCA 1260
AACCAAAGCC TGCCAGAATA AAGAAAGGAC GGACTCGCCA CAGCTGAACT CCTCACTTTT 1320
AGGACACAAG ACTACCATTG TACACTTGAT GGGATGTATT TTTGGCTTTT TTTTGTTGTC 1380
GTTTAAAGAA AGACTTTAAC AGGTGTCATG AAGAACAAAC TGGAATTTCA TTCTGAAGCT 1440
TGCTTTAATC AAATGGATGT GCCTAAAAGC TCCCCTCAAA AAACTGCAGA TTTTGCCTTG 1500
CACTTTTTGA ATCTCTCTTT TTATGTAAAA TAGCGTAGAT GCATCTCTGC GTATTTTCAA 1560
GTTTTTTTAT CTTGCTGTGA GAGCATATGT TGTGACTGTC GTTGACAGTT TTATTTACTG 1620
GTTTCTTTGT GAAGCTGAAA AGGAACATTA AGCGGGACAA AAAATGCCGA TTTTATTTAT 1680
AAAAGTGGGT ACTTAATAAA TGAGTCGTTA TACTATGCAT AAAGAAAAAT CCTAGCAGTA 1740
TTGTCAGGTG GTGGTGCGCC GGCATTGATT TTAGGGCAGA TAAAAGAATT CTGTGTGAGA 1800
GCTTTATGTT TCTCTTTTAA TTCAGAGTTT TTCCAAGGTC TACTTTTGAG TTGCAAACTT 1860
GACTTTGAAA TATTCCTGTT GGTCATGATC AAGGATATTT GAAATCACTA CTGTGTTTTG 1920
CTGCGTATCT GGGGCGGGGG CAGGTTGGGG GGCACAAAGT TAACATATTC TTGGTTAACC 1980
ATGGTTAAAT ATGCTATTTT AATAAAATAT TGAAACTCAC CAAAAAAAAA AAAAAAAAAA 2040
A 2041
(2) INFORMATION FOR SEQ ID NO: 132:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2012 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO 132: TACCAAGCTG CAAGAATCTA CTATATCATG GCAGAAGAAG TAGAGTGGGA CTATTGCCCT 60 GACCGGAGCT GGGAACGGGA ATGGCACAAC CAGTCTGAGA AGGACAGTTA TGGTTACATT 120 TTCCTGAGCA ACAAGGATGG GCTCCTGGGT TCCAGATACA AGAAAGCTGT ATTCAGGGAA 180 TACACTGATG GTACATTCAG GNTCCCTCGG CCAAGGACTG GACCAGAAGA ACACTTGGGA 240 ATCTTGGGTC CACTTATCAA AGGTGAAGTT GGTGATATCC TGACTGTGGT ATTCAAGAAT 300 AATGCCAGCC GCCCCTACTC TGTGCATGCT CATGGAGTGC TAGAATCTAC TACTGTCTGG 360 CCACTGGCTG CTGAGCCTGG TGAGGTGGTC ACTTATCAGT GGAACATCCC AGAGAGGTCT 420 GGCCCTGGGC CAATGACTCT GCTTGTGTTT CCTGGATCTA TTATTCTCCA GTGGATCCCA 480 TCAAGGACAT GTATAGTGGC CTGGTGGGGC CCTTGGCTAT CTGCCAAAAG GGCATCCTGG 540
NAGCCCCATG GAGGACGGAN TGACATGGAT CGGGAATTTG CATTGTTGTT CTTGATTTTT 600
GATGAAAATA AGTCTTGGTA TTTGGAGGAA AATGTGGCAA CCCATGGGTC CCAGGATCCA 660
GGCAGTATTA ACCTACAGGA TGAAACTTTC TTGGAGAGCA ATAAAATGCA TGCAATCAAT 720
GGGAAACTCT ATGCCAACCT TAGGGGTCTT ACCATGTACC AAGGAGAACG AGTGGCCTGG 780
TACATGCTGG CCATGGGCCA AGATGTGGAT CTACACACCA TCCACTTTCA TGCAGAGAGC 840
TTCCTCTATC GGAATGGCGA GAACTACCGG GCAGATGTGG TGGATCTGTT CCCAGGGACT 900
TTTGAGGTTG TGGAGATGGT GGCCAGCAAC CCTGGGACAT GGCTGATGCA CTGCCATGTG 960
ACTGACCATG TCCATGCTGG CATGGAGACC CTCTTCACTG TTTTTTCTCG AACAGAACAC 1020
TTAAGCCCTC TCACCGTCAT CACCAAAGAG ACTGAAAAAG CAGTGCCCCC CAGAGACATT 1080
GAAGAAGGCA ATGTGAAGAT GCTGGGCATG CAGATCCCCA TAAAGAATGT TGAGATGCTG 1140
GCCTCTGTTT TGGTTGCCAT TAGTGTCACC CTTCTGCTCG TTGTTCTGGC TCTTGGTGGA 1200
GTGGTTTGGT ACCAACATCG ACAGAGAAAG CTACGACGCA ATAGGAGGTC CATCCTGGAT 1260
GACAGCTTCA AGCTTCTGTC TTTCAAACAG TAACATCTGG AGCCTGGAGA TATCCTCAGG 1320
AAGCACATCT GTAGTGCACT CCCAGCAGGC CATGGACTAG TCACTAACCC CACACTCAAA 1380
GGGGCATGGG TGGTGGAGAA GCAGAAGGAG CAATCAAGCT TATCTGGATA TTTCTTTCTT 1440
TATTTATTTT ACATGGAAAT AATATGATTT CACTTTTTCT TTAGTTTCTT TGCTCTACGT 1500
GGGCACCTGG CACTAAGGGA GTACCTTATT ATCCTACATC GCAAATTTCA ACAGCTACAT 1560
TATATTTCCT TCTGACACTT GGAAGGTATT GAAATTTCTA GAAATGTATC CTTCTCACAA 1620
AGTAGAGACC AAGAGAAAAA CTCATTGATT GGGTTTCTAC TTCTTTCAAG GACTCAGGAA 1680
ATTTCACTTT GAACTGAGGC CAAGTGAGCT GTTAAGATAA CCCACACTTA AACTAAAGGC 1740
TAAGAATATA GGCTTGATGG GAAATTGAAG GTAGGCTGAG TATTGGGAAT CCAAATTGAA 1800
TTTTGATTCT CCTTGGCAGT GAACTACTTT GAAGAAGTGG TCAATGGGTT GTTGCTGCCA 1860
TGAGCATGTA CAACCTCTGG AGCTAGAAGC TCCTCAGGAA AGCCAGTTCT CCAAGTTCTT 1920
AACCTGTGGC ACTCAAAGGA ATGTTGAGTT ACCTCTTCAT GTTTTAGACA GCAAACCCTA 1980
TCCATTAAAG TACTTGTTAG AACACTGAAA AA 2012
(2) INFORMATION FOR SEQ ID NO: 133:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1669 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 133:
GAGCAGTATT TTAACCAACT TGTATTACAG ATGTTACAGT TCATGTTAGG AAGTCAGAAA 60
AGACTTTGTT TGTCTTTGTT CTGCTGATGT GAGTCATGTT TTGTGGGGTC TTCCATGGCA 120
CATTTACCTG TTGCTCCGTC CAGATGTTGA GGGCCAGTCT AGGCTGACAC ATCCTACCCG 180
AGGACAAGCC TGTTCTCCAT TTCTTCACTC TCCCCTCCCC ATATAGCAAC TCTCCCAGGT 240
TTAGATTACC GTTTTCGACG ACAGATTAAC CAAAAATGCC CCACACAGGT TTTATTACTG 300
TTATATACTA TACTTTTAAC AGTACAGACC CTAAATTTTA TTATTTGTTG CTCCCCCAAT 360
CTGATACCAA ATGTTTAAAG TTGTTTGAAA TCCAAACATG GTAGTGTTCA TGGGTAAATA 420
TTTTCTAGGC TATGTAAGAG TTAGCAGCCC ATAGCATAGA AGTAATCAAG TAGCATCTGA 480
GACTGTTGGA GGCACTAGGG CCTCTCTGGG CCTAACAGCC TCACTTCCCC AGCCTCACCT 540
TGCTGTCCTC TGACACTGCC ATCAGGGCTG TTAGTGGCAC CTGTATGAGG CCAAGTGTGC 600
GTCCAGGGGA ACAGCACAGG TTAATGCGTC TCCCTAGAAC TCATGAAGTC AGTTTAATTC 660
ATGCATGAAC ATGAGTTCAT TTTATCTTTT ATATAGCTTT CTTAGACATA CCAAACCATC 720
ATTCATAAAT CAGATAAATT ATTCAGTTTT TCTGTTTAGA AAGCTAAGTA TGTGTAGCTG 780
GAAACAAAAA TGAGCGTCTT TTCTCTCCTG TTAATCTAGA GTGTGCAGTT ACACATGTGT 840
GGATAATTTC ATGTTCCAGG GGCGCTTGGC ATCTCCCATG GACTGATTCC CAGGAAGAAA 900
AGCCCAAAGG GAAACCCACG ATTCCTTTCG AGTAGATGTG GGAAAGAGCC CATTGGAGGA 960
TATGAGGTCC TGTGAAATTC AGTTGTGTGT GTGGCTCCTT GTTAGCAGTC ATGTTGACAT 1020
GGTGTTAGGA GGCTCCCCAT CCACCCTTTA CATGATGTAG GGACCAGTGT CTTGTGAGAT 1080
TAACCTTGGG ACACAGTGGG TTAGCCTGGA GAAAATGAGA GGCCCTGCCT GGACCCAGGG 1140
AGAGGAGCCA GTGACACAGG GAGAGCGCTG CAGCCCTCCT TCCCTTCCAT TTGGAGGAGG 1200
TGGTGCCAGG AGCCTGCCCG CTTACCTCTG CTGAAGCATA AGTGGACTTT GCTTTTGGGG 1260
CTTATCTCTG ATACATGCTG GAGCCCTGCC TCTCCACTGC TAGATGGAAC CTGGAATCTC 1320
TCATCTACCT CTTAGTCTGT CAGTTTCTAC GTGTGAGAAG CAAGCTTGTG GGCCAGTGTC 1380
CTTGTACATG CTGTAGCACT TAAAAAATAA TTCCAGGGTT CCCTGGAAAA CCAGTCCCAG 1440
GGTTCCTATG ATCTGTAGTT TCTACCTGGA TTATAACTGG TTTTGGGTAC CTGAATTTTG 1500
ATTGGTTAGC CTTAATTATA GTCTGGCGTG ATCATGTAGA ATCTTTTCTG GTGAACAGAT 1560
CATAAAGTTC TATCAAGGAG TTCTATCAAG GCATCCATGT CAGTGGTGCT ATGCTGGTTA 1620
CAACTTGAGA TTTTTGAAAT AAAAAATTTG TCATAAAAAA AAAAAAAAA 1669
(2) INFORMATION FOR SEQ ID NO: 134:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1565 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 134:
CACTTTTGCT ATATAACCTA AGTGATAACC CTCTTTTAGT TACCTGCCAA ACTCTGGNCT 60
TGGTTTATAT TGCAGTTAAC ACAGTTACAA AGCTGTAATG GTGTCTTTTT TTCCTTTGTA 120
ACGGAATGTG TAAATCAAAG TATATACATT GTGTGGTGTT CCTGTTTCTG GAGTTTCATG 180
AGGATTTACA CATGGCATTC AGTGTTCTGT ATAGATCTGC CTACCTTTGT GAATTCATCT 240
GTTAACCCCT CTTCCTTTGA GAGAGCACCG GCGATGGTGG TTAACTCCTT GTGTTTTCTC 300
TCTCTCCTAC TGGTTATTCT TGAATTAAGC ACAGACTCGT CAGCTCGGTT GCTTTATCAT 360
GAATAATGTG TGTGACCTTG CAGTTCTTCC ACAGTTCAGC AAACAAGTGC TAGCTTCACT 420
GACCAAAAAT TAAGGAAGGA AAACACAGTT TTTAAAACGA TCCATCTTTT AACAGCCGAA 480
ACCGATGTGT CTATGGTGCT GCACCTTGCT GTTGTACTTC TGAAATCAGA CGTGTGTGAA 540
CGATCATTTC TGACTTAACC GTGAGATGCT CACGAGTACC CTTCCTGTTG TTTTGTTAGC 600
ATTCAAATCG AGACTATTTA TTTGGAATAT ATACAACAGT GTTTTTCCAC TGTATTTCAT 660
TTGCAAAAGT TGAGAACTGC TTTCTCTACC TTTTGCAAAA TAATTGATAT TCCATATTGG 720
ATTCTCAAAG ACTTCGATAT GGTGAACCTA TTAAACCTAG AAATTGTATT CATCCTTTCA 780
TGACTGTGGC CTGAGTTCCC CAGCCCCTCT CCTCCTTTTT TTTAGATGAG ATTTAGCACA 840
CTCTCAGTTA TTTAAACATG CAACATTTCT TGAGTATGTA TGTTGAGGCC ATCTGAGCTC 900
ATAGCTGATT CAGTAACCAG TTTCATGCTG TGTCATTCAC ACTCACTACT TAATACTGCC 960
ATGGTGAAAA TGTGGAGGAA AAATGTATCC ATGTGTGTCT GGGAAGCATA TACACTTGTA 1020
CATTTTTTAA TACTCTGATT CTGTAACATT TCTGAGTTTT GTTTTGTTTT ACAGNAAAAA 1080
AAAAAAAAGT GATAAAGCAA TCAGAAGACC AAGAGGTTTA CTATTGATGC TTAGGGTCGT 1140
CTGACCTTGG CTGGCCAATA GACCTACACG GCCAAATTAA TTTACGAGAG TAATAATTTT 1200
TCAAAAGCCA ATTTTTTTTC TGTATTTTCT GTATGAAACT GCCAATATCA TGAATAGAAA 1260
GGGAGAACCA TAAAGGAGAA AGAACGTGAT GTTCTGTTAT GTTCATGTAA ACCTAAAGAA 1320
ACAGTGTGGA GGCAGGCGCG ATCAGCCGAA CTCTAGGGAC TTGGTGTTGC TTGGAAGGCA 1380
TCCATACCTG CATTTTGCAT TCTTCGTATG TAATCATATT GCCAAAGACA AACTATTTCA 1440
TCATTTATTG TAAATAACAC TTTTCCCCAG ACCTACCATA AAGTTTCTGT GATGTATTGT 1500
CTTCCAGTTG CAATAAAAAT TACTGAGTTG CATCAATTGA AGAAAAAAAA AAAAAAAAAA 1560 CTCGA 1565
(2) INFORMATION FOR SEQ ID NO: 135:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2007 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 135:
TCTAAAAGCC CCCTTATACC CCACTTTGTG CAGCAAAGAT CCCCGTGCAG GTCACAGCCT 60
GATTTGTGGC CAGGCTGGAC AAATTCCTGA GGCACAACTT GGCTTCAGTT CAGATTTCAA 120
GCTCTCTTGG TGTTGGGACC AGCAGAAGGC AAACGTCCAG CCAACACACA GGACTGTAAG 180
AGGACTCTGA GCTACGTGCC CTGTGAAGAC CCCCAGGCTT TGTCATAGGA GGTCGTTCAG 240
CTTCCCCAAA GTCAGAGGTG ATTTGATTTG GGGAAGACTC AATATTCACA CCTAAGTCGT 300
GAGCATATCC TCΛGTTTTAC TTCCTTATGG CTTGCCCTCC AAGTTCTCTC TCTCATACAC 360
ACACACACCC TTGCTCCAGA ATCACCAGAC ACCTCCATGG CTCCAGCTAT GGGAACAGCT 420
GCATTGGGGC TGCCTTTCTG TTTGGCTTAG GAACTTCTGT GCTTCTTGTG GCTCCACTCG 480
CGAGGCAGCT CGGAGGTGTG GACTCCGATT GGGCTGCAGG CAGCTCTGGG ACGGCACAGG 540
GCGGGCGCTC TGATCAGCTC GTGTAAAACA CACCGTCTTC TTGGCCTCCT GGCAGTTCTT 600
TCTGCGAATA GTCCTCTCCC TGGCCAGTTG AATGGGGGAA GCTGCTGGCA CAGGAAGGAG 660
AGGCGATCCC GGCTGAGGCT TAGGAAATTG CTGGAGCCGG CTCCAAGCAG ATAATTCACT 720
GGGGAGGTTT TCAGAGTCAA ACATCATTCT GCCTGTKTTG GGGGCCAGGT GTGTCACACA 780
AGCATCTCAA AGTCAAAAGC CATCTGGGGC TGCTGCTTCT CTTTCTCAGG CTCTGGGGAA 840
AGGAATCTCC CTCTCCTCTC ACTTGATTCC AAGTGTGGTT GAATTGTCTG GAGCACTGGG 900
ACTTTTTTTC TCTTTTCCTT GATGGACCAA CAGTGCAAAT GCAATCTCGC CATTTAACTT 960
TCAGGTCGAT TTCCTTTCCT GATCAGACAT CTTTGTGCCC CCTTTAGGAA GGAAAAGAAT 1020
ACACCTACGA TGTGCCAGGC ACTGTGTTAG GCGCTTTTAT ATAGATCCTC GTTAGGATGA 1080
GACTAAGGGA TGAGGACATC TCTTTATAAA AGGCCCCTAA GTAATGGATA AACAGAAACA 1140
CTTAGAGGTG AGAAGGTCTG TCTTCAAGAT CCAAGGTAAG ATTGCCTTCA GTCTGATGTT 1200
TGTTCTCAAG GACTTATCCC CTACAATATT CTCCCACTCC ATACTTCTCC TTCTACCCCA 1260
CCATGTGCTC CCGTGCACTC CTCAGATGGT CAGAGGGGTA ACCCAAGTCC TTAGAGAATT 1320
TGGGGACCAA TAGAATATGT GATGTGTGAA TTTTCTTTAA AAAACTTAAG GAGTCTTTGC 1380
TACCTTCTGC TTGTTGAGTT GTTTTGGCAT TCATATTAAA AGCCAGCATC TCACTATTTA 1440
TTGACAGGTT GGGCTGTGTG TGTGCGCATG TCTGTATACA TTTCCAGGCG TGCCTGTGTC 1500
CTGTAGCTTT TTAAAAGGAA ACCCAGTCAT CCCACTATGA ATCTGGCATC TTCTTATGCT 1560
TCTAGTGTTT TGGCCATACA TCAACCAAGG GGTTTAATTT ATCCAATGCT TGACGACATG 1620
TTCAGGAGGG GCTGGATCAA ATTTTGAGAG GGTTATGGGA AAGGGAGGGG GAGAAGAAAT 1680
TGACATTTAT TTTATTATTT ATTTTAAATG TTTACATCTT CTTTATGTTG TATCAAGCCT 1740
GAATAGAAAC TGATAGCATT AAAATACTCC GTTCCTCTCT CTCTTCTCGC TTCCTTTTTT 1800
TTTTTTTTTA AATTTAGGAT AACACATTTT TGTTTCTAAA GTCATTTGTG ATTTGTGCTG 1860
TATAAACTGT ATAAAAGGTT CTGTTTTTAA AGGTGGATTT TCATTCCTCT GGGGACAGTG 1920
GTCGCCAAGA CATCTACATT GTAAGAGAAC ACAGTGGAAG ATCCTGTCCT GATTCTCAAA 1980
AATTATTTTC TCTGTATGAT TAAAAGT 2007
(2) INFORMATION FOR SEQ ID NO: 136:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1291 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 136: CTTTTAACCC TCCCCCTTCA CACACATACA TATCAGGTTG TTTTCTAGTT AAAAACCCAA 60 GTAGCTCAGA TTCTACTTTA ATGTCAGTGC AGATTTCCAT TGAATCATGC CATTATGTTT 120 TTTCTCATTT TTATGCTGTT GGGTCTTAGT TTTTAAATTG ATATAAAGAA CTCAGCAATG 180 GTTTTATTTT CTACTCATAC TTAGGGTTTA GGAAACACTA CCACTAGTTA TCATTTAATC 240 AACTTCAATG GTCTACTGAA ACAAAAATGG TAACTTTTCA TTAGTGGATT ATTTAGAGTT 300 ATAGTAGTTG TTTCCAGAAA ACACTTCCTC ACAATTGTAC TTCCCAATCA AATCATGTGA 360 TCATACAGTT ATTCCCATGA AAGGCAGAAT GTTTGTTTCA AAATTAATCT AGTTTTCTGT 420 ACATTTAAAT TTGAGAAGGT GACAACTGGC TCTTTTCCAG TCTTCCTTCA TGTCAGTTTT 480 CTGATAGACC ACTATTGGCA AACAGTATCT GTCAACTACC AAATGTGTAA AATTTTCTGT 540 ATTTCACTTT GTCTTATTTG TAAATAGTGA ACTAAAACTT TTGGCAGATC AGCAACATTT 600
GCTGAGCCTG TTTTTTAAGC TAATGTGTAT TCTTACTAAT GTTCCTATCA AGAATGGATT 660 TGTAATATAT GCTGTCTATT TCTAATGTTC ACATTCATAT TTTGAGGTTC TATCTTATTT 720 TAATAGAGAA CAGACTTCTC AAAAAATCTT CAGAAGCAGC TTATTATTGA AATATCGAAA 780 TATTGAAATA AACCCGGTGG GTTAGATTAC TCATCTGTCC ACCAAGTGGG ACATTTGCAT 840 GGACTGGGGG CTTAAAGGAC TTAGAAGAGA CCTGTAAGTA AATCCTGAAA ATGAGCCAAT 900 CCCCACTTGA ATGGTTACTG GAGTAAACCC ACCTTTACCA CCCCAATTAC AGCACCCGAG 960 GCCGATAAAC CAACTTGGCT CTGGTTCATT TTTCTTTTCT TCATTTGTGA TCCTCAGATT 1020 CAAAATGTGT GTTCTACACT GTTACAGGCT TCTCTTTTGT TTGATTAAAG ATTTTAGTCC 1080 TACTTTTGTA TGGACACATT AGAATATTCA GAGACCAAAA TAGAAGAATT TGCTGTTAGA 1140 TATTTTTCAG AAGTCAGCAG ATTTGTGGCA AATCATTTAT TTGCCTTTTT AAAAATTCAT 1200 TTAAGCAGTT CAGAGAGTAG ACTACTCAGA AAATTATTTC ACGTAATTGT CTAAGAGGTC 1260 AATATTTTTT AATGCATATT GAATCAAATA A 1291
(2) INFORMATION FOR SEQ ID NO: 137:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1906 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 137:
GGCACGAGGA CCTACTTTTG TAACAGACCA TGGTTGTGTC CAAGGTAAAA CCACAGTGAT 60
ATTTTTGGAT GCTTTGTCTG CAATCTTGAC TTGTTTTTGC AGTATCATTA TTCAGACTTC 120
AAATTGTGAA TCTTTTAAAC ATCTTGATAA TTTCTTCTTG AGAGCTGTTC ATTCTAAAAT 180
GTAATGAAAT TCAGTCTAGT TCTGCTGATA AAGATCATCA GTTTTGAAAG GTTACTGATT 240
TTCCTCTTCC CTCTTAGTTT TTTACCCAAT ATATGGAGAA GAGTAATGGT CAATCTTAAC 300
ATTTTGTTTT AATTGTTTAA TAAAGCTGCT GGGCAGTGGT GCAGCATTCC TACCTAGTGT 360
CATAAAAGCA AAATACTTAC ATAGCTTTCT TAAAATATAG GAATGACATT ACATTTTTAG 420
GAGAAAGTAA GTTGCTTTGC ACCGCCTACT TAATTCCTTT CCATATATTG TGATACAAAC 480
TTTTGAATAT GGAATCTTAC TATTTGAATA GAAATGTGTA TGTATAATAT ACATACATAC 540
ATAAGCATAT ATGTGTGTGT GTGTGTGTAT ATATATATAT ATGCATGCTG TGAAACTTGA 600
CTACACAACA TAAATCACTT TTTAAATTCC AGGAACGGGT AGTCTGACAC GGTGATTATC 660
CTTTTGAGGC TGAATCCGTT ATTAACTTGT TATTTAGGTT TTACTCCCAG TAGCAAGGGA 720
TTCTAAGTTA GTTGCACTTA CATGATTATT GTGATTTAAA ACTAAGAATA AAGGCTGCAT 780
TTTCAAAGAT AAATTGGAAT TGCTGTTGGT GAAATAACAA CCAAAATACT GAATCTGATG 840
TACATACAGG TTTCTACAGG AAGAGATGGT ATAATTTACA ATTTGGAGAT TTAATAACCA 900
GGGCTACCCA GAAAAAGTGA CTTGATAACA TGGTACCAAT AAGTAAGGGA TGCTCTCTCG 960
GTTTGCTTTT GCCACTTTCA AGATTTTAAC TTCTCAGGTT ATTAATCAAA ATTATTGTAT 1020
AAGTTAGCCA ATAGAATTTT TAGGTTAAAA CAACAGATGG GGGGTTTGTG GAGTGTTTAA 1080
TGTCATGGGC ATTTTTAGTA GCATAGACCC TTTGTTCTGC ATTTGAATGT TTCGTATATT 1140
TTTCTTTCAC AGTTAATCTT CCCTCCCCAA GTTTGCTATT CAAATCAACT GCCTGAATGA 1200
CATTTCTAGT AGTCTGATGT ATTTTTCTGA GGAATAGTTT GTGATTCCAA TGCAGGTCTC 1260
TTCATTACCA TTACCTCTAC ACTGCAGAAG AAGCAAAACT CCTTTATTAG AATTACTGCA 1320
CATGTGTATG GGGAAAATAG TTCTGAAAGG CTAGAATGAT ACAAGTGAGC AAAAGTTGGT 1380
CAGCTTGGCT ATGGAGTGGT GGCAATAATC TCTAAACATT CCAAAAGACC ATGAGCTGAA 1440
CCTAAACTCC CTTGGGAATC TGGAACAAAG GAATATGAAA ATTGCCATTT GAAAACTGAC 1500
CAGCTAATCT GGACCTCAGA GATAGATCAG CCAGTGGCCC AAAGCCATTT CAAGTACAGA 1560
AATTATAGAG ACTACAGCTA AATAAATTTG AACATTAAAT ATAATTTTAC CACTTTTTGT 1620
CTTTATAAGC ATATTTGTAA ACTCAGAACT GAGCAGAAGT GACTTTACTT TCTCAAGTTT 1680
GATACTGAGT TGACTGTTCC CTTATCCCTC ACCCTTCCCC TTCCCTTTCC TAAGGCAATA 1740
GTGCACAACT TAGGTTATTT TTGCTTCCGA ATTTGAATGA AAAACTTAAT GCCATGGATT 1800
TTTTTCTTTT GCAAGACACC TGTTTATCAT CTTGTTTAAA TGTAAATGTC CCCTTATGCT 1860
TTTGAAATAA ATTTCCTTTT GTAAAAAAAA AAAAAAAAAA AAAAAA 1906
(2) INFORMATION FOR SEQ ID NO: 138:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1935 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 138: TCTGAACTAA TGCTAACAGA TCCCCCTGAG GGATTCTTGA TGGGCTGAGC AGCTGGCTGG 60 AGCTAGTACT GACTGACATT CATTGTGATG AGGGCAGCTT TCTGGTACAG GATTCTAAGC 120 TCTATGTTTT ATATACATTT TCATCTGTAC TTGCACCTCA CTTTACACAA GAGGAAACTA 180
TGCAAAGTTA GCTGGATCGC TCAAGGTCAC TTAGGTAAGT TGGCAAGTCC ATGCTTCCCA 240
CTCAGCTCCT CAGGTCAGCA AGTCTACTTC TCTGCCTATT TTGTATACTC TCTTTAATAT 300
GTGCCTAGCT TTGGAAAGTC TAGAATGGGT CCCTGGTGCY TTTTTACTTT GAAGAAATCA 360
GTTTCTGCCT CTTTTTGGAA AAGAAAACAA AGTGCAATTG TTTTTTACTG GAAAGTTACC 420
CAATAGCATG AGGTGAACAG GACGTAGTTN AGGCCTTCCT GTAAACAGAA AATCATATCA 480
AAACACTATC TTCCCATCTG TTTCTCAATG CCTGCTACTT CTTGTAGATA TTTCATTTCA 540
GGAGAGCAGC AGTTAAACCC GTGGATTTTG TAGTTAGGAA CCTGGGKTCA AACCCTCTTC 600
CACTAATTGG CTATGTCTCT GGACAAGTTT TTTTTTTTTT TTTTTTTTAA ACCCTTTCTG 660
AACTTTCACT TTCTATGTCT ACCTCAAAGA ATTGTTGTGA GGCTTGAGAT AATGCATTTG 720
TAAAGGGTCT GCCAGATAGG AAGATGCTAG TTATGGATTT ACAAGGTTGT TAAGGCTGTA 780
AGAGTCTAAA ACCTACAGTG AATCACAATG CATTTACCCC CACTGACTTG GACATAAGTG 840
AAAACTAGCC AGAAGTCTCT TTTTCAAATT ACTTACAGGT TATTCAATAT AAAATTTTTG 900
TAATGGATAA TCTTATTTAT CTAAACTAAA GCTTCCTGTT TATACACACT CCTGTTATTC 960
TGGGATAAGA TAAATGACCA CAGTACCTTA ATTTCTAGGT GGGTGCCTGT GATGGTTCAT 1020
TGTAGGTAAG GACATTTTCT YTTTTTCAGC AGCTGTGTAG GTCCAGAGCC TCTGGGAGAG 1080
GAGGGGGGTA GCATGCACCC AGCAGGGGAC TGAACTGGGA AACTCAAGGT TCTTTTTACT 1140
GTGGGGTAGT GAGCTGCCTT TCTGTGATCG GTTTCCCTAG GGATGTTGCT GTTCCCCTCC 1200
TTGCTATTCG CAGCTACATA CAACGTGGCC AACCCCAGTA GGCTGATCCT ATATATGATC 1260
AGTGCTGGTG CTGACTCTCA ATAGCCCCAC CCAAGCTGGC TATAGGTTTA CAGATACATT 1320
AATTAGGCAA CCTAAAATAT TGATGCTGGT GTTGGTGTGA CATAATGCTA TGGCCAGAAC 1380
TCAAACTTAG AGTTATAATT CATGTATTAG GGTTCTCCAG AGGGACAGAA TTAGTAGGAT 1440
ATATGTATAT ATGAAAGGGA GGTTATTAGG GAGAACTGGC TCCCACAGTT AGAAGGCGAA 1500
GTCGCACAAT AGGCCGTCTG CAAGCTGGGT TAGAGAGAAG CCAGTAGTGG CTCAGCCTGA 1560
GTTCAAAAAC CTCAAAACTG GGGAAGCTGA CAGTGCAGCC AGCCTTCAGT CTGTGGCCAA 1620
AGGCCAAGAG CCCCTGGCAA CCAACCCACT GGTGCAAGTC CTAGATTCCA AAGGCTGAAG 1680
AACCTGGAGT CTGATGTCCA AGAGCAGGAA GAGTGGAAGA AAGCCAGAAG ACTCAGCAAA 1740
CAAGGTAGAC AGTGTCTACC ACCAYAGTGG CCATACCAAA GAGGCTACCG ATTCCTTCCT 1800
GCTACCTGGA TCCCTGAAGT TGCCCTGGTC TCTGCACCTT CTAAACCTAG TTCTTAAGAG 1860
CTTTCCATTA CATGAGCTGT CTCAAAGCCC TCCAATWAAT TCTCAGTGTA AGYTTCAAAA 1920
AAAAAAAAAA AAAAA 1935
(2) INFORMATION FOR SEQ ID NO: 139:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1446 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 139:
NGCCCCCTTG GCACAAGTCA GATGAAGCAC GTTCTGCCGG GGAGGCCCTC AMCTTCCAGA 60
GAGGACAGAC ACAGATTTCC TGCTGGGGGA GGGAGGAGTC CACGCATCCT GATGCTGCCT 120
GGAAGCTTAT TTTCCCGTGG CCAGGATCCA TTTCTCTCAG TGGAAACAGG TTCTTGCATG 180
TGGATGTGTG TTTCCCCAGG CAGACGGCCC CTCTYTTCCC AGCACTTCCC TGCCTCCCCC 240
AGGCCTCAGG CCAGCACCCA GTTCCTCCTC ACATGGCAGG TGAGCACAGA CTTCTAGTTG 300
GCAGGAGCTG AGGAGGGTGA ACAAACCCCG AGGGAGGCCC GGCCCTTGCT CCCGAGTTGG 360
GGGGAGGGGG TGTGGCAACG TGCCCCCCGC AGAGGCCACG CATGTTTGAC CAAAGCCCTC 420
ATTGTGGTCC GAGGACAGCC TTTTCCCCAG GCCTCARAGC ATTGCTCATC CGTGCCAAAC 480
TGGGTAGGTG GATTTGAGCG GAAAGACTCC CAAAATGTGC CAAGAATTTC CCRGTCCCAG 540
GCAGGGCAGG GGAAACTAAG GGCAAGCAGG ATACAGGGCG AGGGATGTGG CAGGTGAGGG 600
GGCTCCCGCC TGTGCCCCTT CTCCTCACCA TGTCTCCCCC ACCCTGCCTC AGTTCTCCGT 660
TCCCCTTCAT CTCCGTCCCC CTCTTTGAAG CTGTCCCCAT CTCAGTGTCA GACCAGCCTT 720
CTCCTCAKCT GACCACCCTC CTCTGACCSA CGCCCCCTCC TTGTCTGAAA AAAGGAGCCT 780
TGAATGGTGG AGGGAGGCAG TGGGGAGAAA GGTCTCACCG GACAGGTTGG GAGAATGAGG 840
TCAGCGGTGC TGGGGAACAG ATGGAGGGGG CAGTGGGGAC AGGGCTTGGG CAGACACCAG 900
CAGGAATAAT TTGAAATGTG TGAGGTGACT CCCCGGAGGC CTTGGGCTTG GGCATTTGGG 960
AAAAGAATGA TGTCTGGAAG GGCTTAAGGG ACACAGTGGA CGAGGGGAGA GTCCTCATCT 1020
GCTGGCATTT TGTGGGGTGT TAGTGCCAAA CTTGAATAGG GGCTGGGGTG CTGTCTTCCA 1080
CTGACACCCA AATCCAGAAT CCCTGGTCTT GAGTCCCCAG AACTTTGCCT CTTGACTGTC 1140
CCTTCTCTTC CTACCTCCAT CCATGGAAAA TTAGTTATTT TCTGATCCTT TCCCCTGCCT 1200
GGTCTAGCTC CTCTCCAAAC AGCCATGCCC TCCAAATGCT AGAGACCTGC GCCCTGAACC 1260
CTGTAGACAG ATGCCCTCAG AATTGGGGCA TGGGAGGGGG GSTGGGGGAC CCCATGATTC 1320
AGCCACGGAC TCCAATGCCC AGCTCCTCTC CCCAAAACAA TCCCGACAAT CCCTTATCCC 1380
TACCCCAACC CTTTGCGGCT CTGTACACAT TTTTAAACCT GGCAAAAGAT GAAGAGAATA 1440
TTGTAA 1446
(2) INFORMATION FOR SEQ ID NO: 140:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1109 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 140:
TTTTTTTTTT TTTGATATGA AATTCTCTTT CTCCATTGCA GAAATAAGCT AGGGAAACAC 60
TAACCCAAAA ACTTTCTGTA GAGCTGTTCC TTTGGAGGCA GCATCACTTA TTGGCAGTAA 120
AGACTCAGTA TAAAAGCACC AGCATCCCTA CTTGGGTGAT GGGGATTAAT TTTATAGCAT 180
TCCATTTTCC TAGTGCCACA TGTGAAATTG GATTTTGATG ATCTTAATCT ATATTCTACC 240
CTTATAATAA AAGATCAAAA GATATATCTC CTATGAACAG ATTGGAGATA GGAGATGAAA 300
AGTTGGGAGG ATGTCTTTAT TCTAATCTCA GGGTAGGGAA AATGTGGATA ACATTACTGG 360
GGTCARGGAG GCATTGTTCT TTAGTTGGAG TTCTCATTTT TATTCTCCAG TACTGACTTG 420
TGGGGAAAGC ATACTTTTTC ACTGCCAGGT ACTGAATGCA GAGGCTCAGT GAAGTATATA 480
TGTGGGAAGT GCATGCATTT CGTTTATTAG CAAACATAGC TGGATTAAGA CAAAGTTGTT 540
GGTTTGGAAA GGGGTTAAAG CCTTAAGTGA ACAAATCTAG CTAACAGTGA ATGAACTAGG 600
TAATATAACT TGCATATTTT TAATTTCCTT TGGTTAAAGG TCCCCCATAC TTCTCTGTTC 660
GGAGACATCA GAAGTATGAT TACTTCAGTG TTAGTTTTCT TAATTTTTTT TTTCCCCTAT 720
TTGTCCCTTG TCACTTTGTT GCAAGCTAGA AATCTGTGGG TTATACATAG GGCAGCTCTT 780
TGTGAAAGTG GTTTATTCCA CTGGAGAAAG GGGATTGAAA ATCAGTTAGA ACCAATGTAT 840
TTCTTGCCCC ACGGAACACT ATTCCTATAA GATAGCTGAA AGAAGCTGCT GTGAGGAGCT 900
CAGCTCCAAA CACAGGATCA GCACCTTGTA TAGGAATTCC CATGAATTAT GACTTCTCAT 960
TCTGTTTTAT CAGAGTGCAT ATATCTCCTA CTTCAGGAAA AGTAAAACAG TCATTTACGA 1020
AAGAAAGTCA ATCTGTATCC TAAGCATTTT AATAAAAAGT TAAAACAAAA AATTAAAAGG 1080
GACACTCGAG GGGGGGCCCG AAACCCAAT 1109
(2) INFORMATION FOR SEQ ID NO: 141: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 497 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi ) SEQUENCE DESCRIPTION : SEQ ID NO : 141 :
TAGGACTAAC TTAAATTCTT TTATTCATCT TTTATTTATT AAAAAATTTT ATTTCTTTGA 60 ATTTTCCTGT AATTTCCTTA RGCTCTTCTA TAAAATCTTA TATTCATGTG AACCATACCT 120
CATTATCCTT AACATTTACT CTCAAAAAGC TTTTTATTTT TATTTTTTTG AAGGTAGTTT 180
TTCTGTGTGT ACTCTGTAAC ATGATTTTGC TTTCAAATCA TTGTTGTGCC CCCATACAAA 240
ATGCCTTTTA TTTTTGAGGA TCGTGGACTT TTTAGTATGG CATGAGTGTG CTAAAAGCCA 300
GATATCTTTC CACATTCACT GGTGGCTTTG ACACCTAGTT TTTAATCTCC CATCCTTACT 360 TTAAACCCTG ACAGTGCAGT CCTCAGTCAG GGCCAGGACC GGGCTGAGGC CCTTTGTGGA 420
GATGCTGCAC CACCAGCAGA AGGCTGAGAC CTGGTTACCT GTACCTGTTC ACTTGTAATA 480
AAAAGAATTA TCTAAAA 497
(2) INFORMATION FOR SEQ ID NO: 142:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 269 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 142:
ATGAGGCAGA GGCAAGCTGC CTGCCAACCC CCTCCCTCAA GGAATGGCCT TGCCCAGGAA 60
TGCCCACCAC ACATACCCTC TTCTTTTTTT CTAGTCAAAC TCTTGTTTAT TCCTTGGCTT 120
GCCTCCCTCC TTTCCTCCCC TCTCAACCTT TTACTTCTGG TTTCTATTTC ATGGGATTTG 180 GGGTTGAAGT TAAACTTACA ACAGTGCCGC CAACACCAAG TCTTGCAGGA AAAAAATACA 240
AAGAAATTTA ACAAAAAAAA AAAAAAAAA 269
(2) INFORMATION FOR SEQ ID NO: 143:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 1269 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 143:
TTGATTGACT ATGGTCTCTC CGGCTACCAG GAAGAGTCTG CCGAAGTCAA GGCCATGGAC 60
TTCATCACCT CCACAGCCAT CCTGCCCCTG CTGTTCGGCT GCCTGGGCGT CTTCGGCCTC 120
TTCCGGCTGC TGCAGTGGGT GCGCGGGAAG GCCTACCTGC GGAATGCTGT GGTGGTGATC 180
ACAGGCGCCA CCTCAGGGCT GGGCAAAGAA TGTGCAAAAG TCTTCTATGC TGCGGGTGCT 240
AAACTGGTGC TCTGTGGCCG GAATGGTGGG GCCCTAGAAG AGCTCATCAG AGAACTCACC 300
GCTTCTCATG CCACCAAGGT GCAGACACAC AAGCCTTACT TGGTGACCTT CGACCTCACA 360
GACTCTGGGG CCATAGTTGC AGCAGCAGCT GAGATCCTGC AGTGCTTTGG CTATGTCGAC 420
ATACTTGTCA ACAATGCTGG GATCAGCTAC CGTGGTACCA TCATGGACAC CACAGTGGAT 480
GTGGACAAGA GGGTCATGGA GACAAACTAC TTTGGCCCAG TTGCTCTAAC GAAAGCACTC 540
CTGCCCTCCA TGATCAAGAG GAGGCAAGGC CACATTGTCG CCATCAGCAG CATCCAGGGC 600
AAGATGAGCA TTCCTTTTCG ATCAGCATAT GCAGCCTCCA AGCACGCAAC CCAGGCTTTC 660
TTTGACTGTC TGCGTGCCGA GATGGAACAG TATGAAATTG AGGTGACCGT CATCAGCCCC 720
GGCTACATCC ACACCAACCT CTCTGTAAAT GCCATCACCG CGGATGGATC TAGGTATGGA 780
GTTATGGACA CCACCACAGC CCAGGGCCGA AGCCCTGTGG AGGTGGCCCA GGATGTTCTT 840
GCTGCTGTGG GGAAGAAGAA GAAAGATGTG ATCCTGGCTG ACTTACTGCC TTCCTTGGCT 900
GTTTATCTTC GAACTCTGGC TCCTGGGCTC TTCTTCAGCC TCATGCCTCC AGGGCCAGAA 960
AAGAGCGGAA ATCCAAGAAC TCCTAGTACT CTGACCAGCC AGGGCCAGGG CAGAGAAGCA 1020
GCACTCTTAG GCTTGCTTAC TCTACAAGGG ACAGTTGCAT TTGTTGAGAC TTTAATGGAG 1080
ATTTGTCTCA CAAGTGGGAA AGACTGAAGA AACACATCTC GTGCAGATCT GCTGGCAGAG 1140
GACAATCAAA AACGACAACA AGCTTCTTCC CAGGGTGAGG GGAAACACTT AAGGAATAAA 1200
TATGGAGCTG GGGTTTAACA CTAAAAACTA GAAATAAACA TCTCAAACAG TAAAAAAAAA 1260
AAAAAAAAC 1269
(2) INFORMATION FOR SEQ ID NO: 144:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1944 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 144: AAAAGGCAAA CTATAGGATA ACACAGAGCC CTTTTTGAAA ATAAATTGGC ATTGGAGTGT 60
TTTACCCTCT AGCTGTTTTA CTTAGAATCT AACATATGCT GCCTACCCAC CTCAAAATGT 120
CTGTACTGCA AGAGGGCCCT GGGCCTCTGC TTTCCATATT CACGTTTGGC CAGAGTTGTA 180
GTCCCAAAGA AGAGCATGGG TGGCAGATGG TAGGGAATTG AACTGGCCTG TGCAATGGGC 240
ATGGAGCACA AGGGGTCACA GCATGCCTCC TGCCTTACCG TGGCAGTACG GAGACAGTCC 300
AGAACATGGT CTTCTTGCCA CGGGGTGTTG TTGTCTCTGG TGGTGCTGCA TGTCTGTGGC 360
TCACCTTTAT TCTTGAAACT GAGGTTTACC TGGATCTGGC TACTGAGGCT AGAGCCCACA 420
GCAGAATGGG GTTGGGCCTG TGGCCCCCAA ACTAGGGGGT GTGGGTTCAT CACAGTGTTG 480
CCTTTTGTCT CCTAAAGATA GGGATCTACT TTTGAAGGGA ATTGTTCCTC CCAAATAAAT 540
TTGCTTTACC TTGGTCCTTT CTTTTGTGCC AGTATTCAAG TGGTATAGCT CTGAGCAGGG 600
TCACATTTGG CCAAACCTGA CACTGTCTTG CTGCATTCTC CTTTGGCAAA CATCAGGGTC 660
AGAATTCAGG ATAGCCCTTC CTAGGGCACT GGACTTTCTG GCATGGGGGC TGTGTTTGCA 720
CAAGTTATTT TCATGTTACC TGGAGAGTGT CCAGAGGCTG CTCTGAGGCT GAGGTGTGTT 780
CCCCCTTGCC TGGTTCCAGC TGTCAGAGGG ATACCATCCT AGGGTCTGGG AATCCAAGGC 840
CACGAGACTC CTTGGTTTGT GGTCCGAGAT CCTGTACTAA GGAGGGTCTG GCCAGAGGAA 900
CAGACCAGCT TTTGCACAAT GAAGCGCAAG GGAACAAGTG GTTTGCCTGG TGTCCTACCT 960
GTCCTGAACC TGGTCCTGTG GGCCATTGAA AAGTTAGATC TGTGATCTCT GGGGTTTTTG 1020
TGGCTTTGTT CAATGCTTCC ACTCTAGGGC AGGCAGAGCA GTCTATACTC TCCGAAGCCT 1080
GCTTGACCTC CAAGTAGAGC TGATACAGAG ATCTGTGAAT ATTGTGATAG AAATTCTTTG 1140
GTATTCATAC ATTTCAGCTG CAAGTCAGCA ATTTCCCAGG TACCATGTAA GCTATAAAAC 1200
AGTCATTCTT AAAGACAGAG GATAGCTGTG ACTCATGGGA TCATGAGGTC CATGGCTGGT 1260
TGCAGGTTCC CTTTTTCCTT CCTCAGGTTT TGTCTCTTCC TGTGTTGTCC CCAGCAAGGG 1320
AGAGACTGTG GGGTGGATTG GGAGAACAGA TTAGGAGTAT AGCAAATGAA CCCAGAATGG 1380
AACAGTGGGG AGCTAACTGT GAATGAGGAG AGTACCTGCT GCAGGACCTG GAGGTCAGGT 1440
GTGAATGCTG TATTGGCACA GGGAATAAAT ATCCTGGCGT CTGGAGCCTT CACCTCTCCG 1500
TCAAGTCCTT CCTGTGATAC TGCCATGGCA CAGGATCTGA GTTGCAGCTC TGCACCCTAA 1560
ATCACACCCT GGGCATTGTC TGGGCTGCAG GGCTGCCAGG TTCTGTACTT GTGTCCAGCT 1620
GTGGCCCTGG ATGCTGGAGC TGGAGGGTTT TCTGTGCTCA GACTGTAGCC TGTAGCTCTT 1680
GGCCTGTGTA GAGCCCCCTC CTGTGCCCTC AGTGGCTGTC GTTTGTTAAC ATCATCAGGA 1740
AGATGGGAAA GGTCAGGCAG AATTTTTCTG CCCTACAAAG GGTGGAAGAG AAAGGACACA 1800
GTATTTTCAT GAATTTACCA TATATCTTTC TTTTTCTTCA ACGAAAAAGT TAATTGAGGC 1860
AATGTCATCT GCTCAAAGTT GAGTGGTTTA TTCACAATAA ACTGTAAGTT TCTGATTATA 1920 AAAAAAAAAA AAAAAAAAAA AAAG 1944
(2) INFORMATION FOR SEQ ID NO: 145:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1021 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 145:
TCGACCCACG CGTCCGGGGT GCGCAACGGG GAGTTCCGGC TGGAGACCCG TGCTCTGGGC 60
CGGCGCCTTC ACCATGGCCT CGGCAGAGCT GGACTACACC ATCGAGATCC CGGATCAGCC 120
CTGCTGGAGC CAGAAGAACA GCCCCAGCCC AGGTGGGAAG GAGGCAGAAA CTCGGCAGCC 180
TGTGGTGATT CTYTTGGGCT GGGGTGGCTG CAAGGACAAG AACCTTGCCA AGTACAGTGC 240
CATCTACCAC AAAAGGGGCT GCATCGTAAT CCGATACACA GCCCCGTGGC ACATGGTCTT 300
CTTCTCCGAG TCACTGGGTA TCCCTTCACT TCGTGTTTTG GCCCAGAAGC TGCTCGAGCT 360
GCTCTTTGAT TATGAGATTG AGAAGGAGCC CCTGCTCTTC CATGTCTTCA GCAACGGTGG 420
CGTCATGCTG TACCGCTACG TGCTCGAGCT CCTGCAGACC CGTCGCTTCT GCCGCCTGCG 480
TGTGGTGGGC ACCATCTTTG ACAGCGCTCC TGGTGACAGC AACCTGGTAG GGGCTCTGCG 540
GGCCCTGGCA GCCATCCTGG AGCGCCGGGC CGCCATGCTG CGCCTGTTGC TGCTGGTGGC 600
CTTTGCCCTG GTGGTCGTCC TGTTCCACGT CCTGCTTGCT CCCATCACAG CCNTCTTCCA 660
CACCCACTTC TATGACAGGC TACAGGACGC GGGCTCTCGC TGGCCCGAGC TCTACCTCTA 720
CTCGAGGGCT GACGAAGTAG TCCTGGCCAG AGACATAGAA CGCATGGTGG AGGCACGCCT 780
GGCACGCCGG GTCCTGGCGC GTTCTGTGGA TTTCGTGTCA TCTGCACACG TCAGCCACCT 840
CCGTGACTAC CCTACTTACT ACACAAGCCT CTGTGTCGAC TTCATGCGCA ACTGCGTCCG 900
CTGCTGAGGC CATTGCTCCA TCTCACCTCT GCTCCAGAAA TAAATGCCTG ACACCTCCCC 960
ACAAAAAAAA AAAAAAAAAA ACTCGAGGGG GGGCCCGGTA CCCAATTCGC CCTATAAAGG 1020
T 1021
(2) INFORMATION FOR SEQ ID NO: 146:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1285 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 146:
GGCACGAGGA GGGCCACGGC AGCCATCGCG CTTTGCAGTT CGGTCTCCTG GTGTACGGCC 60
AACGCCAAGT AGGGGATTGC GTTCCCTCCA GTCGCAGACC CTATCAGATT TGGATATGTC 120
CTTCATATTT GATTGGATTT ACAGTGGTTT CAGCAGTGTG CTACAGTTTT TAGGATTATA 180
TAAGAAAACT GGTAAACTGG TATTTCTTGG ATTGGATAAT GCAGGAAAAA CAACATTGCT 240
ACACATGCTA AAAGATGACA GACTTGGACA ACATGTCCCA ACATTACATC CCACTTCCGA 300
AGAACTCACC ATTGCTGGCA TGACGTTTAC AACTTTTGAT CTGGGTGGAC ATGTTCAAGC 360
TCGAAGAGTG TGGAAAAACT ACCTTCCTGC TATCAATGGC ATTGTATTTC TGGTGGATTG 420
TGCAGACCAC GAAAGGCTGT TAGAGTCAAA AGAAGAACTT GATTCACTAA TGACAGATGA 480
AACCATTGCT AATGTGCCTA TACTGATTCT TGGGAATAAG ATCGACAGAC CTGAAGCCAT 540
CAGTGAAGAG AGGTTGCGAG AGATGTTTGG TTTATATGGT CAGACAACAG GAAAGGGGAG 600
TATATCTCTG AAAGAACTGA ATGCCCGACC CTTAGAAGTT TTCATGTGTA GTGTGCTCAA 660
AAGACAAGGT TACGGAGAAG GCTTCCGCTG GATGGCACAG TACATTGATT AACACAAACT 720
CACATTGGTT CCAGGTCTCA ACGTTCAGGC TTACTCAGAG ATTTGATTGC TCAACATGCA 780
TAACTTGAAT TCAATAGACT TTTGCTGGTT ATAAAACAGA TGTTTTTTAG ATTATTAATA 840
TTAAATCAAC TTAATTTGAA TGAGAATTGA AAACTGATTC AAGTAAGTTT GAGTATCACA 900
ATGTTAGCTT TCTAATTCCA TAAAAGTACT T∞TTTTTAC AGTTTATAAT CTGACATCAC 960
CCCAGCGCCA TTTGTAAAGA GCAACTTTCC AGCAGTACAT TTGAAGCACT TTTTAACAAC 1020
ATGAAACTAT AAACCATATT TAAAAGCTCA TCATGTTAAA TTTTTTATGT ACTTTTCTGG 1080
AACTAGTTTT TAAATTTTAG ATTATATGTC CACCTATCKT AAGTGTACAG TTAATAATTA 1140
GCTTATTCAA TGATTGCATG ATGCCTTACA GTTTTCAATA ACTTTTTTTC TTATGCAAAC 1200
GTCATGCAAT AAAACAAACT CTAATCTTTC GCAAAAAAAA AAAAAAAAAA NTCGAGGGGG 1260
GGCCCGTACC CAATTCGCCC TAAAG 1285
(2) INFORMATION FOR SEQ ID NO: 147:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1386 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 147:
GGCACGAGGT GGCGCAGGGG TCAGTGGTTC TCTCGGGTCT CGGGACAGGT GAGCACCCTG 60
ATGAAGGCCA CGGTCCTGAT GCGGCACCTG GGCGGGTGCA GGAGATCGTG GGCGCCCTCC 120
GCAAGGGCGS CGGAGACCGG TTACAGGTGA TTTCTGATTT TRACATGACC TTGAGCAGGT 180
TTGCATATAA TGGAAAGCGA TGCCCTTCTT CTTACAATAT TCTGGATAAT AGCAAGATCA 240
TCAGTGAGGA GTGTCGGAAA GAGCTCACAG CGCTCCTTCA CCACTATTAC CCAATTGAGA 300
TCGACCCACA CCGGACCGTC AAGGAGAAGC TACCTCATAT GGTGGAATGG TGGACCAAAG 360
CGCACAATCT CCTATGTCAG CAGAAGATTC AGAAGTTTCA GATAGCCCAG GTGGTTAGAG 420
AGTCCAATGC AATGCTCAGG GAGGGATATA AGACCTTCTT CAACACACTC TACCATAACA 480
ACATTCCCCT TTTCATCTTT TCTGCGGGCA TTGGTGATAT CCTGGAAGAA ATTATCCGAC 540
AGATGAAAGT GTTCCACCCC AACATCCACA TCGTGTCTAA CTACATGGAT TTTAATGAAG 600
ATGGTTTTCT CCAGGGATTT AAGGGCCAGC TGATACACAC ATACAACAAG AACAGCTCTG 660
TGTGTGAGAA CTSTGGTTAC TTCCAGCAAC TTGAGGGCAA AACCAATGTC ATCCTGCTGG 720
GAGACTCTAT CGGGGACCTC ACCATGGCCG ATGGGGTTCC TGGTGTGCAG AACATTCTCA 780
AAATTGGCTT CCTGAATGAC AAGGTGGAGG AGCGGCGGGA NCGCTACATG GACTCCTATG 840
ACATCGTGCT GGAGAAGGAC GAGACTCTGG ATGTGGTCAA CGGGCTACTG CAGCACATCC 900
TGTGCCAGGG GGTCCAGCTG GAGATGCAAG GCCCCTGAAG GCGCAGGCTN CCAGNCCGCC 960
TGCAGGCCGT GGTGAGGAGG GGCGCCTCCC CAGAGTCTGC TCCCCCGTGA ACACAGAGCA 1020
GANGCCAGGG TGGCCAGCAG TGGCTGGGTC CTTCCGCGCC CCTCCGTCCT CCTTTCCCTG 1080
AGCACCTTCA TCACCAGAGG CTTGAAGGAA CCCCGCCATG TGGCAGGGCA CAGGCACTGT 1140
TCCTGGTGAA CCTTGGACCA CAGCATGTCA GTGCTCTAGG GATTGTCTAC TCCAGGGATT 1200
TTCTTCAAAA TTTTTAAACA TGGGAAGTTC AAACAAATAT AATGTGTGAA ACAGATCAAA 1260
ATTTTTAAAA TGAAAAAAAA GCTGCTCTGA TTCAGGGGAT GTGGGTCGGG GTAGAACCTG 1320
GACCTCTTGG CCTGGGGGCA CATGGGATGC TTCTAGGAAC ACAGTTTGAG AACCACCAAA 1380
AAAAAA 1386
(2) INFORMATION FOR SEQ ID NO: 148:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2098 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 148:
AGCCCTTCTC CCCGCGCTTG GGACTCTGAC ATCTTAAGGC TGCACGGTCG TGTCCTTGTC 60
TGGGTGAGGC CATGTCTGTG ATCCAAGGTT CCTGGAACTG ACACAGGAAG GGGCTGTGAA 120
CCCTAAGTGG GTGTMATCTC CTCCRACCGA GGCTTCTMAC CCTGGAGATG GCAGTTACTC 180
CTGGCCATGG TTGCTGAGCA TGGGCAGACC AGTGGAGGCC ACCCTACTGT GTTATCTGCG 240
CCTTCRATGA AGTGAGACCC TTGGGGAGAA CGGGCTGTGG ATGAAGGAGT GGACTGCAGC 300
CTTGGCCTAG CCACTGGGCT GGGATCTTCT GGGTCATGTG ACTGTGTATC CAGGAGCAGA 360
AACTTGTATT CTCAGGATTC AGGATCTACC CAGCACCAAA GATGTATTTT CAGGAGAACA 420
GACCTAGAAA TGGGCCTCTC TGGCATTTCA GAGTCAGGCA AAGCAGGCAG GGCCAGGGAG 480
CTTCTGTGGG TCTACACAAG AAGGTTCCTG TGAGGGCTAT CAGTTCTTGC CTTCTAGCTT 540
GCTGGTAACT TTGGCGCCTC CGCCAAGCCC TGCCAGACTC CCCTGGCTGT GATGGCATTC 600
TGTGCCATCC TGCCTTGTCC CCAGCCTCTG CAGGATGCCC TCCCTACCCA MCTYTYCCTG 660
GGCCTTCCCT GTCCACTGGG CTGGATTCAT GTTCAAACCA CTGGACTGGC AGGGCAACGA 720
CTTCTTCCCA CCTCAAGATG AGGTCCTCGC CCCCTTGTCT TGGCATAAAA ACACCTTTAA 780
AGCATGAGCC ATGTGCTTCT TTGCCCTTCT CTGTCCTGTT CCAATCTTCT GCCTCCCAGT 840
CACTCCCTGC GGACTATGGG ATCACTGTCC CCCCACCTGT GTGGCCACAC CATGTGTCCT 900
GTCAATCCAG AACTGCCTCT GAGCTCCAGG CTGACCACAG ATCAGCCACA GCCTGATGCC 960
TGCAGCCCCA CTTTGCTCAC CCTTCCCCTC CCCTCCTCCT TCCTTCCACA CAGCAAGCCT 1020
ACCTTTYTCC ATCCATGCTC ACCATAGCCC CCTTCCTTGT GACCTGGACC CTCCATTGTA 1080
CCTCGCTCAG ACTGTCAGCC TCCTGGAGGA GTGGGGTCCA CCTTCTTCTT GCCCTATGCA 1140
GTGCAAGCTT CACTTCTCAC CCAGCAAGGT TGACTCATCT GCCTCCATGT CTCTGGGGCT 1200
TTGCTGTTGC CCTGAAACCT AGCTGGGCTG GTCTTGCTCC CAGCTTGCTT CCCCCTCCTC 1260
GGATGTCCCT TTGCAGGCCC CTGTCGTTCC TCCGGCACCA GTGTCCTTGG CTGCCATGGC 1320
AAGCTCATCA GGGGCTTGTA CCCTGGTCAC CAAGCATGGT AGCAGCTGCC TGCATTGTAT 1380
CTCCATCTGG TCACTGCAGG TGCCAACCCT TCATCCCCCA TGTTTTCCTG GGCCATGGAG 1440
GGCTGACCTC CGTTTCTGGG GAATGTGGCT GAGCTGTGGT AACCAGCTAC ACCCCAGGTG 1500
CTCTTTCCAT GGTGGTGCCT GCTCATCTTG CTGATGCAAA CTAGGAAGTT AGGCTGCATC 1560
TCGGAGTGGC TTTCGCTGGA GAGGTGCTTT GCTGTCTCTC AGACTCAGTC ACTGTGTTCC 1620
CTCCCCGCCT CTCTTATCTC CATGGCTGTT TGCAGCTCTC CCAGGTACTT TGGGGTCTGA 1680
GCTGGAATTC CTTTCTCGTT TGCTCTTCTG CTTCTCACTC TTGTATTAAG AAGGATTCCA 1740 CAAAGGGAGA GTGGCATCCC TGCTGCTGCT GTGCCAGACC AGAGTTTCCT GAGGGGCCCT 1800 GACCCTAACC CTCCAGCTCA GCCCTGTACA CCTGACCCTG TAAATGAGTG GGGTTTGCTG 1860 ACTGTAATCC CTGACACCAG TAAAACCAAA AGGACTCTTG GGGGCTCAGT GTGAGAGCCA 1920 GGGTTACCTA CTCTGCCAAG TGAGGACAAA CTGCTAGGCT GTATCCCATA ATTTCAGGAT 1980 GAGAAACATT AACAATAAAA ATTTGTAGTA AACATAACCT CATGANGACT AAAAAAAAAA 2040 AAAAACTYGG GGGGGGGCCC GTAACCCATT GGGCCCTTNG GGGGGGNGTT TTAAAATT 2098
(2) INFORMATION FOR SEQ ID NO: 149:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1847 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 149:
TCGACCCACG CGTCCGAACT GAGGCGGCGG CGGGAGCCGG TTGGKGTCTG GTCTTCGCGT 60
CGGCCCCGCG GACCAGACGC TGCCCCCGGC GCGGGGAGAA GATGGTGCCK AGCGGCCTCG 120
GGCCCGCCAC GCGCCGCCAC GAGTGAGCCC AGCGCGACCG CGGGCGTCCG CCGAGCAGCT 180
GGCCCGGCTG GGCCCGGGGC GCGCANTGCC CGCGGGGGCG GGGTGGAGCT GATCAGAATA 240
ATGTTCAGCA TCAACCCCCT GGAGAACCTG AAGGTGTACA TCAGCAGTCG GCCTCCCCTG 300
GTGGTCTTCA TGATCAGCGT AANGCCCATG GCCATAGCTT TCCTGACCCT GGGCTACTTC 360
TTCAAAATCA AGGAGATTAA ATCCCCAGAA ATGGCAGAGG ATTGGAATAC TTTTCTGCTA 420
CGGTTCAATG ATTTGGACTT GTCTGTATCA GAGAATGAAA CCCTCAAGCA TCTCACAAAC 480
GACACCACAA CTCCGGAAAG TACAATGACC AGCGGGCAGG CCCGAGCTTC CACCCAGTCC 540
CCCCAGGCCC TGGAGGACTC GGGCCCGGTG AATATCTCAG TCTCAATCAC CCTAACCCTG 600
GACCCACTGA AACCCTTCGG AGGGTATTCC CGCAACGTCA CCCATCTGTA CTCAACCATC 660
TTAGGGCATC AGATTGGACT TTCAGGCAGG GAAGCCCACG AGGAGATAAA CATCACCTTC 720
ACCCTGCCTA CAGCGTGGAG CTCAGATGAC TGCGCCCTCC ACGGTCACTG TGAGCAGGTG 780
GTATTCACAG CCTGCATGAC CCTCACGGCC AGCCCTGGGG TGTTCCCCGT CACTGTACAG 840
CCACCGCACT GTGTTCCTGA CACGTACAGC AACGCCACGC TCTGGTACAA GATCTTCACA 900
ACTGCCAGAG ATGCCAACAC AAAATACGCC CAAGATTACA ATCCTTTCTG GTGTTATAAG 960
GGGCCCATTG GAAAAGTCTA TCATGCTTTA AATCCCAAGC TTACAGTGAT TGTTCCAGAT 1020
GATCACCGTT CATTAATAAA TTTGCATCTC ATCCACACCA GTTACTTCCT CTTTGTGATG 1080 GTGATAACAA TGTTTTGCTA TGCTGTTATC AAGGGCAGAC CTAGCAAATT GCGTCAGAGC 1140 AATCCTGAAT TTTGTCCCGA GAAGGTGGCT TTGGCTGAAG CCTAATTCCA CAGCTCCTTG 1200 TTTTTTGAGA GAGACTGAGA GAACCATAAT CCTTGCCTGC TCAACCCAGC CTGGGCCTGG 1260 ATGCTCTGTG AATACATTAT CTTGCGATGT TGGGTTATTC CAGCCAAAGA CATTTCAAGT 1320 GCCTGTAACT GATTTGTACA TATTTATAAA AATCTATTCA GAAATTGGTC CAATAATGCA 1380 CGTGCTTTGC CCTGGGTACA GCCAGAGCCC TTCAACCCCA CCTTGGACTT GAGGACCTAC 1440 CTGATGGGAC GTTTCCACGT GTCTCTAGAG AAGGATTCCT GGATCTAGCT GGTCACGACG 1500 ATCTTTTCAC CAAGGTCACA GGAGCATTGC GTCGCTGATG GGGTTGAAGT TTGGTTTGGT 1560 TCTTGTTTCA GCCCAATATG TAGAGAACAT TTGAAACAGT CTGCACCTTT GATACGGTAT 1620 TGCATTTCCA AAGCCACCAA TCCATTTTGT GCATTTTATG TGTCTGTGGC TTAATAATCA 1680 TAGTAACAAC AATAATACCT TTTTCTCCAT TTTGCTTGCA GGAAACATAC CTTAAGTTTT 1740 TTTTGTTTTC TTTTTGTTTT TTTGTTTTTT GTTTTCCTTT ATGAAGAAAA AATAAAATAG 1800 TCACATTTTA ATACTACCAA AAAATGGACA AAAAAAGTCG AGGGGGG 1847
(2) INFORMATION FOR SEQ ID NO: 150:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1569 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 150: GACGCTGACG AGAGAAGGCC TCTTCCTTCA GGGTTGGTGC TGTGTTGCAG TGACCGTGGC 60 GGATTACGCC AACTCGGATC CGGCGGTCGT GAGGTCTGGA CGAGTCAAGA AAGCCGTAGC 120 CAACGCTGTT CAGCAGGAAG TAAAATCTCT TTCTCGCTTG GAAGCCTCTC AGGTTCCTGC 180 AGAGGAAGCT CTTTCTGGGG CTGGTGAGCC CTGTGACATC ATCGACAGCA GTCATGAGAT 240 GGATGCCCAG GAGGAAAGCA TCCATGAGAG AACTGTCTCC AGAAAAAAGA AAAGCAAGAG 300 ACACAAAGAA GAACTGGACG GGGCTGGAGG AGAAGAGTAT CCCATGGATA TTTGGCTATT 360 GCTGGCCTCC TATATCCGTC CTGAGGACAT TGTGAATTTT TCCCTGATTT GTAAGAATGC 420 CTGGACTGTC ACTTGCACTG CTGCCTTTTG GACCAGGTTG TACCGAAGCA CTACACGCTG 480 GATGCTTCCC TGCCTTTGCG TCTGCGACCA GAGTCAATGG AGAAGCTGCG CTGTCTCCGG 540
GCTTGTGTGA TCCGATCTCT GTACCATATG TATGAGCCAT TTGCTGCTCG AATCTCCAAG 600 AATCCAGCCA TTCCAGAAAG CACCCCCAGC ACATTAAAGA ATTCCAAATG CTTACTTTTC 660 TGGTGCAGAA AGATTCTTCG GAACAGACAG GAACCAATGT GGGAATTCAA CTTCAAGTTC 720 AAAAAACAGT CCCCTAGGTT AAAGAGCAAG TGTACAGGAG GATTGCAGCC TCCCGTTCAG 780 TACGAAGATG TTCATACCAA TCCAGACCAG GACTGCTGCC TACTGCAGGT CACCACCCTC 840 AATTTCATCT TTATTCCGAT TCTCATGGGA ATGATATTTA CTCTGTTTAC TATCAATGTG 900 AGCACGGACA TGCGGCATCA TCGAGTGAGA CTGGTGTTCC AAGATTCCCC TGTCCATGGT 960 GGTCGGAAAC TGCGCAGTGA ACAGGGTGTC CAAGTCATCC TGGACCCAGT GCACAGCGTT 1020 CGGCTCTTTG ACTGGTGGCA TCCTCAGTAC CCATTCTCCC TGAGAGCGTA GTTACTGCTT 1080 CCCATCCCTT GGGGGCAGCC TCGAGTGTAG TCCATTAGTA ATCAGATTCC AGTTTGGACA 1140 GGGTGGCTGG ATTGTATATC TCGTTAGTAA TGTACATGCT CTTCAGGTTC TAGGGCTCCT 1200 GTTAGGGGAG GGAGAAATGT TGAATCAAGA GGGAAAACAA CTACTATGAT TTATAAACAT 1260 ATTTTAATGT AAAAATTTGC ATTTAAAAGG AGTGGCCCTG TTTTCTGTGT TAAAACCCCA 1320 TTTGGTGCTA TTGAGTTTGT TCTTTATTCT TTTATCCCAG TGAAAATTGT TGATCTTGCT 1380 GTAGGGAAAA ATTAAACTCT TTGAATCTCC AAACAAGGAA GTTTCAGCAT TCCCTTATGG 1440 ATCAGAGGAA CCTTAGAGGC CTGAAATTGT TGCTTCCAGT TTAGCTGCCC CTCAAATTCA 1500 AGTGAATATT TTCCCTTCTC CCTTTACCCT TCTCCAGAAA TAAAGCAGGT GACAGGGTTT 1560 CAGAATCTT 1569
(2) INFORMATION FOR SEQ ID NO: 151:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1540 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 151: CCCACGCGTC CGGAAGGATT GACCAGTTAA CCAACATCTT AGCCCCCATG GCTGTTGGCC 60 AGATTATGAC ATTTGGCTCC CCAGTCATCG GCTGTGGCTT TATTTCGGGA TGGAACTTGG 120 TATCCATGTG CGTGGAGTAC GTCCTGCTCT GGAAGGTTTA CCAGAAAACC CCAGCTCTAG 180 CTGTGAAAGC TGGTCTTAAA GAAGAGGAAA CTGAATTGAA ACAGCTGAAT TTACACAAAG 240 ATACTGAGCC AAAACCCCTG GAGGGAACTC ATCTAATGGG TGTGAAAGAC TCTAACATCC 300 ATGAGCTTGA ACATGAGCAA GAGCCTACTT GTGCCTCCCA GATGGCTGAG CCCTTCCGTA 360
CCTTCCGAGA TGGATGGGTC TCCTACTACA ACCAGCCTGT GTTTCTGGCT GGCATGGGTC 420
TTGCTTTCCT TTATATGACT GTCCTGGGCT TTGACTGCAT CACCACAGGG TACGCCTACA 480
CTCAGGGACT GAGTGGGTTC CATCCTCAGT ATTTTGATGG GAGCATCAGC TATAACTGGA 540
ATAATGGGAA CTGTAGCTTT TACTTGGCTA CGTCGAAAAT GTGGTTTGGT TCGGCAGGTC 600
TGATCTCAGG ATTGGCACAG CTTTCCTGTT TGATCTTGTG TGTGATCTCT GTATTCATGC 660
CTGGAAGCCC CCTGGACTTG TCCGTTTCTC CTTTTGAAGA TATCCGATCA AGGTTCATTC 720
AAGGAGAGTC AATTACACCT ACCAAGATAC CTGAAATTAC AACTGAAATA TACATGTCTA 780
ATGGGTCTAA TTCTGCTAAT ATTGTCCCGG AGACAAGTCC TGAATCTGTG CCCATAATCT 840
CTGTCAGTCT GCTGTTTGCA GGCGTCATTG CTGCTAGAAT CGGTCTTTGG TCCTTTGATT 900
TAACTGTGAC ACAGTTGCTG CAAGAAAATG TAATTGAATC TGAAAGAGGC ATTATAAATG 960
GTGTACAGAA CTCCATGAAC TATCTTCTTG ATCTTCTGCA TTTCATCATG GTCATCCTGG 1020
CTCCAAATCC TGAAGCTTTT GGCTTGCTCG TATTGATTTC AGTCTCCTTT GTGGCAATGG 1080
GCCACATTAT GTATTTCCGA TTTGCCCAAA ATACTCTGGG AAACAAGCTC TTTGCTTGCG 1140
GTCCTGATGC AAAAGAAGTT AGGAAGGAAA ATCAAGCAAA TACATCTGTT GTTTGAGACA 1200
GTTTAACTGT TGCTATCCTG TTACTAGATT ATATAGAGCA CATGTGCTTA TTTTGTACTG 1260
CAGAATTCCA ATAAATGGCT GGGTGTTTTG (CTCTGTTTTT ACCACAGCTG TGCCTTGAGA 1320
ACTAAAAGCT GTTTAGGAAA CCTAAGTCAG CAGAAATTAA CTGGATTAAT TTCCCTTATG 1380
TTGAGGGCCA TGGRAAAAAA ATTGGGAAAA GGAAAAACTC AGTTTTAAAT ACGGGAGACT 1440
ATAATGGATA ACACTGRATT CCCCTATTTC TCATGAGTAG ATACAATCTT ACGTAAAAGA 1500
GTGGTTAGTC ACGTGAATTC AGTTATCATT TGACAGATTC 1540
(2) INFORMATION FOR SEQ ID NO: 152:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1719 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 152: TACTTATGAG GTCAATTGGA AATAAGAACA CCATTTTACT GGGTCTAGGA TTTCAAATAT 60 TACAGTTGGC ATGGTATGGC TTTGGTTCAG AACCTTGGAT GATGTGGGCT GCTGGGGCAG 120 TAGCAGCCAT GTCTAGCATC ACCTTTCCTG CTGTCAGTGC ACTTGTTTCA CGAACTGCTG 180
ATGCTGATCA ACAGGGTGTC GTTCAAGGAA TGATAACAGG AATTCGAGGA TTATGCAATG 240
GTCTGGGACC GGCCCTCTAT GGATTCATTT TCTACATATT CCATGTGGAA CTTAAAGAAC 300
TGCCAATAAC AGGAACAGAC TTGGGAACAA ACACAAGCCC TCAGCACCAC TTTGAACAGA 360
ATTCCATCAT CCCTGGCCCT CCCTTCCTAT TTGGAGCCTG TTCAGTACTG CTG-GCTCTGC 420
TTGTTGCCTT GTTTATTCCG GAACATACCA ATTTAAGCTT AAGGTCCAGC AGTTGGAGAA 480
AGCACTGTGG CAGTCACAGC CATCCTCATA ATACACAAGC GCCAGGAGAG GCCAAAGAAC 540
CTTTACTCCA GGACACAAAT GTGTGACGAC TGAAATCAGG AAGATTTTTC TATCAGCACC 600
CAGGTCTTAG TTTTCACCTC TAGTTCTGGA TGTACATTCC ATTTCCATCC ACAGTGTACT 660
TTAAGATTGT CTTAAGAAAT GTATCTGCAT GAACTCCGTG GGAACTAAAG GAAGTGGGAA 720
CTTAGAACCA GACAGTTTTC CAAAGATGTT ACAATTTCTT TTGAAAAACC TTTTGTTTAT 780
TAGCACCAAT TTCTYGCCAC TAAGCTATTT GTTTTATTAT ACATCCTTTA ATTAAAAACT 840
ATATATGTAA CTTCTTAGAT ATTAGCAAAT GTCTCTGCTA CCATTTCCTT AAGGTGTTGA 900
GCTTTAACTC TATGCTGACT CAGTGAGACA CAGTAGGTAG TATGGTTGTG GACCTATTTG 960
TTTTAACATT GTAAAATTTT GAGTCAGATT TTAATATTGT AAAATCTTGG GTCAAATAAT 1020
TCAAAGCCTT AATGCAGATG CACTAAAACA AAGAAATGGT AAATGAATTG TTTGCATTTA 1080
AAAAAAAAAA CTCTTAAGAA AACTGTACTA AATCTGAATC ATGTTTTGAG CTTCTTTGCA 1140
GTACTTTTAA ACATTATTCA CTACTGTTTT TGAAGTGAGA AAGTATCAGC CATTTAGCAT 1200
TTAAGTTGGG GTATTTAGAG CCTGTAATCT AAATGCTGGC TCAAATTTAT TCCCCAGCTA 1260
CTTCTTATAC CACTATTCTT TTAATGTTTG CATAATCATA AGCACCTCAA CACTTGAATA 1320
CATAATCTAA AAATTATATA GTAAAGCTGG TAGCCTTGAA AATGTCAGTG TGATATCTAT 1380
TATGTAGATA AATATATATA GTGGCCTTTC AGGACTGTCA CAGTAACACT TTATTTACAG 1440
AGCTAATGTT TGTCCTAAAT TTTCAGGACC CTAGAGGAGA GCTTTATACA ATTACCGATG 1500
TGAATTTCTC TAAAGTGTAT ATTTTTGTGT CCAGTTATAT TATTTAAAAA AGTGTTACTT 1560
TGTAAAAATT GTATATAAAG AACTGTATAG TTTACACTGT TTTCATCTTG TGTGTGGTTA 1620
TTGCTTAATG CTTTTTAAAC TTGGAACACT CACTATGGTT AAATAAGGTC TTAAAAGAAA 1680
TGTAAATATT YTGTTAATAA AGTTAAATAT TTTAATGAT 1719
(2) INFORMATION FOR SEQ ID NO: 153:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 863 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 153:
GGCACGAGGG AAGCCGGGAC GATGTCCGCA TGACAACCGA CGTTGGAGTT TGGAGGTGCT 60
TGCCTTAGAG CAAGGGAAAC AGCTCTCATT CAAAGGAACT AGAAGCCTCT CCCTCAGTGG 120 TAGGGAGACA GCCAGGAGCG GTTTTCTGGG AACTGTGGGA TGTGCCCTTG GGGGCCCGAG 180
AAAACAGAAG GAAGATGCTC CAGACCAGTA ACTACAGCCT GGTGCTCTCT CTGCAGTTCC 240
TGCTGCTGTC CTATGACCTC TTTGTCAATT CCTTCTCAGA ACTGCTCCAA AAGACTCCTG 300
TCATCCAGCT TGTGCTCTTC ATCATCCAGG ATATTGCAGT CCTCTTCAAC ATCATCATCA 360
TTTTCCTCAT GTTCTTCAAC ACCTTCGTCT TCCAGGCTGG CCTGGTCAAC CTCCTATTCC 420 ATAAGTTCAA AGGGACCATC ATCCTGACAG CTGTGTACTT TGCCCTCAGC ATCTCCCTTC 480
ATGTCTGGGT CATGAACTTA CGCTGGAAAA ACTCCAACAG CTTCATATGG ACAGATGGAC 540
TTCAAATGCT GTTTGTATTC CAGAGACTAG CAGCAGTGTT GTACTGCTAC TTCTATAAAC 600
GGACAGCCGT AAGACTAGGC GATCCTCACT TCTACCAGGA CTCTTTGTGG CTGCGCAAGG 660
AGTTCATGCA AGTTCGAAGG TGACCTCTTG TCACACTGAT GGATACTTTT CCTTCCTGGA 720 TAGRAGGCCA C^TTTGCTGC TTTGCAGGGG AGAGTTGGGC CCTATGCATG GGGCAAAACA 780
GGTGGGATTT TCCAAGGGAA GGGTTCAGAA TTAGGCNTGT TGTTTCAGCC ATTTCCAAGG 840
AAGGGGAAGG GTTTCCCTNC CCT 863
(2) INFORMATION FOR SEQ ID NO: 154:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1101 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 154:
AACAGCAAAA AAGAATGATT TCTTCTGAAA TTGTGGAACA TGAGGATTCA AGTTTTTATT 60
TTGTTACTAG GTGCTGGAGG AACATCCCAG TTCACAAAGC CCCCATCTCT TCCTCTGGAG 120
CCAGAGCCTG CGGTGGAATC AAGTCCAACT GAAACATCAG AACAAATAAG AGAGAAATAA 180 GAATAGAATG AATGACCCCA AAATARGGTT TTCTTGGGCG AGGATGTCCT GGATTAGGAA 240
AGGTGACATG ACACAGGCAG AGCAGAGTGG CACCCACCAC AGAATACAGT GTGTGTTATT 300
ACGAGGAGCC AGCAGTTGAG CCTAAGGTCC TTCTACCTAC CTGGTATTGG CATTTGAGGT 360
CGGAAACCCT CTACTGCCCC ATAAGCCAGG AAAAGTGAAA AGAGAACACA GTTCCTTTAA 420
GAACTGGCAG CAAGGCTTGA GGCCTTATGT ATGTAGCTGA GTCAGCAAGG TACATGATGC 480 TGTCTGCTTT CAAAAGGACT TTTCTCTCCT AGCTGACTGA CTCCTTCCTT AGTTCAAGGA 540
ACAGCTGAGA CAGACCTCTG CTGAGTAGCT CTGTGATGAC AAAGCCTTGG TTTAACTGAG 600
GTGATCCTCA GGTTGTGAGG TTTATTAGTC CCCAAGGCAA ACACAAATAT TAGATTAATA 660
ATCCAACTTT AATAGTATAC ATTTAAAAGA AAAAAAACAA AAGCCCTGGA AGNTTGAGGC 720
CAAGCCTGCT GAGTATTGCA GCTGCATTTG CCCAAAGGGA ATCCAGAACA AGTCCCTCCC 780 TGTATTTTGT TCTTGAGAGG GGTCAGTCTA GAAGCTAGAT CCTATCAGGA TGAGGAGCAG 840
CAGCCCAGGG CTTGTCTGGA TCAGCACCAA CGATTTTAAA GAAAAAAGGA AGAGTTTCTT 900
AGATGAGTAA TTGTTATTGA AGATAGTCAG TGATAACCAC TGACCAGATG CTATCAATAC 960
ACTATGTGTC CTTTTTAGAA TAAAGATTAC ATATCATCAT TCCTTTGGGG AAAATTGTTA 1020
TTCAGGTATA AAAACAAGAG ATTATAATAA AAAANTAAAA GAACCCTAAA AAAAAAAAAC 1080 CTCGTGCCGA ATTCCCTGCA G 1101
(2) INFORMATION FOR SEQ ID NO: 155:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2031 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 155: CAATTAACCC GTTTGAGGCC TAGGTTGTTT GGCAAGCCCC NGGCCTAAAG TTTTAATTCG 60
GCAGAGCCAA GGGCCTGAAA GGAAGGGAAA GGGGAGGGTA GCGGGAGGGT AGCAGGTGAG 120
TTCCTAGGGC TGGAAGGTTT AGCAGCAGCC TGGTGCAGTG CCCTGTCATC AAGACAAACC 180
CACGGTCCTC CTGGGTGCCT ACCAAGCTTG GTTTGTACAA AAGCAAGGTG GGAGTCTATT 240
TTTGTACATG AGATACATCA CACTTACCTG TGGGCCAGTA TTGTGAAGTG AGTCTGAGTT 300 GTTTACACTG ATGCCTTCCC TGCCCACCAC AAATTGTGTA CATAGTCTTC AGAATGATAC 360
CACCCCTTTC CCCAGCTCCC AACCAAGAGC TGGTTCTAGG CCTGTGTTAT ATGTCATATT 420
TAGCGTTTTT ATATATGACC TTTGATTTCT GTTCTTTGTA TTTTAGCACA GTGTATGCAC 480
CTTCATTTAA ATACATCTGT GTGCATACAG ATACGCATAT ATGTGTGTGC GTATGCATAT 540
ATCTCTCATC TGTAGTTTCC AAGAGTTCAG CTGAAGCAGA TGGAGTCCTG CAGCCCAGGA 600 GACACCCTGC ATCCCTGCTA ATAGTGTTTG CCACAAGTAT TAGTGAGTCT TCCTTATTAA 660
TATTTTCATT TCAGAAGACT GAAGCAAAGC TGATAGTGTT TGCTGTTTCT TTGGCAGCTA 720
AGTGAGGGTC TTGGGATGAC TTGCTGTGTT CCTCAAGCTG CACTTTGGGG CCATCTCTGC 780
AGTATTAAGC CCCCTTTTTG CTTGGTGGTA CTCTGTCTGT GCCTGTGTGT GTGTGTGATA 840
GTCACTCTTG CATGGCTTCC ATGTCTGGTT TGTGGCATTT GGGGATAAGT GCTGAACCAG 900
AGCATTTGCA GTTTGTTTGA GGCCTCGTTG CCAATGATAG ATCACTCCTG TTGACCTGGT 960
ATGTCTGCTT GCTTGCTGCT TTTCCTTGCT TTCTCTTGGA AGAGGAAAGG ACTCTGGTCA 1020
GGCCCAGGCT GAGTGAGATG AGCTGCAGCT GGCTCATGGC CTTCTTAGAG CAGAGAGAGG 1080
AGTATGTCAT TTTACTAAGT TCCTAAACAA ACATTTATGC AGGCAACACT CCTTGCAGAT 1140
CCAGAAACTG AGGCACAATA GGGTTATGAC TTGCTCAAGA ATATGTAGCT GCTAGGGGGT 1200
AAATCAAGGC ATCACAATTT CTGTTCAGCG GGCAGGAATA GGCTGTGAAT TGCTAGCACT 1260
TTTTTTTTAA GCAATTACTT TTTGACTTGT TCCTCTGAAA GTGCAAGAGG CGTACACCTT 1320
TCCCAAATGT AGACTAGAAT CTGCAGGATG CCACCCACTG TATAGTTCTG CTTTCCCAGA 1380
GAGGAAGAAC TTTTAGAAAC CAAATGATCT TAATTGTTAT TGCCCACCCC TCC^TTTTCC 1440
GGGTAGAAAA TTCACAGTAG GAATGATTGT TAAGAGAGAG TGCTTGGAAC CATGGGTTAA 1500
CAGGAAAGGC TACCTAACTT CACATATCTG CAACCAGAGC AGCCACCAAG CATTACTTAG 1560
CAGCAGGAAA ATGATTGTAT TTGAGTTCCT GTGTGTCCAA AACTGAGGCA CCATGTTCTT 1620
TGAAAACATG CCACCTCAAG GCTGGGCGCG GTGGCTCACA CCTGTTAATC CCAGCACTTT 1680
GGGAGGCCGA GGCGGGCGGA TCACCGGAGT CGGGGAGTTT GAGACCAGCC TGGACCAACA 1740
TGGGAGAAAC CCCATCTCTA CCTAAAAATA CAAAATTAGC CGGGCGTGGT GGCATGCGCC 1800
TATAATCTCA GCTACTTGGG AGGGYTGAGG CAGGRGAATT GCTTGAACCC RGGANGGCGG 1860
AGGTTTGCGG TTGAGTTGAG GATCGTGCCA TTGCACTTCC GGGCCTTGGG GCAACAACAG 1920
CAAAAAYTCC GTCTTCAAMW MRTGCCGAAT TCGATATCAA GCTTATCGAT ACCGTCGACC 1980
TCGAGGGGGG GCCCGGTACC CAATTCGCCC TATAGNGATC GTATTACAAT C 2031
(2) INFORMATION FOR SEQ ID NO: 156:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1981 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 156:
CCTGCACCCT GAGCCCTTCA CCCCTCCGAG TTCCCCCCAG GTTGGCTTCC TTCGATTCCT 60
TTTCTTGGTA TCAACGTTTG ATTGGAAGAA CAACCCCCTC TTTGTCAACC TCAATAATGA 120
GCTCACTGTG GAGGAGCAGC TCGGGCACAG CTCMCCGTYA TGGTCATTGT TACCCCCCAA 180
GACCGCAAAA ACTCTGTGTG GACACAGGAT GGACCCTCAG CCCAGATCCT GCAGCAGCTT 240
GTGGTCCTGG CAGCTGAAGC CCTGCCCATG TTAGAGAAGC AGCTCATGGA TCCCCGGGGA 300
CCTGGGGACA TCAGGACAGT GTTCCGGCCG CCCTTGGACA TTTACGACGT GCTGATTCGC 360
CTGTYTCCTC GCCATATCCC GCGGCACCGC AGGCTTGTGG ACTCGCCAGY TGCCTCCTTC 420
TGCCGGGGCC TGCTCAGCCA GCCGGGGCCC TCATCCCTGA TGCCCGTGCT GGGTNATGAT 480
CCTNCTCAGC TCTATCTGAC GCAGCTCAGG GAGGCCTTTG GGGATCTGGC CCTTTTCTTC 540
TATGACCAGC ATGGTGGAGA GGTGATTGGT GTCCTCTGGA AGCCCACCAG CTTCCAGCCG 600
CAGCCCTTCA AGGCCTCCAG CACAAAGGGG CGCATGGTGA TGTCTCGAGG TGGGGAGCTA 660
GTAATGGTGC CCAATGTTGA AGCAATCCTG GAGGACTTTG CTGTGCTGGG TGAAGGCCTG 720
GTGCAGACTG TGGAGGCCCG AAGTGAGAGG TGGACTGTGT GATCCCAGCT CTGGAGCAAG 780
CTGTAGACGG ACAGCAGGAC ATTGGACCTC TAGAGCAAGA TGTCAGTAGG ATGACCTCCA 840
CCCTCCTTGG ACATGAATCC TCCATGGAGG GCCTGCTGGC TGAACATGCT GAATCATCTC 900
CAACAAAACC CAGCCCCAAC TTTCTCTCTG ATGCTCCAGC ATTGGGGCAG GGGCATGGTG 960
GCCCATGTAG TCTCCTGGGC CTCACCATCC CAGAAGAGGA GTGGGAGCCA GCTCAGAGAA 1020
GGAACTGAAC CCAGGAGATC CATCCACCTA TTAGCCCTGG GCCTGGACCT CCCTGCGATT 1080
TCCCACTCCT TTCTTAGTCT TCTTCCAGAA ACAGAGAAGG GGATGTGTGC CTGGGAGAGG 1140
CTCTGTCTCC TTCCTGCTGC CAGGACCTGT GCCTAGACTT AGCATGCCCT TCACTGCAGT 1200
GTCAGGCCTT TAGATGGGAC CCAGCGAAAA TGTGGCCCTT CTGAGTCACA TCACCGACAC 1260
TGAGCAGTGG AAAGGGGCTA TATGTGTATG AATAGACCAC ATTGAAGGAG CACAATGCCC 1320
TCCTGTGTTG ATGCCACTTC CCAGGGTGGA GACAGTGGAA AAGAACCGAG GACAGGAAAG 1380
GATTGGGTAG GTGAAGGGGT CAGGGGACTG GTAGTCACCC AATCTTGGAG AGGTGCAAAA 1440
AGCACTGGGG GCTACCCGTT AGCTGCATCT GCCCTGGCTG TTTGCCCGTT CATGTCACAA 1500
ACTGCCACTA CTATGTACCT GCAGTGGGGT TGCAGAGATG GGGGAGACTC AAGTCTTACT 1560
CCCCAGGAGC TCCCAGGGCC CAAGGAGGAG AATGCTGCCT CCTTTCAGTC TGGTCTACAC 1620
CCACTTTCTG GTAGCCTCTC TGCTTCCTGT AATTCTGGCT GTTTTTCCAG ACTCAGCTCA 1680
AATAGTGCCC CTCCTTAAGC CCATCCCTCG CCCCCAGCCT GAGGTGATCT TTCCCTCCTC 1740
TGAACTATTA GAGCAGTTAC TGTCTGTTCA GTTCGTTTGG CAGGCACACA CAGTGGCATA 1800
AATTCTATTG TTTTGAACTC TGATTTAAAA TTAAATTGCA GCTGGGCGTG GTGGCTCATG 1860
CTTGTAATCC CAACACTTAG GGAGTMAGGR GAATCACTTG ASCYCAGGAG TYCTAGACCA 1920 ATCTCGGCAA MAGAGAGACC CCATCTCTTT TAAATAAAAA GTTAAATTGC TTAAAAAAAA 1980
A 1981
(2) INFORMATION FOR SEQ ID NO: 157:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 915 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 157:
GAATTCGGCA CGAGCGCGGC CATGGCGCTC CTGCTTTCGG TGCTCCGTGT ACTCCTGGGC 60 GGCTTCTTCG CGCTCGTGGG GTTGGCCAAG CTCTCGGAGG AGATCTCGGC TCCAGTTTCG 120
GAGCGGATGA ATGCCCTGTT CGTGCAGTTT GCTGAGGTGT TCCCGCTGAA GGTATTTGGC 180
TACCAGCCAG ATCCCCTGAA CTACCAAATA GCTGTGGGCT TTCTGGAACT GCTGGCTGGG 240 TTGCTGCTGG TCATGGGCCC ACCGATGCTG CAAGAGATCA GTAACTTGTT CTTGATTCTG 300
CTCATGATGG GGGCTATCTT (^CCTTGGCA GCTCTGAAAG AGTCACTAAG CACCTGTATC 360
CCAGCCATTG TCTGCCTGGG GTTCCTGCTG CTGCTGAATG TCGGCCAGCT CTTAGCCCAG 420
ACTAAGAAGG TGGTCAGACC CACTAGGAAG AAGACTCTAA GTACATTCAA GGAATCCTGG 480
AAGTAGAGCA TCTCTGTCTC TTTATGCCAT GCAGCTGTCA CAGCAGGAAC ATGGTAGAAC 540 ACAGAGTCTA TCATCTTGTT ACCAGTATAA TATCCAGGGT CAGCCAGTGT TGAAAGAGAC 600
ATTTTGTCTA CCTGGCACTG CTTTCTCTTT TTAGCTTTAC TACTCTTTTG TGAGGAGTAC 660
ATGTTATGCA TATTAACATT CCTCATGTCA TATGAAAATA CAAAATAAGC AGAAAAGAAA 720
TTTAAATCAA CCAAAATTCT GATGCCCCAA ATAACCACTT TTAATGCCTT GGTGTAAGTA 780
TACCTCTGAA CTTTTTTCTG TGCCTTTAAA CAGATATATA TTTTTTTTWA ATGAAAATAA 840 AACCATATAT CCTATTTTAT TTCCTCCTTT TAAAACCTTA TAAACTATAA MAAAAAAAAA 900
AAAAAAAAAA CTCGA 915
(2) INFORMATION FOR SEQ ID NO: 158:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 2117 base pairs
(B) TYPE : nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 158:
AGAGCGAAGC GAGGGTGGCG CGGGTCCGGG CATGAAGCTG GGCCGGGCCG TGCTGGGCCT 60
GCTGCTGCTG GCGCCGTCCG TGGTGCAGGC GGTGGAGCCC ATCAGCCTGG GACTGGCCCT 120
GGCCGGCGTC CTCACCGGCT ACATCTACCC GCGTCTCTAC TGCCTCTTCG CCGAGTGCTG 180
CGGGCAGAAG CGGAGCCTTA GCCGGGAGGC ACTGCAGAAG GATCTGGACG ACAACCTCTT 240
TGGACAGCAT CTTGCAAAGA AAATCATCTT AAATGCCGTG TTTGGTTTCA TAAACAACCC 300
AAAGCCCAAG AAACCTCTCA CGCTCTCCCT GCACGGGTGG ACAGGCACCG GCAAAAATTT 360
CGTCAGCAAG ATCATCGCAG AGAATATTTA CGAGGGTGGT CTGAACAGTG ACTATGTCCA 420
CCTGTTTGTG GCCACATTGC ACTTTCCACA TGCTTCAAAC ATCACCTTGT ACAAGGATCA 480
GTTACAGTTG TGGATTCGAG GCAACGTGAG TGCCTGTGCG AGGTCCATCT TCATATTTGA 540
TGAAATGGAT AAGATGCATC CAGGCCTCAT AGATGCCATC AAGCCTTTCC TCGACTATTA 600
TGACCTGGTG GATGGGGTCT CCTACCAGAA AGCCATGTTC ATATTTCTCA GCAATGCTGG 660
AGCAGAAAGG ATCACAGATG TGGCTTTGGA TTTCTGGAGG AGTGGAAAGC AGAGGGAAGA 720
CATCAAGCTC AAAGACATTG AACACGCGTT GTCTGTGTCG GTTTTCAATA ACAAGAACAG 780
TGGCTTCTGG CACAGCAGCT TAATTGACCG GAACCTCATT GATTATTTTG TTCCCTTCCT 840
CCCCCTGGAA TACAAACACC TAAAAATGTG TATCCGAGTG GAAATGCAGT CCCGAGGCTA 900
TGAAATTGAT GAAGACATTG TAAGCAGAGT GGCTGAGGAG ATGACATTTT TCCCCAAAGA 960
GGAGAGAGTT TTCTCAGATA AAGGCTGCAA AACGGTGTTC ACCAAGTTAG ATTATTACTA 1020
CGATGATTGA CAGTCATGAT TGGCAGCCGG AGTCACTGCC TGGAGTTGGA AAAGAAACAA 1080
CACTCAGTCC TTCCACACTT CCACCCCCAG CTCCTTTCCC TGGAAGAGGA ATCCAGTGAA 1140
TGTTCCTGTT TGATGTGACA GGAATTCTCC CTGGCATTGT TTCCACCCCC TGGTGCCTGC 1200
AGGCCACCCA GGGACCACGG GCGAGGACGT GAAGCCTCCC GAACACGCAC AGAAGGAAGG 1260
AGCCAGCTCC CAGCCCACTC ATCGCAGGGC TCATGATTTT TTACAAATTA TGTTTTAATT 1320
CCAAGTGTTT CTGTTTCAAG GAAGGATGAA TAAGTTTTAT TGAAAATGTG GTAACTTTAT 1380
TTAAAATGAT TTTTAACATT ATGAGAGACT GCTCAGATTC TAAGTTGTTG GCCTTGTGTG 1440
TGTGTTTTTT TTTAAGTTCT CATCATTATT ACATAGACTG TGATGTATCT TTACTGGAAA 1500
TGAGCCCAAG CACACATGCA TGGCATTTGT TCCACAGGAG GGCATCCCTG GGGATGTGGC 1560
TGGAGCATGA GCCAGCTCTG TCCCAGGATG GTCCCAGCGG ATGCTGCCAG GGGCAKTGAA 1620
GTGTTTAGGT GAAGGACAAG TAGGTAAGAG GACGCCTTCA GGCACCACAG ATAAGCCTGA 1680 AACAGCCTCT CCAAGGGTTT TCACCTTAGC AACAATGGGA GCTGTGGGAG TGATTTTGGC 1740 CACACTGTCA ACATTTGTTA GAACCAGTCT TTTGAAAGAA AAGTATTTCC AACTTGTCAC 1800 TTGCCAGTCA CTCCGTTTTG CAAAAGGTGG CCCTTCACTG TCCATTCCAA ATAGCCCACA 1860 CGTGCTCTCT GCTGGATTCT AAATTATGTG AATTTTGCCA TATTAAATCT TCCTCATTTA 1920 TACTATTATT TGTTACGTTC AATCAGAATC CCCGAAACCT CCTATAAAGC TTAGCTGCCC 1980 CTTCTGAGGA TGCTGAGAAC GGTGTCTTTC TTTATAAATG CAAATGGCTA CCGTTTTACA 2040 ATAAAATTTT GCATGTGCAA AAAAAAAAAA ANAAAAAAAA AAAATCCCGG GGGGGGGCCG 2100 GTAACCAATT TGNCCCC 2117
(2) INFORMATION FOR SEQ ID NO: 159:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2395 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 159:
TGTTCCTTAA TCCCTTTTCT AAAAAGGGGG GAAAATCCGG ATGGATTTTA GGGATTGGTC 60
TGGTGTCAGC TGTGTTTTAT TGCACACCTA AATCCTGATT ATAGGCTTTT CATTTCTCCG 120
CAAAGCCTTT ATTTTGCCAG TTAAGCCAAA TGTGTTTTCC AGAAAGTTAG TTATTTTCTC 180
CTCTTTCTTT CCTTTCTTTC CTCCCTTTTT CCCGTCTGAC CCCAAACGTT ATTGTCCAAA 240
CATGACTGGA CAGCAGCTTT TGTTTCTTGA CCCTGTAATA TGACAGTCTG CTAATATTGA 300
CAGAAGGTCC AGTTTTTGGG TTATAGTCGT GATTTTCGCT AATCAATCAT ATTAGCAGGA 360
AAAAAAAKGA CTTGTTTCTG TTGTACTTGA GTCTTAAGAA AAAGTGGCCC ATAGTTTAGT 420
GGACAATTTC CAAAGGCTTT AGTACCACCT GTATTTCAAA ATGGGGGACC CAAACTCCCG 480
GAAGAAACAA GCTCTGAACA GACTACGTGC TCAGCTTAGA AAGAAAAAAG AATCTCTAGC 540
TGACCAGTTT GACTTCAAGA TGTATATTGC CTTTGTATTC AAGGAGAAGA AGAAAAAGTC 600
AGCACTTTTT GAAGTGTCTG AGGTTATACC AGTCATGACA AATAATTATG AAGAAAATAT 660
CCTGAAAGGT GTGCGAGATT CCAGCTATTC CTTGGAAAGT TCCCTAGAGC TTTTACAGAA 720
GGATGTGGTA CAGCTCCATG CTCCTCGATA TCAGTCTATG AGAAGGGATG TAATTGGCTG 780
TACTCAGGAG ATGGATTTCA TTCTTTGGCC TCGGAATGAT ATTGAAAAAA TCGTCTGTCT 840
CCTGTTTTCT AGGTGGAAAG AATCTGATGA GCCTTTTAGG CCTGTTCAGG CAAATTTGAG 900
TTTCATCATG GTGACTATGA AAAACAGTTT CTGCATGTAC TGAGCCGCAA GGACAAGACT 960
GGAATCGTTG TCAACAATCC TAACCAGTCA GTGTTTCTCT TCATTGACAG ACAGCACTTG 1020
CAGACTCCAA AAAACAAAGC TACAATCTTC AAGTTATGCA GCATCTGCCT CTACCTGCCA 1080
CAGGAACAGC TCACCCACTG GGGCAGTTCG CACCATAGAG GRTCACCTCC GTCCTTATAT 1140
GCCAGAGTAG AGTACTGACC AGCAAAATGG AGAAGATCAG AGAATGCAGC AGCAGTTTTT 1200
TTTCTTGTTT TCTTACCACT TTATTCTTTC AGAGTTTAAA GAAAATGGAC TCATGCACAG 1260
AACACTATGC ATTTTGAAAC TTGTTCATCC TGCATTTTTT TAAATCATTT TTATCTCAGA 1320
ACTTAAACAA AAATTAGATG TCGTGCACGG ACTGTGTGAA AGAAGATGCT TTGCATATTT 1380
GCTGCACTGC ATCAGTATCT TACTAAAAAT GTGAAATGAA AGGACTATTG TACACTGAAA 1440
TGCTTAAATG TATCTGAAAG CACAAGGTGA TACTCATTTT TATGGTCTTC CCATTTGTGC 1500
T∞TTTTTGC CTCTTTGACA TCTGTCATCA GTATTTAGAG GGTGAGAAGT GAATGTAACA 1560
GGTATAAATA ACATTTTTAA AAACAATAAC TTTGCTATAA TCACAGTTGT TCCAGAGCAC 1620
TGTCAGATAC ATTCTAATGA CCAGAACTGG TTTAAAAAAA GAAAATACAA CCATGGGAAA 1680
GAAATCTTAA ATGAAAAACG CATCTCATTG TAGGCATTTT TGCCTCATAT TTTACTGGGC 1740
CATGTTTGTT TCCTGGTACT CATGTATTTT TTTTTTCCAG ATCTCTTTCC CCAAGTTGCT 1800
ATTGTAAGAG TATTCTGCTG CGTGTGGATC CAGTTATACA CATTAAAGCA GATCTGGAGT 1860
CTCAAGTAGC TATAAAGCAG CTATAAAACA GAAATACATG CATAGCTGCA GAAACCATGA 1920
TAGGTAGAGG ACTTTTCTTT TGGTTTTGTT TTGTTTTGTT TTGTTTTGTT TTTGGTTTTA 1980
CAGAGAAGAG ATTTTTATTA CAAAGAAAAA AATTCCAGTG AATTGTGCAG AAATGCTGGT 2040
TTTTACACCA TCCTAAAGAA AAACTTTACA AGGGTGTTTT GGAGTAGAAA AAAGGTTATA 2100
AAGTTGGAAT CTTAAATTGT AAAATTAACC ATTGAGTGTC AAAGTTCTAA AAGCAGAACT 2160
CATTTTCTGC AATGAACATA AGGAAAGACT ACTGTATAGG TTTTTTTTTT TTCTCCTTTT 2220
AAATGAAGAA AAGCTTTGCT TAAGGGTTGC ATACTTTTAT TGGAGTAAAT CTGAATGATC 2280
CTACTCCTTT GGAGTAAAAC TAGTGCTTAC CAGTTTCCAA TTGTATTTAG CTTCTGGTTG 2340
GAATTTGAAA AAAAAAGAAA AAAAGAAAAA GAAAACCTAA ATAAAATAGG TGAAA 2395
(2) INFORMATION FOR SEQ ID NO: 160:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2120 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 160:
CCCCGGATAC CGCCTGACGT AGTGCCAATC ACACCTCTCG CGTCTCGGCG CCTCGGAGGC 60
TAATGAGGAC GCCTGGCGAA ACGCAGTAAC GGATTTCCGG GTGGACCTTC GCTTTACGGC 120
TCGTGAGTTC TTCCGCCCAA CCCAGAGGAA GCGGGAGAGC AGTTTACGAC AGCGCCGGTC 180
GTGTTTACGG CGGCGCCCGC TGCGCGCGCA TGTTTCCTCT TTTCCTGGTT TCTCAAGAGT 240
GCTGCTGCTA ACGCGGTCCC CGGCACGCAC CATCTGTTGC CATCCCGGCC GGCCGAGGCA 300
TTGCAGATTT TGGAAGATGG CAAAGTTCAT GACACCCGTG ATCCAGGACA ACCCCTCAGG 360
CTGGGGTCCC TGTGCGGTTC CCGAGCAGTT TCGGGATATG CCCTACCAGC CGTTCAGCAA 420
AGGAGATCGG CTAGGAAAGG TTGCAGACTG GACAGGAGCC ACATACCAAG ATAAGAGGTA 480
CACAAATAAG TACTCCTCTC AGTTTGGTGG TGGAAGTCAA TATGCTTATT TCCATGAGGA 540
GGATGAAAGT AGCTTCCAGC TGGTGGATAC AGCGCGCACA CAGAAGACGG CCTACCAGCG 600
GAATCGAATG AGATTTGCCC AGAGGAACCT CCGCAGAGAC AAAGATCGTC GGAACATGTT 660
GCAGTTCAAC CTGCAGATCC TGCCTAAGAG TGCCAAACAG AAAGAGAGAG AACGCATTCG 720
ACTGCAGAAA AAGTTCCAGA AACAATTTGG GGTTAGGCAG AAATGGGATC AGAAATCACA 780
GAAACCCCGA GACTCTTCAG TTCAAGTTCG TAGTGATTGG GAAGTGAAAG AGGAAATGGA 840
TTTTCCTCAG TTGATGAAGA TGCGCTACTT GGAAGTATCA GAGCCACAGG ACATTGAGTG 900
TTGTGGGGCC CTAGAATACT ACGACAAAGC CTTTGACCGC ATCACCACGA GGAGTGAGAA 960
GCCACTGCGG ASATNCAAGC GCATCTTCCA CACTGTCACC ACCACAGACG ACCCTGTCAT 1020
CCGCAAGCTG GCAAAAACTC AGGGGAATGT GTTTGCCACT GATGCCATCC TGGCCACGCT 1080
GATGAGCTGT ACCCGCTCAG TGTATTCCTG GGATATTGTC GTCCAGAGAG TTGGGTCCAA 1140
ACTCTTCTTT GACAAGAGAG ACAACTCTGA CTTTGACCTC CTGACAGTGA GTGAGACTGC 1200
CAATGAGCCC CCTCAAGATG AAGGTAATTC CTTCAATTCA CCCCGCAACC TGGCCATGGA 1260
GGCAACCTAC ATCAACCACA ATTTCTCCCA GCAGTGCTTG AGAATGGGGA AGGAAAGATA 1320
CAACTTCCCC AACCCAAACC CGTTTGTGGA GGACGACATG GATAAGAATG AAATCGCCTC 1380
TGTTGCGTAC CGTTACCGCA GTGGNAAGCT TGGAGATGAT ATTGACCTTA TTGTCCGTTG 1440
TGAGCACGAT GGCGTCATGA CTGGAGCCAA CGGGGAAGTG TCCTTCATCA ACATCAAGAC 1500
ACTCAATGAG TGGGATTCCA GGCACTGTAA TGGCGTTGAC TGGCGTCAGA AGCTGGACTC 1560
TCAGCGAGGG GCTGTCATTG CCACGGAGCT GAAGAACAAC AGCTACAAGT TGGCCCGGTG 1620
GACCTGCTGT GCTTTGCTGG CTGGATCTGA GTACCTCAAG CTTGGTTATG TGTCTCGGTA 1680
CCACGTGAAA GACTCCTCAC GCCACGTCAT CCTAGGCACC CAGCAGTTCA AGCCTAATGA 1740 GTTTGCCAGC CAGATCAACC TGAGCGTGGA GAATGCCTGG GGCATTTTAC GCTGCGTCAT 1800 TGACATCTGC ATGAAGCTGG AGGAGGGCAA ATACCTCATC CTCAAGGACC CCAACAAGCA 1860 GGTCATCCGT GTCTACAGCC TCCCTGATGG CACCTTCAGC TCTGATGAAG ATGAGGAGGA 1920 AGAGGAGGAG GAAGAAGAGG AAGAAGAAGA GGAAGAAACT TAAACCAGTG ATGTGGAGCT 1980 GGAGTTTCTC CTTCCACCGA GACTACGAGG GCCTTTGATG CTTAGTGGAA TGTGTGTCTA 2040 ACTTGCTCTC TGACATTTAG CAGATGAAAT AAAATATATA TCTGTTTAGT CTTAAAAAAA 2100 AAAAAAAAAA AAAAAAAAAN 2120
(2) INFORMATION FOR SEQ ID NO: 161:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 900 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 161:
GGAAGCTGAA GTCCTTCCAG ACCAGGGACA ACCAGGGCAT TCTCTATGAA GCTGCACCCA 60
CCTCCACCCT CACCTGTRAC TCAGCACCAC AGAAGCAAAA GTTCTCACTC AAACTGGATG 120
CCAAGGATGG GCGCTTGTTC AATGAGCAGA ACTTCTTCCA GCGGGCCGCC AAGCCTCTGC 180
AAGTCAACAA GTGGAAGAAG CTGTACTCGA CCCCACTGCT GGCCATCCCT ACCTGCATGG 240
GTTTCGGTGT TCACCAGGAC AAATACAGGT TCTTGGTGTT ACCCAGCCTG GGGAGGAGCC 300
TTCAGTCGGC CCTGGATGTC AGCCCAAAGC ATGTGCTGTG CAGAGAGGTC TGTGCTGCAG 360
GTGGCCTGCC GGCTGCTGGA TGCCCTGGAG TTCCTCCATG AGAATGAGTA TGTTCATGGA 420
AATGTGACAG CTGAAAATAT CTTTGTGGAT CCAGAGGACC AGAGTCAGGT GACTTTGGCA 480
GGCTATGGCT TCGCNTTCCG CTATTGCCCA AGTGGCAAAC ACGTGGCCTA CGTGGAAGGC 540
AGCAGGAGCC CTCACGAGGG GGACCTTGAG TTCATTAGCA TGGACCTGCA CAAGGGATCC 600
GGGCCCTCCC GCCGCRGCGA CCTCCAGAGC CTGGGCTACT GCATGCTGAA GTGGCTCTAC 660
GGGTTTCTGC CATGGACAAA TTGCCTTCCC AAMAMTGAGG ACATCATGAA GCAAAAACAG 720
AAGTTTGTTG ATAAGCCGGG GCCCTTCGTG GGACCCTGCG GTCACTGGAT CAGGCCCTCA 780
GAGACCCTGC AGAAGTACCT GAAGGTGGTG ATGGCCCTCA CGTATGAGGA GAAGCCGCCC 840
TACGCCATGC TGAGGAACAA CCTAGAAGCT TTGCTGCAGG ATCTGCGTGT GTCTCCATAT 900
(2) INFORMATION FOR SEQ ID NO: 162:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1003 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 162:
GGCACGAGAT GAGGGGCACC CAGTGCTTCT AGGGCAGGCT GGGTGGTGGT CCCCTAGGTA 60 TCAGCCTCTC TTACTGTACT CTCCGGGAAT GTTAACCTTT CTATTTTCAG CCTGTGCCAC 120
CTGTCTAGGC AAGCTGGCTT CCCCATTGGC CCCTGTGGGT CCACAGCAGC GTGGCTGCCC 180
CCCAGGGCCA CCGCTTCTTT CTTGATCCTC TTTCCTTAAC AGTGACTTGG GCTTGAGTCT 240
GGCAAGGAAC CTTGCTTTTA GCTTCACCAC CAAGGAGAGA GGTTGACATG ACCTCCCCGC 300
CCCCTCACCA AGCCTGGGAA CAGAGGGGAT GTGGTGAGAG CCAGGTTCCT CTGGCCCTCT 360 CCAGGGTGTT TTCCACTAGT CACTACTGTC TTCTCCTTGT AGCTAATCAA TCAATATTCT 420
TCCCTTGCCT GTGGGCAGTG GAGAGGCTGC TGGGTGTACG CTGCACCTGC CCACTGAGTT 480
GGGGAAAGAG GATAATCAGT GAGCACTGTT CTGCTCAGAG CTCCTGATCT ACCCCACCCC 540
CTAGGATCCA GGACTGGGTC AAAGCTGCAT GAAACCAGGC CCTGGCAGCA AACCTGGGAA 600
TGGCTGGAGG TGGGAGAGAA CCTGAACTTC TCTTTCCCTC TCCCTCCTCC AACATTACTG 660 GAACTCTATC CTGTTAGGAT CTTCTGAGCT TGTTTCCCTG CTGGGTGGGA CAGAGGACAA 720
AGGAGAAGGG AGGGTCTAGA AGAGGCAGCC CTTCTTTGTC CTCTGGGGTA AATGAGCTTG 780
ACCTAGAGTA AATGGAGAGA CCAAAAGCCT CTGATTTTTA ATTTCCATAA AATGTTAGAA 840
GTATATATAT ACATATATAT ATTTCTTTAA ATTTTTGAGT CTTTGATATG TCTAAAAATC 900
CATTCCCTCT GCCCTGAAGC CTGAGTGAGA CACATGAAGA AAACTGTGTT TCATTTAAAG 960 ATGTTAATTA AATGATTGAA ACTTGAAAAA AAAAAAAAAA AAA 1003
(2) INFORMATION FOR SEQ ID NO: 163:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2196 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 163: AAGAAGCGGC ACACGGATGT GCAGTTCTAC ACAGAAGTGG GAGAGATAAC CACGGACTTG 60
GGGAAACATC AGCATATCCA TGACCGAGAT GACCTCTATG CTGAGCAGAT GGAACGAGAA 120
ATGAGGCACA AACTGAAAAC AGCCTTTAAA AATTTCATTG AGAAAGTAGA GGCTCTAACT 180
AAGGAGGAAC TGGAATTTGA AGTGCCTTTT AGGGACTTGG GATTTAACGG AGCTCCCTAT 240
AGGAGTACCT GCCTCCTTCA GCCCACTAGT AGTGCGCTGG TAAATGCTAC GGAATGGCCA 300
CCTTTTGTGG TGACATTGGA TGAGGTAGAG CTGATCCACT TTRAGCGGGT CCAGTTTCAC 360
CTGAAGAACT TTGATATGGT AATCGTCTAC AAGGACTACA GCAAGAAAGT GACCATGATC 420
AACGCCATTC CTGTAGCCTC TCTTGACCCC ATCAAGGAAT GGTTGAATTC CTGCGACCTG 480
AAATACACAG AAGGAGTACA GTCCCTCAAC TGGACTAAAA TCATGAAGAC CATTGTTGAT 540
GACCCTGAGG GCTTCTTCGA ACAAGGTGGC TGGTCTTTCC TGGAGCCTGA GGGTGAGGGG 600
AGTGATGCTG AAGAAGGGGA TTCAGAGTCT GAAATTGAAG ATGAGACTTT TAATCCTTCA 660
GAAGATCACT ATGAAGAGGA AGAGGAGGAC AGTGATGAAG ATTATTCATC AGAAGCAGAA 720
GAGTCAGACT ATTCTAAGGA GTCATTGGGT AGTGAAGAAG AGAGTGGAAA GGATTGGGAT 780
GAACTGGAGG AAGAAGCCCG AAAAGCGGAC CGAGAAAGTC GTTACGAGGA AGAAGAAGAA 840
CAAAGTCGAA GTATGAGCCG GAAGAGGAAG GCATCTGTGC ACAGTTCGGG CCGTGGCTCT 900
AACCGTGGTT CCAGACACAG CTCTGCACCC CCCAAGAAAA AGAGGAAGTA ACTTCTGAAC 960
TTTGGCCCTG AGCTCCATTC TTCCTCCAGC CAACCCCTGA AAATTTTACA TGACATAGAA 1020
ACTGTATTTT TCCTTTCGTT TTCATTTGAA GTTTTGCCAT TTGTGTTTAT GGGTTTAGGG 1080
GGCCATTTGT GTGGACCAAT CTACTCGGGG AATTCCAGGC CCACCAGGAC ACGTGCCAAT 1140
GGCCCCATTC AGATCGCAAG GGAGGAGGTG TTCTTGAAGA CAGGAGGAGG CTCCCGCTGT 1200
TAATAAATAT TGTTTCATTC TTCTCTCTTC CTGTCACCTT CTGCCAAGAC ATTGATGGCT 1260
TCTGACATCT TATTTGGTGT CTCAAAGCTG TATTTCCAAG ACAGTGGTAC AAGGTGACCC 1320
TTAATTACCC GTATCATGGT TCTTGACCAG CACATTCAAT CCTCCAACCT ACCCTACTGC 1380
CATGACCTTC CGCACATCTC TAAGTTTTAT CTTTGCAATA CTCAAGGTTC TCGGAAATTT 1440
GCTAATGGTT GTGATAAACC ATACAGCTTG AGCCAGTGAG GCAGATTGGG CTGGTGCCTT 1500
CGTCTGAGTT TTCCTGCTTT CCTGCCTCGT GCAGATTCTG AGGTATATCT GCTGCCTTGG 1560
AAGACATAAG AAGCAGTGAT ACTCCCTGGC TCGGTTATTT TCTCCATACA ATGCACACAT 1620
GGTACAATGA TAGAAGGCAA AATTGCCACT GTCTTCTTTT TTTTCTCATA TATCTAAGGA 1680
AGATATATCA GGTTGTGCCT CATGTACCGC TTCTAGTGAA ATGTAGAGGA AGGCTCAAAG 1740
GAGTCAACAT TTAGATCTGG AAGGGACAAG TCATGCCTTG GGCCTAGAAT ACCCTGATGA 1800
GAAAAGAGAA GAGGAAGGGA GGCCATATCT ACAACANCAN CCTCTCGGCA CTGCTGCTCC 1860
TTATTTTAAC TTTGTCTTGC ATTGTCCTGT ATTTATCACA GTTTCTGTTG AACΛC :TTTT 1920
CAAGTATTTG GGGAGTTTAT CTTGCCATCC TCCCCTTCTG GTTCTCTGCA CCCACCTGTC 1980
CCACTGCAGT TCCTTCCGTG CTCTGTGACT TTAAGAGAAG AAGGGGGGAG GGGTCCCGGA 2040
TTTTATGTTT GTTTGTTTTT TCTCCTTAGC AGTAGGACTT GATATTTTCA ATTTTGGAAG 2100 AACTAAAAGA TGAATAAACT GGGTTTTTTT TGTTGTTTGT TTTTGTAAAA AAAAAAAAAA 2160
AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAA 2196
(2) INFORMATION FOR SEQ ID NO: 164:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 1945 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 164:
GCACAGAGTC GGGCGGACGG ACAGGGAGAG GAGGAGAGGG GGTCTGCGCG CGGCCGCTAC 60 CCAGAAGCCA GCGGACGGCA GCACGGAGTG GGCTGTCCCC GAGCCCAGCC CCGAGCGAGC 120
CCCCCCCCCG CCCCCGMAGG ACGCGCCTYC CAGCCAGCCC GACTYCTAGG AGGAGGGGAG 180
GCGGGAAAGC AGCTCAAGCC TCACCCACCG CCCTGCCCCC AGCCCCGCCA CTCCCAGGCT 240 CCTCGGGACT CGGCGGGTCC TCCTGGGAGT CTCGGAGGGG ACCGGCTGTG CAGACGCCAT 300
GGAGTTGGTG CTGGTCTTCC TCTGCAGCCT GCTGGCCCCC ATGGTCCTGG CCAGTGCAGC 360
TGAAAAGGAG AAGGAAATGG ACCCTTTTCA TTATGATTAC CAGACCCTGA GGATTGGGCG 420
ACTGGTGTTC GCTGTGGTCC TCTTCTCGGT TGGGATCCTC CTTATCCTAA GTCGCAGGTG 480
CAAGTGCAGT TTCAATCAGA AGCCCCGGGC CCCAGGAGAT GAGGAAGCCC AGGTGGAGAA 540 CCTCATCACC GCCAATGCAA CAGAGCCCCA GAAAGCAGAG AACTGAAGTG CAGCCATCAG 600
GTGGAAGCCT CTGGAACCTG AGGCGGCTGC TTGAACCTTT GGATGCAAAT GTCGATGCTT 660
AAGAAAACCG GCCACTTCAG CAACAGCCCT TTCCCCAGGA GAAGCCAAGA ACTTGTGTGT 720
CCCCCACCCT ATCCCCTCTA ACACCATTCC TCCACCTGAT GATGCAACTA ACACTTGCCT 780
CCCCACTGCA GCCTGCGGTC CTGCCCACCT CCCGTGATGT GTGTGTGTGT GTGTGTGTGT 840 GTGACTGTGT GTGTTTGCTA ACTGTGGTCT TTGTGGCTAC TTGTTTGTGG ATGGTATTGT 900
GTTTGTTAGT GAACTGTGGA CTCGCTTTCC CAGGCAGGGG CTGAGCCACA TGGCCATCTG 960
CTCCTCCCTG CCCCCGTGGC CCTCCATCAC CTTCTGCTCC TAGGAGGCTG CTTGTTGCCC 1020
GAGACCAGCC CCCTCCCCTG ATTTAGGGAT GCGTAGGGTA AGAGCACGGG CAGTGGTCTT 1080
CAGTCGTCTT GGGACCTGGG AAGGTTTGCA GCACTTTGTC ATCATTCTTC ATGGACTCCT 1140
TTCACTCCTT TAACAAAAAC CTTGCTTCCT TATCCCACCT GATCCCAGTC TGAAGGTCTC 1200
TTAGCAACTG GAGATACAAA GCAAGGAGCT GCTGAGCCCA GCGTTGACGT CAGGCAGGCT 1260
ATGCCCTTCC GTGGTTAATT TCTTCCCAGG GGCTTCCACG AGGAGTCCCC ATCTCCCCCG 1320
CCCCTTCACA GAGCGCCCGG GGATTCCAGG CCCAGGGCTT CTACTCTGCC CCTGGGGAAT 1380
GTGTCCCCTG CATATCTTCT CAGCAATAAC TCCATGGGCT CTGGGACCCT ACCCCTTCCA 1440
ACCTTCCCTG CTTCTGAGAC TTCAATCTAC AGCCCAGCTC ATCCAGATGC AGACTACAGT 1500
CCCTGCAATT GCGTCTCTGG CAGGCAATAG TTGAAGGACT CCTGTTCCGT TGGGGCCAGC 1560
ACACCGGGAT GGATGGAGGG AGAGCAGAGG CCTTTGCTTC TCTGCCTACG TCCCCTTAGA 1620
TGGGCAGCAG AGGCAACTCC CGCATCCTTT GCTCTGCCTG TCRGTGGTCA GAGCGGTGAG 1680
CGAGGTGCGT TGGAGACTCA GCAGGCTCCG TGCAGCCCTT GGGAACAGTG AGAGGTTGAA 1740
GGTCATAACG AGAGTGGGAA CTCAACCCAG ATCCCGCCCC TCCTGTCCTC TGTGTTCCCG 1800
CGGAAACCAA CCAAACCGTG CGCTGTGACC CATTGCTGTT CTCTGTATCG TGATCTATCC 1860
TCAACAACAA CAGAAAAAAG GAATAAAATA TCCTTTGTTT CCTAGTGAAA AAAAAAAAAA 1920
AAAAAAAAAA AAAAAAAAAA CTCGA 1945
(2) INFORMATION FOR SEQ ID NO: 165:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2933 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: ; ib : GGGTCGACCC ACGCGTCCGG CAGCCGTCGT TTGAGTCGTT GCTGCCGCTG CCCCCTCCCG 60 GATCAGGAGC CAGTGTATAC CGCCCGCCCA CCGCCTTGGT GCCGCTAGAG GAAACGAGAA 120 GGAGGCCGCC TGCGGTTTGT CGCCGCAGCT CGCCCMCYGY CYGGRAGAGC CGAGCCCCGG 180 CCCAGTCGGT CGCYTGCCAC CSCTCGTAGC CGTTACCCGC GCGCCGCCAC AGCCGCCGGC 240 CGGGAGAGCC GCGCGCCATG GCYTCTGGAG CCGATTCAAA AGGTGATGAC CTATCAACAG 300 CCATTCTCAA ACAGAAGAAC CGTCCCAATC GGTTAATTGT TGATGAAGCC ATCAATGAGG 360 ACAACAGTGT GGTGTCCTTG TCCCAGCCCA AGATGGATGA ATTGCAGTTG TTCCGAGGTG 420 ACACAGTGTT GCTGAAAGGA AAGAAGAGAC GAGAAGCTGT TTGCATCGTC CTTTCTGATG 480
ATACTTGTTC TGATGAGAAG ATTCGGATCA ATAGAGTTGT TCGGAATAAC CTTCGTGTAC 540
GCCTAGGGGA TGTCATCAGC ATCCAGCCAT GCCCTGATGT GAAGTACGGC AAACGTATCC 600
ATGTGCTGCC CATTGATGAC ACAGTGGAAG GCATTACTGG TAATCTCTTC GAGGTATACC 660
TTAAGCCGTA CTTCCTGGAA GCGTATCGAC CCATCCGGAA AGGAGACATT TTTCTTGTCC 720
GTGGTGGGAT GCGTGCTGTG GAGTTCAAAG TGGTGGAAAC AGATCCTAGC CCTTATTGCA 780
TTCTTGCTCC AGACACAGTG ATCCACTGCG AAGGGGAGCC TATCAAACGA GAGGATGAGG 840
AAGAGTCCTT GAATGAAGTA GGGTATGATG ACATTGGTGG CTGCAGGAAG CAGCTAGCTC 900
AGATAAAGGA GATGGTGGAA CTGCCCCTGA GACATCCTGC CCTCTTTAAG GCAATTGGTG 960
TGAAGCCTCC TAGAGGAATC CTGCTTTACG GACCTCCTGG AACAGGAAAG ACCCTGATTG 1020
CTCGAGCTGT AGCAAATGAG ACTGGAGCCT TCTTCTTCTT GATCAATGGT CCTGAGATCA 1080
TGAGCAAATT GGCTGGTGAG TCTGAGAGCA ACCTTCGTAA AGCCTTTGAG GAGGCTCAGA 1140
AGAATGCTCC TGCCATCATC TTCATTGATG AGCTAGATGC CATCGCTCCC AAAAGAGAGA 1200
AAACTCATGG CGAGGTGGAG CGGCGCATTG TATCACAGTT GTTGACCCTC ATGGATGGCC 1260
TAAAGCAGAG GGCACATGTG ATTGTTATGG CAGCAACCAA CAGACCCAAC AGCATTGACC 1320
CAGCTCTACG GCGATTTGGT CGCTTTCACA GGGAGCTAGA TATTGGAATT CCTGATGCTA 1380
CAGGACGCTT AGAGATTCTT CAGATCCATA CCAAGAACAT GAAGCTGGCA GATGATGTGG 1440
ACCTGGAACA GTAGCCAATG AGACTCACGG GCATGTGGGT GCTGACTTAG CAGCCCTGTG 1500
CTCAGAGGCT GCTCTGCAAG CCATCCGCAA GAAGATGGAT CTCATTGACC TAGAGGATGA 1560
GACCATTGAT GCCGAGGTCA TGAACTCTCT AGCAGTTACT ATGGATGACT TCCGGTGGGC 1620
CTTGAGCCAG AGTAACCCAT CAGCACTGCG GGAAACCGTG GTAGAGGTGC CACAGGTAAC 1680
CTGGGAAGAC ATCGGGGGCC TAGAGGATGT CAAACGTGAG CTACAGGAGC TGGTCCAGTA 1740
TCCTGTGGAG CACCCAGACA AATTCCTGAA GTTTGGCATG ACACCTTCCA AGGGAGTTCT 1800
GTTCTATGGA CCTCCTGGCT GTGGGAAAAC TTTGTTGGCC AAAGCCATTG CTAATGAATG 1860
CCAGGCCAAC TTCATCTCCA TCAAGGGTCC TGAGCTGCTC ACCATGTGGT TTGGGGAGTC 1920
TGAGGCCAAT GTCAGAGAAA TCTTTGACAA GGCCCGCCAA GCTGCCCCCT GTGTGCTATT 1980
CTTTGATGAG CTGGATTCGA TTGCCAAGGC TCGTGGAGGT AACATTGGAG ATGGTGGTGG 2040
GGCTGCTGAC CGAGTCATCA ACCAGATCCT GACAGAAATG GATCGCATGT CCACAAAAAA 2100
AAATGTGTTC ATCATTGGCG CTACCAACCG GCCTGACATC ATTGATCCTG CCATCCTCAG 2160
ACCTGGCCGT CTTGATCAGC TCATCTACAT CCCACTTCCT GATCAGAAGT CCCGTGTTGC 2220
CATCCTCAAG GCTAACCTGC GCAAGTCCCC AGTTGCCAAG GATGTGGACT TGGAGTTCCT 2280
GGCTAAAATG ACTAATGGCT TCTCTGGAGC TGACCTGACA GAGATTTGCC AGCGTGCTTG 2340
CAAGCTGGCC ATCCGTGAAT CCATCGAGAG TGAGATTAGG CGAGAACGAG AGAGGCAGAC 2400
AAACCCATCA GCCATGGAGG TAGAAGAGGA TGATCCAGTG CCTGAGATCC GTCGAGATCA 2460
CTTTGAAGAA GCCATGCGCT TTGCGCGCCG TTCTGTCAGT GACAATGACA TTCGGAAGTA 2520
TGAGATGTTT GCCCAGACCC TTCAGCAGAG TCGGGGCTTT GGCAGCTTCA GATTCCCTTC 2580
AGGGAACCAG GGTGGAGCTG GCCCCAGTCA GGGCAGTGGA GGCGGCACAG GTGGCAGTGT 2640
ATACACAGAA GACAATCATG ATGACCTGTA TGGCTAAGTG GTGGTGGCCA GCGTGCAGTG 2700
AGCTGGCCTG CCTGGACCTT GTTCCCTGGG GGTGGGGGCG CTTGCCCAGG AGAGGGACCA 2760
GGGGTGCGCC CACAGCCTGC TCCATTCTCC AGTCTGAACA GTTCAGCTAC AGTCTGACTC 2820
TGGACAGGGG GTTTCTGTTG CAAAAATACA AAACAAAAGC GATAAAATAA AAGCGATTTT 2880
CATTTGGTAA AAAAAAAAAA AAAAAAAAAT CCGGGG GGG GCCCGAACCA TTT 2933
(2) INFORMATION FOR SEQ ID NO: 166:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2243 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 166:
TCGGAGAGCC GGCGGGCGNG CGCCTCTCGG CCAGGAAGCG CCTCTTGGAC GCGTGTNACC 60
GATGCCCAGA AGTGGCCTTG GGCTGGGGAT CACCATAGCT TTTCTAGCTA CGCTGATCAC 120
GCAGTTTCTC GTGTATAATG GTGTCTATCA GTATACATCC CCAGATTTCC TCTATATTCG 180
TTCTTGGCTC CCTTGTATAT TTTTCTCAGG AGGCGTCACG GTGGGGAACA TAGGACGACA 240
GTTAGCTATG G3TGTTCCTG AAAAGCCCCA TAGTGATTGA GTCTTCAAAA CCACCGATTC 300
TGAGAGCAAG GAAGATTTTG GAAGAAAATC TGACTGTGGA TTATGACAAA GATTATCTTT 360
TTTCTTAAGT AATCTATTTA GATCGGGCTG ACTGTACAAA TGACTCCTGG AAAAAACTCT 420
TCACCTAGTC TAGAATAGGG AGGTGGAGAA TGATGACTTA CCCTGAAGTC TTCCCTTGAC 480
TGCCCGCACT GGCGCCTGTC TGTGCCCTGG AGCATTCTGC CCAGGCTACG TGGGTTCAGG 540
CAGGTGGCAG CTTCCCAAGT ATTCGATTTC ATTCATCTGA TTAAAACAAG TTGCCATATT 600
TCAAAGCCTT GAACTAAGAC TCAATTACCA ACCCGCAGTT TTGTGTCAGT GCCCAAAGGA 660
GGTAGGTTGA TGGTGCTTAA CAAACATGAA GTATGGTGTA ATAGGAATAA TATTTATCCA 720
AAAGATTTTT AAAAATAGGG CTCTGTTTAA AAAAAAAAAC AAAACARGAA AAGCAGCAGT 780
GATTATAGAG AGGTCACACT CTAAGTGGGG TCGCGGCGTG GCCACGCTTC ACGGTCACGC 840
TCGTCCGTCC TGCAGTGGCG TGTTTACATG GTCACACGTG TGTGTATCAC CAGTGGGTCA 900
ACTGCTTGTC ATTCCTCCCG TGGCAGTTTG TGTAGACAAT CTTACTGAGC AAAAGGCAAT 960
GAAAAGTCTT GGTTCCCACA CTGCGATATA TTGGAATTTT CACCTCAGTT TATGAAGTTT 1020
ATTTCGAAAT CCATAGTCAT CTAAGAATGA ATACCTGTCT GCCATGTATT TCAATCTTAG 1080
TGAGCCAAAA TTCTTTGTTT GTTACTACAG AATAGAGATG ACTGTTTTTT GCCACAGCCC 1140
TATGGRATTT GCAATCTGTG ATTGCCTTGT AAAAAGGAGA GTGCATATGG CACTGCATTA 1200
AACGTGTGGT GTTTCTAGTC AATGATATTG GTGAGCACAA TGTATTCATT TAATGGCATA 1260
GACCATACCA GACCTAATTT GCAAGTATTG GGTCTTAAAC TTCAAGTGCA ATGTATATGA 1320
AAACCAATCT GAGCCTTGTA TCTCTTAAAT ATTTATTTTT TTTAACGTGT GAGATGTTCG 1380
AGAGAAGGTT CTCCATTCAT TTCAGTGCTG CCTGGAGGAA ACTCGGCAAT GATTTCTTTC 1440
AGTTGTGAAG TTCCTTTCGT GTTACACCCT CCACTGAACC CTCAACCTTC GAAATACTCC 1500
AGTTTTGTGG GTTTGGTCAT TTTTACTTAT AAATTTACCT TTTTGTATTT TGCftATTTAC 1560
ATGTGTTTGG TTTGTTTTAA ATTCTGTGAA AGTGGCTTGA TTAAAAGACT CCTTTTAAAT 1620
GGAAGCCACC AGTCAGCAGA ATGGAAGCTT AGAGGAACTT GCCTGTGAGC GCTCX3TCTTT 1680
GTCTTTGGTT TTGTGATGTA ACGATCTTTG CTGGGGTTTT TTGCTTTGTT TTGAGGGAAA 1740
TGTCTTGGAG TAAATTTTAA GTTCCTGGAG TTAATTTGTT TTACAGGAAT TTTGTTTTTT 1800
AAAAAAATAG GATCATTCTG AACTTTGGAA TGACCCCCTT ATATATTTTC TGAAAATGAA 1860
AACAGTTACA TGAAAAAAAT TTCCAATGAA GATGTCAGCA TTTTATGAAA AACCAGAAGT 1920
TATTAGATGA AAGCAGCGAG TGAATCTTTA AAACAGACTT GATCACGCAC ACACAATAAG 1980
TCTTTCTCTC CGAAACCGGA AGTAAATCTA TATCTGTTAG AAATAATGTA GCCAAAAGAA 2040
TGTAAATTTG AGGATTTTTT TGCCAATAGT TTATAGAAAA TATATGAACC AAAGTGATTT 2100
GAGTTTGTAA AAATGTAAAA TAGTATGAAC AAAATTTGCA CTCTACCAGA TTTGAACATC 2160
TAGTGAGGTT CACATTCATA CTAAGTTTTC AACATTGTGT TCTTTTTGCA TTCATTTTTT 2220
ACTTTTATTA AAGGTTCAAA ACC 2243
(2) INFORMATION FOR SEQ ID NO: 167:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1816 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 167:
GGTGGGNAGC TTTNAATTTC CCCTTACWGG GGCGCTNTAA GGGGAAACCT TCCCGGAATT 60
TTCGGGTCGA CCCACGCGTC CGGCCAGCCT AGGAGAAGAA GTTCGTAGTC CCAGAGGTGA 120
GGCAGGAGGC GGCAGTTTCT GGCGGGTGAG GGCGGAGCTG AAGTGACAGC GGAGGCGGAA 180
GCAACGGTCG GTGGGGCGGA GAAGGGGGCT GGCCCCAGGA GGAGGAGGAA ACCCTTCCGA 240
GAAAACAGCA ACAAGCTGAG CTGCTGTGAC AGAGGGGAAC AAGATGGCGG CGCCGAAGGG 300
GAGCCTCTGG GTGAGGACCC AACTGGGGCT CCCGCCGCTG CTGCTGCTGA CCATGGCCTT 360
GGCCGGAGGT TCGGGGACCG CTTCGGCTGA AGCATTTGAC TCGGTCTTGG GTGATACGGC 420
GTCTTGCCAC CGGGCCTGTC AGTTGACCTA CCCCTTGCAC ACCTACCCTA AGGAAGAAGA 480
GTTGTACGCA TGTCAGAGAG GTTGCAGGCT GTTTTCAATT TGTCAGTTTG TGGATGATGG 540
AATTGACTTA AATCGAACTA AATTGGAATG TGAATCTGCA TGTACAGAAG CATATTCCCA 600
ATCTGATGAG CAATATGCTT GCCATCTTGG KTGCCAGAAT CAGCTGCCAT TCGCTGAACT 660
GAGACAAGAA CAACTTATGT CCCTGATGCC AAAAATGCAC CTACTCTTTC CTCTAACTCT 720
GGTGAGGTCA TTCTGGAGTG ACATGATGGA CTCCGCACAG AGCTTCATAA CCTCTTCATG 780
GACTTTTTAT CTTCAAGCCG ATGACGGAAA AATAGTTATA TTCCRGTCTA AGCCCAGRAA 840
TCCCAGGTAC GCACCACATT TGGAGCCAGG AGCCCTACCA AATTTGRGRG RAWCMTCTCT 900
AAGCAAAATG TCCNTCAKMT CGSMAATGAG AAATTCACAA GCGCACAGGA ATTTTCTTGA 960
AGATGGAGAA AGTGATGGCT TTTTAAGATG CCTCTCTCTT AACTCTGGGT GGATTTTAAC 1020
TACAACTCTT GTCCTCTCGG TGATGGTATT GCTTTGGATT TGTTGTGCAA CTTGTTGCTA 1080
CACGCTGTTG GACGCAGTAT AGTTTCCCTC TGAGAAGCTG AGTATCTATG GTGACTTGGA 1140
GTTTATGAAT GAACAAAAGC TAAACAGATA TCCAGCTTCT TCTCTTGTGG TTGTTAGATC 1200
TAAAACTGAA GATCATGAAG AAGCAGGGCC TCTACCTACA AAAGTGAATC TTGCTCATTC 1260
TGAAATTTAA GCATTTTTCT TTTAAAAGAC AAGTGTAATA GACATCTAAA ATTCCACTCC 1320
TCATAGAGCT TTTAAAATGG TTTCATTGGA TATAGGCCTT AAGAAATCAC TATAAAATGC 1380
AAATAAAGTT ACTCAAATCT GTGAAAAAAA AAAAAAAAAA AAAAAAAAAC TCGAGGGGGG 1440
GCCCGTTACC AAKTCGCCCT ATWGTGADTB GTATTMTTAT TTTACTAATA TCTGTAGCTA 1500
TTTTCTTTTT KGCTTKGGTT ATKGTTTTTY TCCCTTYTCT WAGCTATRAG CTGATCATKG 1560
CYSCTTCTCA CCTCCTGCCA TGATACTGTC AGTTACCTTA GTTAACAAGC TGAATATTTA 1620
GTAGAAATGA TGCTTCTGCT CAGGAATGGC CCACAAATCT GTAATTTGAA ATTTAGCAGG 1680
AAATGACCTT TAATGACACT ACATTTTCAG GAACTGAAAT CATTAAAATT TTATTTGAAT 1740 AATTATGTCC TGAAAAAAAA AAAAAAAAAA AMWMRARASK RRWWACTCGA GGGGGGGCCC 1800 GGTACCCNAT TCGCCG 1816
(2) INFORMATION FOR SEQ ID NO: 168:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 945 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 168:
AGAAACCGTT GATGGGACTG AGAAACCAGA GTTAAAACCT CTTTGGAGCT TCTGAGGACT 60
CAGCTGGAAC CAACGGGCAC AGTTGGCAAC ACCATCAACT TCTCCCAAGC AGAGAAACCC 120
GAACCCACCA ACCAGGGGCA GGATAGCCTG AAGAAACATC TACACGCAGA AATCAAAGTT 180
ATTGGGACTA TCCAGATCTT GTGTGGCATG ATGGTATTGA GCTTGGGGAT (-ATTTTGGCA 240
TCTGCTTCCT TCTCTCCAAA TTTTACCCAA GTGACTTCTA CACTGTTGAA CTCTGCTTAC 300
CCATTCATAG GACCCTTTTT TTTTATCATC TCTGGCTCTC TATCAATCGC CACAGAGAAA 360
AGGTTRACCA AGCTTTTGGT GCATAGCAGC CTGGTTGGAA GCATTCTGAG TGCTCTGTCT 420
GCCCTGGTGG GTTTCATTAT CCTGTCTGTC AAACAGGCCA CCTTAAATCC TGCCTCACTG 480
CAGTGTGAGT TGGACAAAAA TAATATACCA ACAAGAAGTT ATGTTTCTTA CTTTTATCAT 540
GATTCACTTT ATACCACGGA CTGCTATACA GCCAAAGCCA GTCTGGCTGG AWCTCTCTCT 600
CTCATGCTGA TTTGCACTCT GCTGGAATTC TGCCTAGCTG TGCTCACTGC TGTGCTGCGG 660
TGGAAACAGG CTTACTCTGA CTTCCCTGGG AGTGTACTTT TCCTGCCTCA CAGTTACATT 720
GGTAATTCTG GCATGTCCTC AAAAATGACT CATGACTGTG GATATGAAGA ACTATTGACT 780
TCTTAAGAAA AAAGGGAGAA ATATTAATCA GAAAGTTGAT TCTTATGATA ATATGGAAAA 840
GTTAACCATT ATAGAAAAGC AAAGCTTGAG TTTCCTAAAT GTAAGCTTTT AAAGTAATGA 900
ACATTAAAAA AAACCATTAT TTCACTGTCA TTTAAAGATA ATGTG 945
(2) INFORMATION FOR SEQ ID NO: 169:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 902 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 169:
GGCAGAGCCA CAGGAAGGAT GAGGAAGACC AGGCTCTGGG GGCTGCTGTG GATGCTCTTT 60
GTCTCAGAAC TCCGAGCTGC AACTAAATTA ACTGAGGAAA AGTATGAACT GAAAGAGGGG 120 CAGACCCTGG ATGTGAAATG TCACTACACG CTAGAGAAGT TTGCCAGCAG CCAGAAAGCT 180
TGGCAGATAA TAAGGGACGG AGAGATGCCC AAGACCCTCG CATGCACAGA GAGGCCTTCA 240
AAGAATTCCC ATCCAGTCCA AGTGGGGAGG ATCATACTAG AAGACTACCA TGATCATGGT 300
TTACTGCGCG TCCGAATGGT CAACCTTCAA GTGGAAGATT CTGGACTGTA TCAGTGTGTG 360
ATCTACCAGC CTCCCAAGGA GCCTCACATG CTGTTCGATC GCATCCGCTT GGTGGTGACC 420 AAC^TTTTT CAGGGACCCC TGGCTCCAAT GAGAATTCTA CCCAGAATGT GTATAAGATT 480
CCTCCTACCA CCACTAAGGC CTTGTGCCCA CTCTATACCA GCCCCAGAAC TGTGACCCAA 540
GCTCCACCCA AGTCAACTGC CGATGTCTCC ACTCCTGACT CTGAAATCAA CCTTACAAAT 600
GTGACAGATA TCATCAGGGT TCCGGTGTTC AACATTGTCA TTCTCCTGGC TGGTGGATTC 660
CTGAGTAAGA GCCTGGTCTT CTCTGTCCTG TTTGCTGTCA CGCTGAGGTC ATTTGTACCC 720 TAGGCCCACG AACCCACGAG AATGTCCTCT GACTTCCAGC CACATCCATC TGGCAGTTGT 780
GCCAAGGGAG GAGGGAGGAG GTAAAAGGCA GGGAGTTAAT AACATGAATT AAATGTGTAA 840
TCACCRGCTA AAAAAAAAAA AAAAAAAACN CGANCCTNGG TTTTCAGCTC CATCAGCTCC 900
TT 902
(2) INFORMATION FOR SEQ ID NO: 170:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1883 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 170:
AGAAAACAAC TGAAAAACCA CATTTTTCTA CATACAGCTG GGGAGGTAGC TGAGAACTTG 60
GCACTGCGCA CACATACTAG GTTGAAAGAG AGTTGAGGAA ACCAGAAGGC CAAGTGGATC 120 TGCTGGCAAA CCCTGAACCT GTCTCCTGCG CTTGCTCTAC AGTTCTGAAG TTGAAAATCC 180
TTTTCATGCC TAGCATCTGC TTGAGTTATA AACCCCAAGG CAGCCATGTC ATAGACTAGT 240
GTTTACTCTT GTTTTGACTT TGTTTTAATG CTTCCTAAGA CCCAAGTGCC TCCTGCTGTT 300
TCCTCCTTTG TGGTAGCCTC TCGCCATCTG GGACCTCAAT CCCCAGCTTT CCCACTTTCA 360
GCAGTCCTTT GCTCTCTTTG CTTCTACCTC AAATAGCCCC AGGAGTGGGC TTTAGTCTCC 420
AATATGGAGC ATYTCAAGCT TCTCCTGGGG GATGGGGATT GGGATGGGCA GAATCTGTTT 480
TGGWTCTCCG GGTTATTTCC AGTGGGTGTA AAAGCAGAGC TGGGCCTTTC CCTCTCTTAT 540
CCCTGAGGCT GGGTAAGAAG GACTGTATCT ACACCTGTTC TTCCCTACCT TCTCTTTTGT 600
TAGGGAGGCC TCATTCTAAG TTCCTCAAGA GAGTCCTTGG CTTAAAGCTG TAGCAAGGGT 660
GTGCTAGGTG GGGGATTTGG AGCAAAACCG TCGAGTAGGC ATGATACTGG TATGGAGTGG 720
GCCTGCAAAA TCAGACAGAA ATGGCTTGAG AAGCCGCAGG GGAGCATGCC TGTCTCTCAG 780
TGATAGAGTA TGGGAGGGAC CTCCCTAGCT TGGAAAATGA GAATTGAAGG GGTTATGAAC 840
AAATAGGATG CCTAGTTGAG GATGTTCCCA AAGTTTTCTC CAATCTTATC ATTAGTAGAT 900
TTTATAAGCC ACAGAGACAA ACCAGAAACG GAATAATGTT ACTTTGGATG CTTTATTTTT 960
TTGTTCTAGG TGTGGCTTTG TACATGCAGA AGAATGCTAT ATGCTGCACA TTTTGCCTTT 1020
AAAGTCTTAC GACTTTCCCC ATTTTAGTCT AATGGGAAGA TACAGATGTG CAAGTCTGCT 1080
TTTTTCTTTT TTGTTATTAT TTTTTTTTTT TTGCTCTGTG TTATGGACAT TTTCAGACAT 1140
GCACAGAAGT GGAGAGGATG GTCCTTGGAC CCCATCTGTC CATCACCTAG CTGCATCACT 1200
TATCAGCTAT GGTCAACCTG GTTTCATCTG TATCTCTCTC TTTTCACCTG TATTGTTTAT 1260
TGAAAATCCA AGACACTATG CCAATGCAAC CGTGACTACT TTGGGAGATT GGTAGTCTCT 1320
TTTGATGGTG ATAGTGATGG GGTGCACTAT CATAATCACA TCAGGTCTGC TTTTTGCTTT 1380
TAATGTTAAC TAATGAAGTT CCAGAGATGG GCCTTAGAAA TGTGTTTTAA GAATTAACAA 1440
GGAGTCTCAA AAAGAAATGA GAGGGATGCT TCCTTTCCCC TTGCATCTAC AAAACAAGAG 1500
AGAGACTGTT CTGTTGTAAA ACTCTTTCAA AAATTCTGAT ATGGTAAGGT ACTTGAGACC 1560
CTTCACCAGA ATGTCAATCT TTTTTTCTGT GTAACATGGA AACTTGTGTC ACCATTAGCA 1620
TTGTTATCAG CTTGTACTGG TCTCATAACT CTCGTTTTGG AAGAATAATT TGGAAATTGT 1680
TGCTGTGTTC TGTGAAAATA ACCTCCCCAA AATAATTAGT AACTGGTTGT TCTACTTGGT 1740
AATTTGACAC CCTGTTAATA ACGCAATTAT TTCTGTGTTC TTAAACAGTA TAAATAGTTG 1800
TAAGTTTGCA TGCATCATGG AAAAATAAAA ACCTGTATCT CTGTTAAAAA AAAAAAAAAA 1860
AAAAAAAAAA AAAAAAAAAA AAA 1883
(2) INFORMATION FOR SEQ ID NO: 171: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2100 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 171:
TACTTTTAGA TTTACTGCCT TCAAAAAGTG CCTATTCTGA GCAACATAAA CGTTATTCCT 60
TACATATGTA TGTACACACG GTACCCAGAG TCGTACTGTG GCAGCCTTCA AAAACATACC 120
ATCAGAAAGA GTAGGTGCTC AGATAAGGNA ACTTTGCCAA ATGNAAGAAA GTCACTCACT 180
TCCAATATCC CCTCTTCAAG CGGCTACCGT GRAASGGGCT GCAAACACAT TCCCTGAGCA 240
TCCCTTGCTG ATACAGCTTC TTTATATTTA TATCCTACTG GATGGTAGCA TATTGCTAAG 300
GTTTCCTGTA CTCTGCTTCA AGGGAATGTA AGYTTTATGG CATTGAAACA TTTAGGAAAA 360
AAAAAGATGT TTAAGAGAAT TAATAGAGCC GTAGTCTGTA TTAGGATGTG TGTCATATGT 420
GTGTTCTATA AACTAAGCAT CGGTGGGTTT AGAGTGTTAA AGTGTCAGCA CATTCCTTCT 480
CCTTTTGTCT CTCAGGCTAA CATGAGAGAA AATAGAAAAG TCTTGGCTGT GGGGATTGGA 540
AGCTCAGGGG GCCAAATGTC CTTGCCAGAT CCTTAGAGCA TTACTTTGAC TCCTAAAAAT 600
AGTAGTGTAT GTTATTTGAT GGCTTTTGTT TCCATAGTTC CATCACTGAC AAAACTGTCA 660
ATACTGTTGA TGGAGCAGCA GCATAGCCTA GAGTGATGCA TTCTTACCCA GAGGTGGCAA 720
TAGGAGAGGG TCCATGTAAA TAGGACGAGG TAGACAGTGC ATGATTGTAG GAGAAGGGTT 780
GAAGGGAGGA CATGATTCCA AAAAAGATCG TTCTCAATGT GTCGTCTGAC TCAACCAGCT 840
GGCAGATTAC ACTTGCCAAG TCGTTCCCTT TCCTTCTAAG TCAGTTGGCT CCATATTCAC 900
TTGAATATGC CTCTGTTTGG GCAAAGCAAG ATACCTCCAC TTAACCTTTA TCCAAGGAAG 960
CTCTTGGTGT CCTCTTGGTC ATAAAGTTGT CTCCTACCTA ACCCAGTTTT ACCAAATGGA 1020
AGTAAAAGGG GACAAACTAT GGAAGATGGA CTCCATGCCA TTGCAGTCAG CCACCATTCT 1080
CTTTTCCATA TAAGGAGCCC CATTACATAA GCTACGGGTG AGGTTGGAAC AGCTATGTTT 1140
CATAATTTCA AGAGTGTGAC CACCCTGCTC TAGTCATCAT CATTGGATGA ATCCAGTTGA 1200
CTCTTTGGCA AAAGGGTGAT ACTTTTCACT AAAAATGCCT ACTCTTCCTG TTGATGTTCC 1260
TTTTCTGTTT TTACCTTGTC CAATTTCCAC ACTAGTCATT TTTTTTATTT TTTAGAGGAT 1320
CAGATTTTAG CGCTGGAAAA TGAGTTCAAA AATTTCAGTG TAATGTCATA AGGATGTTGG 1380
GATACAGAGA TTTTTTTTTT CCTTGGAAAC AAATGGACTG GGAAGAAACA CAGCATGGCT 1440
TTGCTCTGAG TTTCAATCTG ATGATTATGA CCATGGAAGA TAGTCTTATG TAAAGGTTAA 1500
ATCGTGTTTA CAAGTGGATA GATAAGGCGG AGATGGTGAG AAGCCGGGTT TTCTCTATGC 1560
TAAATCTCTC TACTAAGAGC AGCACTTCCT ACTAGCTAAG CACAATCATA GCCCCACCGT 1620
GATGAGCTGC TAGTCTGAAT AACATTCCCT GACTTAGGGA AAGGCACACA AAAACATATA 1680
AAGAATATGT CTATTTTCAT ATGTGTGATA CTGACAGAGC CATGGTATTC CTAAAATATA 1740
GGTTTCTCTT TTTCTTGTA TTCTTAGCAA ATTGCATTTA TTCACTACAT TACAAACCAT 1800
CACTGATGTA TCCAAAATAG CACACATAGT TCAGTATGAA AATAAGAGAA TAAAATCTGT 1860
TATAAGCAAG TGATTTAGGT ATTTTCTTTT GTGTTTATGC ATTATCTGAC TATATTAAAA 1920
CCTGTTTTTC TATTTACCTT CTATCAGTTT TCTCTACCAA TTATGTTTTT TCAATGCTCT 1980
ATAAGAATGA ATATGGAAAT TATATTTCTT TTTTCTGTAA AAGAGTTGCA ACTACTTTAT 2040
TATATTTAGA AATCCAATAA ACTTCTTATT ACATTTAAAA AAAAAAAAAA AAAACTCGAA 2100
(2) INFORMATION FOR SEQ ID NO: 172:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1930 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 172:
CCTTTGANTG TGGTCCCGGG TGCNGATTCG CAGCGCCTCC GCCGCGGCTC GTGGTTGTCC 60
CGCCATGGCA CTGTCGCGGG GGCTGCCCCG GGAGCTGGCT GAGGCGGTGG CCGGGGGCCG 120
GGTGCTGGTG GTGGGGGCGG GCGGCATCGG CTGCGAGCTC CTCAAGAATC TCGTGCTCAC 180
CGGTTTCTCC CACATCGACC TGATTGATCT GGATACTATT GATGTAAGCA ACCTCAACAG 240
ACTGTTTTTG TTTCAAAAGA AACATGTTGG AAGATCAAAG GCACAGGTTC CCAAGGAAAG 300
TGTACTGCAG TTTTACCCGA AAGCTAATAT CGTTGCCTAC CATGACAGCA TCATGAACCC 360
TGACTATAAT GTGGAATTTT TCCGACAGTT TATACTGGTT ATGAATGCTT TAGATAACAG 420
AGCTGCCCGA AACCATGTTA ATAGAATGTG CCTGGCAGCT GATGTTCCTC TTATTGAAAG 480
TGGAACAGCT GGGTATCTTG GACAAGTAAC TACTATCAAA AAGGGTGTGA CCGAGTGTTA 540
TGAGTGTCAT CCTAAGCCGA CCCAGAGAAC CTTTCCTGGC TGTACAATTC GTAACACACC 600
TTCAGAACCT ATACATTGCA TCGTTTGGGC AAAGTACTTG TTCAACCAGT TGTTTGGGGA 660
AGAAGATGCT GATCAAGAAG TATCTCCTGA CAGAGCTGAC CCTGAAGCTG CCTGGGAACC 720
AACGGAAGCC GAAGCCAGAG CTAGAGCATC TAATGAAGAT GGTGACATTA AACGTATTTC 780
TACTAAGGAA TCGGCTAAAT CAACTGGATA TGATCCAGTT AAACTTTTTA CCAAGCTTTT 840
TAAAGATGAC ATCAGGTATC TGTTCACAAT GGACAAACTA TGGCGGAAAA GGAAACCTCC 900
AGTTCCGTTG GACTGGGCTG AAGTACAAAG TCAAGGAGAA GAAACGAATG CATCAGATCA 960 ACAGAATGAA CCCCAGTTAG GCCTGAAAGA CCAGCAGGTT CTAGATGTAA AGAGCTATGC 1020 ACGTCTTTTT TCAAAGAGCA TCGAGACTTT GAGAGTTCAT TTAGCAGAAA AGGGGGATGG 1080 AGCTGAGCTC ATATGGGATA AGGATGACCC ATCTGCAATG GATTTTGTCA CCTCTGCTGC 1140 AAACCTCAGG ATGCATATTT TCAGTATGAA TATGAAGAGT AGATTTGATA TCAAATCAAT 1200 GGCAGGGAAC ATTATTCCTG CTATTGCTAC TACTAATGCA GTAATTGCTG GGTTGATAGT 1260 ATTGGAAGGA TTGAAGATTT TATCAGGAAA AATAGACCAG TGCAGAACAA TTTTTTTGAA 1320 TAAACAACCA AACCCAAGAA AGAAGCTTCT TGTGCCTTGT GCACTGGATC CTGCCAACCC 1380 CAATTGTTAT GTATGTGCCA GCAAGCCAGA GGTGACTCTG CGGCTGAATG TCCATAAAGT 1440 GACTGTTCTC ACCTTACAAG ACAAGATAGT GAAAGAAAAA TTTGCTATGG TAGCACCAGA 1500 TGTCCAAATT GAAGATGGGA AAGGAACAAT CCTAATATCT TCCGAAGAGG GAGAGACGGA 1560 AGCTAATAAT CACAAGAAGT TGTCAGAATT TGGAATTAGA AATGGCAGCC GGCTTCAAGC 1620 AGATGACTTC CTCCAGGACT ATACTTTATT GATCAACATC CTTCATAGTG AAGACCTAGG 1680 AAAGGACGTT GAATTTGAAG TTGTTGGTGA TGCCCCGGAA AAAGTGGCGS CCAAACAAGC 1740 TGAAGATGCT GCCAAAAGCA TAACCAATGG GCAGTGATGA TGGGAGCTTC AGCCCTCCAC 1800 CTYCACAGCT TCAAGCAGGC AAGATGGACG TYTCYCATAG TTGATYCGGR TGAAGAAGRT 1860 TCTCCAATAA TTGCCCGACG TTCATTGAAG GAAGGAGGAG GAGGCCCGCC AAGAGGGGAA 1920 TTTAGCNTTG 1930
(2) INFORMATION FOR SEQ ID NO: 173:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1509 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO : 173 : GGCCCTGGCC TCTGGGCTGA GGCTTGCTAG GGACTCGGGG TGGCTCTAAG GGGCAGGGAT 60 AGGGCTGGGG AGCGCCGGCC TGTGGCCCTG ACCAGCCCCT TCTCGTGCRG GTTCCACCCC 120 GATGCAGGTG GTCACGTGCT TGACGCGGGA CAGCTACCTG ACGCACTGCT TCCTCCAGCA 180 CCTCATGGTC GTGCTGTCCT CTCTGGAACG CACGCCCTCG CCGGAGCCTG TTGACAAGGA 240 CTTCTACTCC GAGTTTGGGA ACAAGACCAC AGGGAAGATG GAGAACTACG AGCTGATCCA 300 CTCTAGTCGC GTCAAGTTTA CCTACCCCAG TCAGGAGGAG ATTGGCGACC TGACGTTCAC 360
TCTGGCCCAA AAGATGGCTG AGCCAGAGAA GGCCCCAGCC CTCAGCATCC TGCTGTACGT 420 GCAGGCCTTC CAGGTGGGCA TGCCACCCCC TGGGTGCTGC AGGGGCCCCC TGCGCCCCAA 480 GACACTCCTG CTCACCAGCT CCGAGATCTT CCTCCTGGAT GAGGACTGTG TCCACTACCC 540 ACTCCCCGAG TTTGCCAAAG AGCCGCCGCA GAGAGACAGG TACCGGCTGG ACGATGGCCG 600 CCGCGTCCGG GACCTGGACC GAGTGCTCAT GGGCTACCAG ACCTACCCGC AGCCCTCACC 660 CTCGTCTTCG ATGACGTGCA AGGTCATGAC CTCATGGGCA GTGTCACCCT GGACCACTTT 720 GGGGAGGTGC CAGGTCGCCC GGCTAGAGCC AGCCAGGGCC GTGAAGTCCA GTGGCAGGTG 780 TTTGTCCCCA GTGCTGAGAG CAGAGAGAAG CTCATCTCGC TCTTGGCTCG CCAGTGGGAG 840 GCCCTGTGTG GCCGTGAGCT GCCTGTCGAG CTCACCGGCT AGCCCAGGCC ACAGCCAGCC 900 TGTCGTGTCC AGCCTGACGC CTACTGGGCC AGGGCAGCAG GCTTTTGTGT TCTCTAAAAA 960 TGTTTTATCC TCCCTTTGGT ACCTTAATTT GACTGTCCTC GCAGAGAATG TGAACATGTG 1020 TGTGTGTTGT GTTAATTCTT TCTCATGTTG GGAGTGAGAA TGCCGGGCCC CTCAGGGCTG 1080 TCGGTGTGCT GTCAGCCTCC CACAGGTGGT ACAGCCGTGC ACACCAGTGT CGTGTCTGCT 1140 GTTGTGGGAC CGTTGTTAAC ACGTGACACT GTGGGTCTGA CTTTCTCTTC TACACGTCCT 1200 TTCCTGAAGT GTCGAGTCCA GTCCTTTGTT GCTCTTGCTG TTGCTGTTCC TGTTGCTGTT 1260 GGCATCTTGC TGCTAATCCT GAGGCTGGTA GCAGAATGCA CATTGGAAGC TCCCACCCCA 1320 TATTGTTCTT CAAAGTGGAG GTCTCCCCTG ATCCAGACAA GTGGGAGAGC CCGTGGGGGC 1380 AGGGGACCTC GAGCTGCCAG CACCAAGCGT GATTCCTGCT GCCTGTATTC TCTATTCCAA 1440 TAAAGCAGAG TTTGACACCG TGAAAAAAAA AAAAAAAAAA AAAAAAAAAA ATTNCTGCGG 1500 CCTCAAGGG 1509
(2) INFORMATION FOR SEQ ID NO: 174:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 3173 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 174: TCGACCCCAS GCGTCCGTGC TTTTCCACAG AAGGTTAGAC CCTGAAAGAG ATGGCTCAGC 60 ACCACCTATG GATCTTGCTC CTTTGCCTGC AAACCTGGCC GGAAGCAGCT GGAAAAGACT 120 CAGAAATCTT CACAGTGAAT GGGATTCTGG GAGAGTCAGT CACTTTCCCT GTAAATATCC 180
AAGAACCACG GCAAGTTAAA ATCATTGCTT GGACTTCTAA AACATCTGTT GCTTATGTAA 240
CACCAGGAGA CTCAGAAACA GCACCCGTAG TTACTGTGAC CCACAGAAAT TATTATGAAC 300
GGATACATGC CTTAGGTCCG AACTACAATC TGGTCATTAG CGATCTGAGG ATGGAAGACG 360
CAGGAGACTA CAAAGCAGAC ATAAATACAC AGGCTGATCC CTACACCACC ACCAAGCGCT 420
ACAACCTGCA AATCTATCGT CGGCTTGGGA AACCAAAAAT TACACAGAGT TTAATGGCAT 480
CTGTGAACAG CACCTGTAAT GTCACACTGA CATGCTCTGT AGAGAAAGAA GAAAAGAATG 540
TGACATACAA TTGGAGTCCC CTGGGAGAAG AGGGTAATGT CCTTCAAATC TTCCAGACTC 600
CTGAGGACCA AGAGCTGACT TACACGTGTA CAGCCCAGAA CCCTGTCAGC AACAATTCTG 660
ACTCCATCTC TGCCCGGCAG CTCTGTGCAG ACATCGCAAT GGGCTTCCGT ACTCACCACA 720
CCGGGTTGCT GAGCGTGCTG GCTATGTTCT TTCTGCTTGT TCTCATTCTG TCTTCAGTGT 780
TTTTGTTCCG TTTGTTCAAG AGAAGACAAG ATGCTGCCTC AAAGAAAACC ATATACACAT 840
ATATCATCGC TTCAAGGAAC ACCCAGCCAG CAGAGTCCAG AATCTATGAT GAAATCCTGC 900
AGTCCAAGGT GCTTCCCTCC AAGGAAGAGC CAGTGAACAC AGTTTATTCC GAAGTGCAGT 960
TTGCTGATAA GATGGGGAAA GCCAGCACAC AGGACAGTAA ACCTCCTGGG ACTTCAAGCT 1020
ATGAAATTGT GATCTAGGCT GCTGGGCTGA ATTCTCCCTC TGGAAACTGA GTTACAACCA 1080
CCAATACTGG CAGGTTCCCT GGATCCAGAT CTTCTCTGCC CAACTCTTAC TGGGAGATTG 1140
CAAACTGCCA CATCTCAGCC TGTAAGCAAA GCAGGAAACC TTCTGCTGGG CATAGCTTGT 1200
GCCTAAATGG ACAAATGGAT GCATACCCTT CCTGAAATGA CTCCCTTCTG AATGAATGAC 1260
AAAGCAGCTT ACCTAGTATA GTTTTCCCAA ACTTCTTCCC ATCATAGCAC ATGTAGAAAA 1320
TAATATTTTT ATGGCACACT GGGATAAACA AGCAAGATTG CTCACTTCTG GAAGCTGCAT 1380
ATGACTAGAG GCCTCTTGTG ACTGGAGGTA ACAACCCTGC CCAGTAACTG TGGGAGAAGG 1440
GGATCAATAT TTTGCACACC TGTAATAGGC CATGGCACAC CAGCCAAGAT GCTCTGCTCA 1500
CAGTCAGTAT GTCTGAAGAT CCCTGGTGCG TGGCCTTCAC CACGCATCTT GAGCAAATTA 1560
GGAAAATGTA CCCTTCGCTT GAGGCAGATG CAGCCCTTCC CCCGAGTGCA TGGCTTGGAG 1620
AGCAGAATGT GGGCTGCATA TAAGCACACT CATCCCTTTG TCTGGGAATC TTTGTGCAGG 1680
GCATAACAGG CTTAGTAAGT CCAAACACAG ATGACAGTGC TGTGTGGGTC TCTGTCAGAG 1740
TTGTGGCTCT CAGCCATGTA GACACACTCT CCAAATGGAG TGTTGGAAAA TGTTCTTTCT 1800
GCAGGGTCTA GAGACTGCTG GGACACTTTT CTTGGAGTGC TACTTCAGAA GCCTTATAGG 1860
ATTTTCTTTC TGGCCAAGAT TTCCTTCTGT ATCACTCCAA GCAGCCTCAG CAGAAGAAGC 1920
AGCCATGCCC AGTATTCCCA CTCTCCAAAA GGAACTGACC AGCTTATATT TCTCACACTT 1980
CTGGGGAACT GGGTATAATC CAACCATCAA AATAGAAGAC CTTGCAAGAA GCAGAGTCAT 2040
TCTCCAGAAG GAACTTGGGA GATGATGGTG CAGATGATGA AACTGGGTTC ATCCCAGTTC 2100
CAAAGACTCA GAGAACTAGA GTTTAAGCTG AGGCAGAGTG CCGCCACCCT GGCATGCCCC 2160
ACAAACAGAT CACCAGCCAG CTTACACAGG CATTAACTCT CCTCAATGAG GAAGAATCAT 2220
TCACAACTGA GCAAGACATT CATATGATCA TTTAAGGAAG TGTTTCCCTT ATGTGTTAGC 2280
AAGTATAATC GGCTAACTCC TAAATCCCAA TGAATAGTCC TAGGCTGGAC AGCAATGGGC 2340
TGCAATTAGG CAGATAAAGA CATCAGTCCC AGTAAATGAA TCCATAGACT CATCTAGCAC 2400
CAACTACCAT TAGCACTATG TTAGGAGCTG CAAGGCCCCA AAGTAGAAGA TCTGGATAAT 2460
GTCTGCTCTT GTGTAGCTCA GGAGACAATT CCAGCACAGA CACTACAGTT AACGCTGAAC 2520
TGCAGCTGCA AGTAATAGCA TGAACAGTCA GAAAAATACC TTATGAGGGG GCAGGGCTGA 2580
AGCTGGGCCT TGAAGGATGG ATGAAATTTG GATAGAGAAT GAGGAAGACA GAGGGCCTCC 2640
AAGTGAGAGA AGCATGAAAA ATGAGCAGGG GCCTGGATCA GTGGGGTGTA TTCAGAGCAC 2700
CTCTCCAGAT GCACCATGCA TGCTCACAGT CCCTTGCCTA TGTGTGGCAG AGTGTCCCAG 2760
CCAGATGTGT GCCCCCACCC CATGTCCATT TACATGTCCT TCAATGCCCA CCTCAAAAGG 2820
TACCTCTTCT GTAAAGCTTT CCCTGGTATC AGGAATCAAA ATTAATCAGG GATCTTTTCA 2880
CACTGCTGTT TTTTCCTCTT TGCTCCTTCT ATCACTAAAA CTCATCTCAT TCAGCCTTAC 2940
AGCATAACTA ATTATTTGTT TTCCTCACTA CATTGTACAT GTGGGAATTA CAGATAAACG 3000
GAAGCCKGCT GGGGTGGTGG CTCACGCCTG TAATCCCAAC ACTTTGGGAG GCCAAGGCAG 3060
GCGGATCACC TGAGGTCAGG ARTTCGAGAT TARTCTGCCC AACATGGTGA AACCCCATNT 3120
NTACTAAAAA TACGAAATTA GCCAGGTGTG GTGGCACACA TCTGTAGTCC CAG 3173
(2) INFORMATION FOR SEQ ID NO: 175:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 991 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 175: AAATTCGCCA CAGCTGAGAG GAGACACAAG GAGCAGCCCG CAAGCACCAA GTGAGAGGCA 60 TCAAGTTACA GTGTGTTTCC CTTTGGCTCC TGGGTACAAT ACTGATATTG TGCTCAGTAG 120 ACAACCACGG TCTCAGCAGA TGTCTGATTT CCACAGACAT GCACCATATA GAAGAGAGTT 180 TCCAAGAAAT CAAAAGAGCC ATCCAAGCTA AGGACACCTT CCCAAATGTC ACTATCCTGT 240
CCACATTGGA GACTCTGCAG ATCATTAAGC CCTTAGATGT GTGCTGCGTG ACCAAGAACC 300
TCCTGGCGTT CTACGTGGAC AGGGTGTTCA AGCATCATCA GGAGCCAAAC CCCAAAATCT 360
TGAGAAAAAT CAGCAGCATT GCCAACTCTT TCCTCTACAT GCAGAAAACT CTGCGGCAAT 420
GTCAGGAACA GAGGCAGTGT CACTGCAGGC AGGAAGCCAC CAATGCCACC AGAGTCATCC 480 ATGACAACTA TGATCAGCTG GAGGTCCACG CTGCTGCCAT TAAATCCCTG GGAGAGCTCG 540
ACGTCTTTCT AGCCTGCATT AATAAGAATC ATGAAGTAAT GTCCTCAGCT TGATGACAAG 600
GAACCTGTAT AGTGATCCAG GGATGAACAC CCCCTGTGCG GTTTACTGTG GGAGACAGCC 660
CACCTTGAAG GGGAAGGAGA TGGGGAAGGC CCCTTGCAGC TGAAAGTCCC ACTGGCTGGC 720
CTCAGGCTGT CTTATTCCGC TTGAAAATAG CCAAAAAGTC TACTGTGGTA TTTGTAATAA 780 ACTCTATCTG CTGAAAGGGC CTGCAGGCCA TCCTGGGAGT AAAGGGCTGC CTTCCCATCT 840
AATTTATTGT GAAGTCATAT ACTCCATCTC TGTGATGTGA GCCAAGTGAT ATCCTGTAGT 900
ACACATTGTA CTGAGTGGTT TTTCTGAATA AATTCCATAT TTTACCTAAA AAAAAAAAAA 960
AAAAACTCGA GGGGGGGCCC GTACCCAATT T 991
(2) INFORMATION FOR SEQ ID NO: 176:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1290 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 176:
ACAGCCCTCT TCGGAGCCTG AGCCCGGCTC TCCTCACTCA CCTCAACCCC CAGGCGGCCC 60
CTCCACAGGG CCCCTCTCCT GCCTGGACGG CTCTGCTGGT CTCCCCGTCC CCTGCAGAAG 120 AACAAGGCCA TGGGTCGGCC CCTGCTGCTG CCCCTRCTGC YCCTGCTGCW GCCGCCAGCA 180
TTTCTGCAGC CTRGTGGCTC CACAGGATCT GGTCCAAGCT ACCTTTATGG GGTCACTCAA 240
CCAAAACACC TCTCAGCCTC CATGGGTGGC TCTGTCGAAA TCCCCTTCTC CTTCTATTAC 300
CCCTGGGAGT TAGCCAYAGY TCCCRACGTG AGAATATCCT GGAGACGGGG CCACTTCCAC 360
GCGCAGTCCT TCTACAGCAC AAGGCCGCCT TCCATTCACA AGGATTATGT GAACCGGCTC 420 TTTCTGAACT GGACAGAGGG TCAGGAGAGC GGCTTCCTCA GGATCTCAAA CCTGCGGAAG 480
GAGGACCAGT CTGTGTATTT CTGCCGAGTC GAGCTGGACA CCCGGAGATC AGGGAGGCAG 540
CAGTTGCAGT CCATCAAGGG GACCAAACTC ACCATCACCC AGGCTGTCAC AACCACCACC 600
ACCTGGAGGC CCAGCAGCAC AACCACCATA GCCGGCCTCA GGGTCACAGA AAGCAAAGGC 660 CACTCAGAAT CATCGCACCT AAGTCTGGAC ACTGCCATCA GGGTTGCATT GGCTCTCGCT 720 GTGCTCAAAA CTGTCATTTT GGGACTGCTG TGCCTCCTCC TCTGTGGTGG AGGAGAAGGA 780 AAGGTAGCAG GGCGCCAAGC AGTGACTTCT GACCAACAGA GTGTGGGGAG AAGGGATGTG 840 TATTAGCCCC GGAGCACGTC ATGTGAGACC CGCTTCTGAG TCCTCCACAC TCGTTCCCCA 900 TTGGCAAGAT ACATGGAGAG CACCCTGAGG ACCTTTAAAA GGCAAAGCCG CAAGGCAGAA 960 GGAGGCTGGG TCCCTGAATC ACCGACTGGA GGAGAGTTAC CTACAAGAGC CTTCATCCAG 1020 GAGCATCCAC ACTGCAATGA TATAGGAATG AGGTCTGAAC TCCACTGAAT TAAACCACTG 1080 GCATTTGGGC GCTGTTYATT ATAGCAGTGC AAAGAGTTCC TTTATCCTCC CCAAGGATGG 1140 AAAATACAAT TTATTTTGCT TACCATACAC CCCTTTTCTC CTCGTCCACA TTTTCCAATC 1200 TGTATGGTGG CTGTCTTCTA TGGCAGAAGG TTTTGGGGAA TAAATAGCGT GANATGNTNC 1260 TGACTNAAAA AAAAAAAAAA AAAAACTCGA 1290
(2) INFORMATION FOR SEQ ID NO: 177:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2290 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi ) SEQUENCE DESCRIPTION : SEQ ID NO : : 177: TGGGGCCCCT TTTGCATGCT CTGCGTGTTT TTGCCAAGAG TTACAGGATG TCAAGTGTGG 60 GGAGCTCAGC ACCCTTGCTG TGGACCAGTG AAGGCTGTTC CAGACCAGGT GCTTCCAGAC 120 ATTTCCAGGC TCCAGGAGAG AGGCTGGGAG CCCCCACAGA AAGCACAGGA AAATGCAAAA 180 AAAAAACAGT CTTTTTTTTT TTTTTGCTTT TTATTATGAA AACAAAACAA ATGCCCCAGC 240 AGAAGGGTCC ATGATTACCA GAAACATCAA AGAGTACTTT CTACCATTTT TATTCTGTTG 300 TGTTGAGGCC AGCATTGCAA TAAACAAGCT AAACTACTTA CATTGGACTC ATTTTCAGTA 360 ACTGACATTT ACAGGAATAT ACTAGAAACG GCACTAAAAA GTTTAAGAAA AGTTACGGTA 420 AACTTCCATC CACATCATAC AGAAAAGTAA CATTTTAAAT ATAAAAAAGA AAAACTTCCT 480 GGAAGCATTA TCCCAGTATT AAGGAACAGT GCTACTCTGC ATGTGACAAA TTCTGTATGT 540 GGGTGTTACT CTTTCCCAAA AGACTGTCAG AGGCGTGAGT GCTGCAAAAG AACAACAACA 600 AAAACAAACA CACAAAAAAA TGTGTCTTAC AGTTTGTAAG CAAGATGACA CTGCCCAACA 660 CAAAGAGGGG TCTGCAGTTC AGTTCACGCC CGAAGCCTGC CCCCTCGGCC TCCAGGGGTC 720
ATTCAGAGTC TTCTCAAATC CAATTCCGAC ACACGACTTC TCACTACTCC TCTCCCCTTG 780
AAAAAAGCAT GTTAGAAGCT GCCCTACAGG TCTCAGCAGT GGGACAATCT AATTGAATCA 840
CCGCAGCCTT CTAATACAGA AGAAACGGAC GTGACTGTCA CCCTCAGCCC GCCAGCAAGG 900
GCGCTGAGGA AGTCATTAAT CCTTCGAAAC TCTGAAAAGA AACCAGTGTT GAAGTCTGGA 960
CAGAAAGCCT TAAAAAAGTG ACAGCACCAA TGCAGCTGCT CAGTGTACCC NCCGTGGGCT 1020
GTCAGGGTCA GTGGCTTCTT TCTAGATGAA AGGAGCAGAG GCGAGCCGAC GCCACCGTCA 1080
CAGAGAACCA GCCGAGAAGG AAAGGCCCCA CGATGCTCCC TGTGCGCTGC CCCCACAGCC 1140
GGCCGCTCCC CCGACGGCTC ACACAGGCAG CACCTCACTG CCCTGTGGCT GGAGGGGCAT 1200
TGCAAGGAGC GCCCCCCAGC CCCAGGCACC CCCGGCTTAG GGTGTACGTA TCACCCAGCC 1260
CTGTGCTGGC AGCACGTTAC CAACCAGCCT GCGTGAAGAC CTGTCAACTG TCGTGTGTGA 1320
ATTCCTTAAA TTCGGTTTAA ATAGTCCATT AAAGATCTGT TTAGAAAATA CCTTTGAAAA 1380
CGAGGGTAAC TTTAAAAAAT GCAAACTTTC AAATCCATTT ATATTTTTAT TATAAACAAA 1440
ACTTAATTAA AAGTTTAACA AACTGGCTGA AAACTCACCA AGTGTCAGAC TCACCAGCAA 1500
TTTAAAAAAT GATAATTTAC CAGCATCTCC TCATCAGAGT TCCCTCTCCA GTAAGGGTAT 1560
ACCTACATCT GTAAGGGTCA GTGGACTCTG AATCAATTTT ATGGTTGTTT TAAAATCACC 1620
GTGTATTAGG ATACTAATGA TAGTCCCTAT ATCCATCCAG AAATGCTGGC AGAAAGCACT 1680
GGCCACCATA CAGGACAGAC CACACCACAG CTCCATACCC AGCGTCTGCC TGGAGGCTCC 1740
CCCACGCTGA GGTCCGGGAG AATGCCTGGT TTCAGTCATT TCCGGACTAA CTGTGACAAC 1800
GCGTGAGCAG GGAGCACCGT GCGAGTCTCC GGGAGGGAAT CCTCCTGCGG CCCAGAGACT 1860
CCTCCACCCC TGGGGAGGGC AGACAGCCTC GGGARGGCCT GGCCAGGCCA CTGGAGGCTG 1920
GCAGGGAGCA GGCATGTCCA CCCGCAAGCC TGGGAGGCTA ACTCTGGCAT TCCTGGCCGG 1980
AGCCGCCATG CTCATTGGTG GGCCAGTTTG GGACATCCCC GTACTCAAAG ACCATATGGC 2040
AGCCTCTGCG AAAACAAAAC CAAAACATCA CCTTCTATTA AACTCTGTAT ATTATTATTT 2100
TTTACAATAG AAAGTTAAAA ATCAAGACTT AGATTTACTA TACATTTTTT CTCTCAGATT 2160
ACAAAGTTTA TATTATATAA CTGGGGTTCC CTAAATTGAT TTCTTTTAAA ACAGTCTTAA 2220
AGAGACCAGA AGTGAATACA AAAGAACTAA ACAAAATAAA AAATTAGAAT GTGCTGTAGC 2280
TGAAAGCTGT 2290
(2) INFORMATION FOR SEQ ID NO: 178:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 549 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 178:
GGCACGAGCC ATGCCTGGCC TCTCCTTGAT TCTTACAGTC ACTTTGTTGG CTGTTTCTGA 60
CTCAGCAGCT ACCTGCATTG TGGCCAAAGG ATGACCTATT CCTTCTCAGG AGGGCAAAAA 120
TGTGGAATAG TGTCTGTCCA TGCCTCTCCT CATGGGCTAC CACCTCTGCC ACCGTGGTTA 180 ATCAGTAACA ACCAGGAGAG AAGCTGCTGG AACTGACCTC TGGGAACTCC CTGCGATGCT 240
TTGGTGCAGG AATGTAGTAG GCATACACGT GGTTGCGTGG ATCTGGGCCC TCCTGATGTG 300
AGTAGAGAGG TAAAAGGCCA CCATCTCCTT GACCTCTGGG GAACTCATCC ACAAAGAAGA 360
TGTTTCCAAG ATGCTTCTGA AGATTGCCTA AAAATAGCCG GTTTCCACCC CCGTGAATGC 420
ATCCATTCTA GAATGCTCCT TCACCAGGAC CAGAGAACTG ATTTACAGAA GTGACATGAA 480 AACATTCCAT CCCAGAATTT GCAGTAGCTC AAATTAAGTT TCTAGCTATT AAAAAGAAAA 540
AAAAAAAAA 549
(2) INFORMATION FOR SEQ ID NO: 179:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 1509 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 179:
GGCACGAGGG CTCATTCATT CCGCGCCGGG CCTCCCAGAC ACCTGCGCCC TTCTGCAGCC 60 GCCCGCCGCA TCCGCCGCCG CAGCCCCCAG CATGTCGGGC CCAGACGTCG AGACGCCGTC 120
CGCCATCCAG ATCTGCCGGA TCATGCGCCC AGATGATGCC AACGTGGCCG GCAATGTCCA 180
CGGGCGGACC ATCCTGAAGA TGATCGAGGA GGCAGGCGCC ATCATCAGCA CCCGGCATTG 240 CAACAGCCAG AACGGGGAGC GCTGTGTCGC CGCCCTGGCT CGTGTCGAGC GCACCGACTT 300
CCTGTCTCCC ATGTGCATCG GTGAGCTGGC GCATGTCAGC GCGGAGATCA CCTACACCTC 360
CAAGCACTCT GTGGAGGTGC AGGTCAACGT GATGTCCGAA AACATCCTCA CAGGTGCCAA 420
AAAGCTGACC AATAAGGCCA CCCTGTGGTA TGTGCCCCTG TCGCTGAAGA ATGTGGACAA 480
GGTCCTCGAG GTGCCTCCTG TTGTGTATTC CCGGCANGAG CAGGAGGAGG AGGGCCGGAA 540 GCGGTATGAA GCCCAGAAGC TGGAGCGCAT GGAGACCAAG TGGAGGAACG GGGACATCGT 600
CCAGCCAGTC CTCAACCCAG AGCCGAACAC TGTCAGCTAC AGCCAGTCCA GCTTGATCCA 660
CCTGGTGGGG CCTTCAGACT GCACCCTGCA CC^GCTTTGTG CACGGAGGTG TGACCATGAA 720
GCTCATGGAT GAGGTCGCCG GGATCGTGGC TGCACGCCAC TGCAAGACCA ACATCGTCAC 780
AGCTTCCGTG GACGCCATTA ATTTTCATGA CAAGATCAGA AAAGGCTGCG TCATCACCAT 840
CTCGGGACGC ATGACCTTCA CGAGCAATAA GTCCATGGAG ATCGAGGTGT TGGTGGACGC 900
CGACCCTGTT GTGGACAGCT CTCAGAAGCG CTACCGGGCC GCCAGTGCCT TCTTCACCTA 960
CGTGTCGCTG AGCCAGGAAG GCAGGTCGCT GCCTGTGCCC CAGCTGGTGC CCGAGACCGA 1020
GGACGAGAAG AAGCGCTTTG AGGAAGGCAA AGGGCGGTAC CTGCAGATGA AGGCGAAGCR 1080
ACAGGGCCAC GCGGASCYTC AGCCCTAGAC TCCCTCCTCC TGCCACTGCT GCCTCGAGTA 1140
GCCATGGCAA CGGGCCCAGT GTCCAGTCAC TTAGAAGTTC CCCCCTTGGC CAAAAACCCA 1200
ATTCACATTG AGAGCTGGTG TTGTCTGAAG TTTTCGTATC ACAGTGTTAA CCTGTACTCT 1260
CTCCTGCAAA CCTACACACC AAAGCTTTAT TTATATCATT CCAGTATCAA TGCTACACAG 1320
TGTTGTCCCG AGCGCCGGGA GGCGTTGGGC AGAAACCCTC GGCAATGCTT CCGAGCACGC 1380
TGTAGGGTAT GGGAAGAACC CAGCACCACT AATAAAGCTG CTGCTTGGCT GGAAAAAAAA 1440
AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA 1500
AGAAAAAAN 1509
(2) INFORMATION FOR SEQ ID NO: 180:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1316 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO 180: AGCTGTATCA TAGGAAAGAT GGCCACACCG GCGGTACCAG TAAGTCCTCC TCCGGCCACG 60 CCAACCCCAG TCCCGGCGGC GGCCCCAGCC TCAGTTCCAG CGCCAACGCC AGCACCGGCT 120 GCGGCTCCGG TTCCCGCTGC GGCTCCAGCC TGCATCCTCA GACCCTGCGG CAGCAGCGGC 180 TGCAACTGCG GCTCCTGGCC AGACCCCGGC CTCAGCGCAA NTCCAGCGCA GACCCCAGCG 240 CCCGCTCTGC CTGGTCCTGC TCTTCCAGGG CCCTTCCCCG GCGGCCGCGT GGTCAGGCTG 300 CACCCAGTCA TTTTGGCCTC CATTGTGGAC AGCTACGAGA GACGCAACGA GGGTGCTGCC 360 CGAGTTATCG GGACCCTGTT GGGAACTGTC GACAAACACT CAGTGGAGCT CACCAATTGC 420
TTTTCAGTGC CGCACAATGA GTCAGAAGAT GAAGTGGCTG TTGACATGGA ATTTGCTAAG 480
AATATGTATG AACTGCATAA AAAAGTTTCT CCAAATGAGC TCATCCTGGG CTGGTACGCT 540
ACGGGCCATG ACATCACAGA GCACTCTGTG CTGNATCCAT GAGTACTACA GCCGAGAGGC 600
CCCCAACCCC ATCCACCTCA CTGTGGACAC AAGTCTCCAG AACGGCCGCA TGAGCATCAA 660
AGCCTACGTC AGCACTTTAA TGGGAGTCCC TGGGAGGACC ATGGGAGTGA TGTTCACGCC 720
TCTGACAGTG AAATACGCGT ACTACGACAC TGAACGCATC GGAGTTGACC TGATCATGAA 780
GACCTGCTTT AGCCCCAACA GAGTGATTGG ACTCTCAAGT GACTTGCAGC AAGTAGGAGG 840
GGCATCAGCT CGCATCCAGG ATGCCCTGAG TACAGTGTTG CAATATGCAG AGGATGTACT 900
GTCTGGAAAG GTGTCAGCTG ACAATACTGT GGGCCGCTTC CTGATGAGCC TGGTTAACCA 960
AGTACCGAAA ATAGTTCCCG ATGACTTTGA GACCATGCTC AACAGCAACA TCAATGACCT 1020
TTTGATGGTG ACCTACCTGG CCAACCTCAC ACAGTCACAG ATTGCACTCA ATGAAAAACT 1080
TGTAAACCTG TGAATGGACC CCAAGCAGTA CACTTGCTGC TCTAGGTATT AACCCCAGGA 1140
CTCAGAAGTG AAGGAGAAAT GGGTTTTTTG TGGTCTTGAG TCACACTGAG ATAGTCAGTT 1200
GTGTGTGACT CTAATAAACG GAGCCTACCT TTTGTAAATT AAAAAAAAAA AAAAAAACCN 1260
SGRGGGGGGG CCCGGTCCCA TTSSCCCTTT NGTAATTCGT NTTACAATCC CCNGGC 1316
(2) INFORMATION FOR SEQ ID NO: 181:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 777 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 181: GGCATGWKCA GACATGACTT CTATTGCCAG GCTGGTCAAG TGGCAGGGTC ATGAGGGAGA 60 CATCGATAAG GGTGCTCCTT ATGCTCCCTG CTCTGGAATC CACCAGCGGG CTATCTGCGT 120 TTATGGGGCT GGGGACTAGA ATTGGATGCT TCAAAACCAT CACCTGTTGG CCAACAAGTT 180 TGACCCAAAG GTAGATGATA ATGCTCTTCA GTGCTTAGAA GAATACCTAC GTTATAAGGG 240 CCATTCTATT GGGACCTGAA CTTTGAAGAC CACAMTATTG AAGAGGCGTT GCTTACCYGT 300 TGGGGCCCAA GAGGCATGTT ACCAAACATG GYYCARGAAM YTTGGYKGGG AMCARKKKKG 360 GKKGGGARRM CMRGCGYTTC SCAAWTTCSK KGGCMWCCYT TTAGGCTAAR RRGGGCKGTW 420 ATTAGATTGT GGGTAAAGTA GGATCTTTTG CCCTTGCAAA TTTGCTGCCT GGGTGAATGY 480 TGCTTGTTCC TTCTCMACCC CTAACCCTAG TAGTTCCTCC ACTAACTTTC TCACTAAGTG 540
AGAATGAGAA CTGCTGTGAT AGGGAGAGTG AAGGAGGGAT ATGTGGTAGA GCACTTGATT 600
TCAGTTGAAT GCCTGCTGGT AGCTTTTCCA TTCTGTGGAG CTGCCGTTCC TAATAATTCC 660
5
AGGTTTGGTA GCGTGGAGGA GAACTTTGAT GGAAAGAGAA CCTTCCCTTC TGTACTGTTA 720
ACTTAAAAAT AAATAGCTCC TGATTCAAAG TAAAAAAAAA AAAAAAAAAA AAAAAAA 777
10
(2) INFORMATION FOR SEQ ID NO: 182:
15 (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 791 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
20
(xi ) SEQUENCE DESCRIPTION : SEQ ID NO : 182 :
GGCACAGATA ACTATGTACA TGTATTCCTT AAATGTTTTT TTAAGTTTTA TATTCTTGGC 60
25 ACTGGTCTTC AAATGTGTAC ATGTGTGCCA GGGAGCAAAT GCCTTCTTGT TTCTGAAATT 120
GGTCTTTTAG ACTGTTCTTT TTTCCCATCT TCTCACCTCC TGCCCCTCCT TCAGGGTACT 180
TCCGTGGCCA GAACCCCTCC AGGTCAGAGG CAGAAGAGAA GCCTCATGGG TCACAGCAGC 240
30
AGATGTGGGC TGGAGATCTA TTCATTTGGT TTTGGCTTGA ATTTTCTGRA TGGTTTACTT 300
GATCYTGGGA AAGANATATC TTGCCAGGAA AAATGATAGN CCTTGACAAT GTTGAATGAT 360
35 CCTGCACCAC CTTGAAAGAC ATTTCTAATA TGGTTTGTCA GGCAAAGTGC TTAGTAGTCA 420
TTTGTGGCCT GAGGTAGAAG TCCTCAGAAA TCAGCAGACT TCACTGATAA AATGCTGACT 480
TGCCCCTGGA CTGCGCTCTG TGAGAGTGCC CTTCTGCACT GTGCACAGTA GGTGTGAACA 540 0
CACCACACCT ACAGGGACCA CGTGGTGGGC TGTGGACTAG CGGCCAAGCT CCCTGCAGGC 600
CCACTAATAG AATTCAGCTT TTAGCATGGG CTGTTTCATA CTGTTCTGAT GAAACTGATT 660
45 TGGTTTCTTT CCTCCATACC CCTTCTGCAT TTCAGTGTTT TTGTTTAGTT TTCCTGGTTT 720
TTAATTATAA CTACAAAATA AAATCTTTAG GCTATTCACC TTAGCTTAGT AAAAAAAAAA 780
AAAAAAAACT C 791
DU
(2) INFORMATION FOR SEQ ID NO: 183:
55
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1405 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double 60 (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 183:
AAATTGATTA ACAGCTTGAA AGAAGGCTCT GCTTTTGAAG GCCTAGATAG CAGCACTGCC 60
AGTAGCATGG AGCTGGAAGA ACTTCGGCAT GAGAAAGAGA TGCAGAGGGA GGAAATACAG 120
AAGCTGATGC GCCAGATACA TCAGCTCAGA TCCGAATTAC AGGATATGGA GGCACAGCAA 180
GTTAATGAAG CAGAATCAGC AAGAGAACAG TTACAGGWTC TGCATGACCA AATAGCTGGG 240
CAGAAAGCAT CCAAACAAGA ACTAGAGACA GAACTGGAGC GACTGAAGCA GGAGTTCCAC 300
TATATAGAAG AAGATCTTTA TCGAACAAAG AACACATTGC AAAGCAGAAT TAAAGATCGA 360
GACGAAGAAA TTCAAAAACT CAGGAATCAG CTTACCAATA AAACTTTAAG CAATAGCAGT 420
CAGTCTGAGT TAGAAAATCG ACTCCATCAG CTAACAGAGA CTCTCATCCA GAAACAGACC 480
ATGCTGGAGA GTCTCAGCAC AGAAAAGAAC TCCCTGGTCT TTCAACTGGA GCGCCTCGAA 540
CAGCAGATGA ACTCCGCCTC TGGAAGTAGT AGTAATGGGT CTTCGATTAA TATGTCTGGA 600
ATTGACAATG GTGAAGCCAC TCGTCTGCGA AATGTTCCTG TTCTTTTTAA TGACACAGAA 660
ACTAATCTGG CAGGAATGTA CGGAAAAGTT CGCAAAGCTG CTAGTTCAAT TGATCAGTTT 720
AGTATTCGCC TGGGAATTTT TCTCCGAAGA TACCCCATAG CGCGAGTTTT TGTAATTATA 780
TATATGGCTT TGCTTCACCT CTGGGTCATG ATTGTTCTGT TGACTTACAC ACCAGAAATG 840
CACCACGACC AACCATATGG CAAATCAACC AAGCCCAGTT GTTGCAGTGA TTXX5TTGTCT 900
TTTTCTAGAC TTGGGATCTG CAAGAAGGCC AATTGCCTAA AATTTCTGAG AACAGTGCAC 960
AAGATTATTT TATCACTACA AGCTTTTAAC TTTTTAAGTT ATTGTACAAG TATTCTACCT 1020
AAATCTTCCA ATTTCCTTTA AATCGTAAGA GTTTCTAAAA CAGACAATAA TTTAACAAGC 1080
TCAGCTCTGC TTTATCTGAG TTTAGTGGTC CTAATATATA TGTAGAGAAA GATGGTGGGG 1140
TTGTTCACCT CTGTACAGAC CATCTCTATG TTAGGTGACA TTGATTATGG GTTATAATCA 1200
GGCAAACTAA TTGTATTTAG TGACAAAAAT AAAAAGTTTT TTTTTTATAA TTCAGTCTGC 1260
TTTTGGATTT TCATATATTT AACTTTGCAA AAAGATTTAC TTTGTACATG TTACAGGCTT 1320
GATTGGTGTA AATCTTTTTA TAAATACATA AATAAAAGNA AAATATGCAT TTTTCTTTTC 1380
TAAAAAAAAA AAAAAAAAAA CTCGA 1405
(2) INFORMATION FOR SEQ ID NO: 184:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1596 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 184:
GTCATGCAGT GCGCCGCAGA ACTGTGCTCT TTGAGGCCGA CGCTAGGGGC CCGGAAGGGA 60
AACTGCGAGG CGAAGGTGAC CGGGGACCGA GCATTTCAGA TCTGCTCGGT AGACCTGGTG 120
CACCACCACC ATGTTGCCTG CAAGGCTGGT GTGTCTCCGG ACACTACCTT CTAGGGTTTT 180
CCACCCAGCT TTCACCAAGG CCTCCCCTGT TGTGAAGAAT TCCATCACGA AGAATCAATG 240
GCTGTTAACA CCTAGCAGGG AATATGCCAC CAAAACAAGA ATTGGGATCC GGCGTGGGAG 300
AACTGGCCAA GAACTCAAAG AGGCAGCATT GGAACCATCG ATGGAAAAAA TATTTAAAAT 360
TGATCAGATG GCAAGATGGT TTGTTGCTGG AGCGCCTGCT GTTGGTCTTG GAGCATTGTG 420
CTACTATGGC TTGGGACTGT CTAATGAGAT TGGAGCTATT GAAAAGGCTG TAATTTGGCC 480
TCAGTATGTC AAGGATAGAA TTCATTCCAC CTATATGTAC TTAGCAGGGA GTATTGGTTT 540
AACAGCTTTG TCTGCCATAG CAATCAGCAG AACGCCTGTT CTCATGAACT TCATGATGAG 600
AGGCTCTTGG GTGACAATTG GTGTGACCTT TGCAGCCATG GTTGGAGCTG GAATGCTGGT 660
ACGATCAATA CCATATGACC AGAGCCCAGG CCCAAAGCAT CTTGCTTGGT TGCTACATTC 720
TGGTGTGATG GGTGCAGTGG TGGCTCCTCT GACAATATTA GGGGGTCCTC TTCTCATCAG 780
AGCTGCATGG TACACAGCTG GCATTGTGGG AGGCCTCTCC ACTGTGGCCA TGTGTGCGCC 840
CAGTGAAAAG TTTCTGAACA TGGGTGCACC CCTGGGAGTG GGCCTGGGTC TCGTCTTTGT 900
GTCCTCATTG GGATCTATGT TTCTTCCACC TACCACCGTG GCTGGTGCCA CTCTTTACTC 960
AGTGGCAATG TACGGTGGAT TAGTTCTTTT CAGCATGTTC CTTCTGTATG ATACCCAGAA 1020
AGTAATCAAG CGTGCAGAAG TATCACCAAT GTATGGAGTT CAAAAATATG ATCCCATTAA 1080
CTCGATGCTG AGTATCTACA TGGATACATT AAATATATTT ATGCGAGTTG CAACTATGCT 1140
GGCAACTGGA GGCAACAGAA AGAAATGAAG TGACTCAGCT TCTGGCTTCT CTGCTACATC 1200
AAATATCTTG TTTAATGGGG CAGATATGCA TTAAATAGTT TGTACAAGCA GCTTTCGTTG 1260
AAGTTTAGAA GATAAGAAAC ATGTCATCAT ATTTAAATGT TCCGGTAATG TGATGCCTCA 1320
GGTCTGCCTT TTTTTCTGGA GAATAAATGC AGTAATCCTC TCCCAAATAA GCACACACAT 1380
TTTCAATTCT CATGTTTGAG TGATTTTAAA ATGTTTTGGT GAATGTGAAA ACTAAAGTTT 1440
GTCTCATGAG AATGTAAGTC TTTTTTCTAC TTTAAAATTT AGTAGGTTCA CTGAGTAACT 1500
AAAATTTAGC AAACCTGTGT TTGCATATTT TTTKGGAGTG CAGMMTAWTG TAATTARAGC 1560
ATTCCAGTAA NAGTGTNTTT AAAGTTGNTC TATATN 1596
(2) INFORMATION FOR SEQ ID NO: 185:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 2293 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 185:
GCGCAGAGCC CGYACGAGCA GGACGACGAC GACAAGGGCG ACTCCAAGGA AACGCGGCTC 60
ACCCTGATGG AGGAAGTGCT CCTGCTGGGC CTCAAGGACC GCGARGGTTA CACATCATTT 120
TGGAATGACT GTATATCATC TGGATTACGT GGCTGTATGT TAATTGAATT AGCATTGAGA 180
GGAAGGTTAC AACTAGAGGC TTGTGGAATG AGACGTAAAA GTCTATTAAC AAGAAAGGTA 240 ATCTGTAAGT CAGATGCTCC AACAGGGGAT GTTCTTCTTG ATGAAGCTCT GAAGCATGTT 300
AAGGAAACTC AGCCTCCAGA AACGGTCCAG AACTGGATTG AATTACTTAG TGGTGAGACA 360
TGGAATCCAT TAAAATTGCA TTATCAGTTA AGAAATGTAC GGGAACGATT AGCTAAAAAC 420
CTGGTGGAAA AGGGTGTATT GACAACAGAG AAACAGAACT TCCTACTTTT TGACATGACA 480
ACACATCCCC TCACCAATAA CAACATTAAG CAGCGCCTCA TCAAGAAAGT ACAGGAAGCC 540 GTTCTTGACA AATGGGTGAA TGACCCTCAC CGCATGGACA GGCGCTTGCT GGCCCTCATT 600
TACCTGGCTC ATGCCTCGGA CGTCCTGCAG AATGCTTTTG CTCCTCTTCT GGACGAGCAG 660
TATGATTTGG CTACCAAGAG AGTGCGGCAG CTTCTCGACT TAGACCCTGA AGTGGAATGT 720
CTGAAGGCCA ACACCAATGA GGTTCTGTGG GCGGTGGTGG CGGCGTTCAC CAAGTAACTC 780
TGCTCGGCGT GAACCATTCT CCTTTCTCTC AAGTAAACCA GTAGTTTTTC TTCTGTTGAC 840 TTCTGGTTTT CTGTAATTTG TACTTTCCCA CACTATAATT GGCTTCTGTT TTACAAAATG 900
GTGCGTGGCT TTTTCTTTTT TGTACGTGTA CAGGATTCTG CTGGTACGAG AGGCCTTCCT 960
CTTTCTGTTT TTAAAAAAAG TTTTACTGCC ATATTGGCAT TCCATTCCCT GTTGCCATCC 1020
TCACTGTTAC CTGTTTTGGG TTTCTGGTCT ACTTTGACTT TCAAAGTACC TCCAGCCTCC 1080
TCATACGCAC AGCTTTTGGA TGACCTCAGC TTGAGTTTCT CCATATGTGC ATGTACATCT 1140 AGCATTCTGC CTACAGTTCA GACAGAAGTC ACAAAAAGGC CTTCAACTCA CCAAAGGTAA 1200
ATATCTGTAT CTATTAGGAC ATTTTTTACA TAGACTTCAG TTGAGATGTA TACTTAGCAA 1260
AATTATTTTT AAATTGAAAC AGCACAGTAA ATACTTAATA TAAAATGTCC CTTGGATTTT 1320
GCTTCCCATG TAAATCTATT GTATTATTAC ACTTGTTATA ATTTTAACTA TAAAGGTCCA 1380
ATTGTTTCAC AGAGCCAGTT TGGGATGGGC TGCATTCCAT TTATGCTGTA TATAGTTTGA 1440 ATTATATATA AATTACCCCT TCTTCTGGCC ACCCCTGCTC CCATCTTAGT ATTTTGCAAG 1500
ATCTAATCAG TTGTACACCT GCTGCCCCTC GCTTGCTTCA ATCATGGTTA TTTGATGGCA 1560 AAATCGACCT CTTCTCGCTG AAGGAGAGAG AAAAGATGTG TGTCTGATTG GTCCTGGGAT 1620 TTTTTGAGCT GTGCCATTTA TGGTACTCTT TGCCTATGCA TCCCCTTTTT AGATTTTTTT 1680 TAAATTTTAT CTTACTGTTT TTATAATTTC TATTGGGAAG AGGCTTGTGA CCAGTACCAA 1740 TCTTGAGTTT CTTTTTCTGT CCACAAGTAA ATTAATATCT GCTCTGAAAT GTCATTTATC 1800 TACTCACACA TTCTTGGGGA AAAAAATCAA ATGTCAGTCC TAGCAGATGT TGCATGTAAA 1860 TTGGTAGCAA GTAATGATTA CAACCCAGAG GATTAAGAAT TTTGTAACAG AAAGCTCTAT 1920 GTTTTAATTT TTTATATACA ATTAGGATAA TTAGCATTGT CAGACTATAA ACCTTTGCTT 1980 TTTAAAGTTT ATTTTTACTA TTTCTTTATC ACTTTATTGT ATCATCACCA TTGCTTTCAT 2040 AATGTAAATA CTATATGTTG AACAAATTAA ATGTCAAAAT TTTTTATTAC CATAGTCCAT 2100 GTTAATAGTG GGGCTTTCAG GTGTTTAGAG ATTTTTTTTC TTGTTCTTAA CATTCATTGC 2160 AAAAGTACTA GATGGTGTAT AACTCTAGAG TTGAATTTTA AGGGATTCCC TAATATGTAT 2220 ACTATCTTTT TATCTGAAGT AATAAATAAA CAATGATCTT GAAAGTGCCY RAAAMAAAAA 2280 AAAAAAAAAA AAA 2293
(2) INFORMATION FOR SEQ ID NO: 186:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1212 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi ) SEQUENCE DESCRIPTION : SEQ ID NO : 186: GGCACGAGGC GAGCCGGCGC ACCGTACGCT GGGACGTGTG GTTTCAGCTC GTGCGCCTCC 60 CCGTGGGTTT GCGACGTTTA GCGACTATTG CGCCTGCGCC ACGCCGGCTG CGAGACTGGG 120 GCCGTGGCTG CTGGTCCCGG GTGATGCTAG GCGGCTCCCT GGGCTCCAGG CTGTTGCGGG 180 GTGTAGGTGG GAGTCACGGA CGCTTCGGGG CCCGAGGTGT CCGCGAAGGT GGCGCACATG 240 GGCGGCAGGG GAGAGCATGG CTCAGCGGAT GGTCTGCCTG GACCTGGAGA TCACAGGATT 300 GGACATTGAG AAGGACCAGA TTATTGAGAT GGCCTGTCTG ATAACTGACT CTGATCTCAA 360 CATTTTGGCT GAAGGTCCTA ACCTGATTAT AAAACAACCA GATGAGTTGC TGGACAGCAT 420 GTCAGATTGG TGTAAGGAGC ATCACGGGAA GTCTGGCCTT ACCAAGGCAG TGAAGGAGAG 480 TACAATTACA TTGCAGCAGG CAGAGTATGA ATTTCTGTCC TTTGTACGAC AGCAGACTCC 540
TCCAGGGCTC TGTCCACTTG CAGGAAATTC AGTTCATGAA GATAAGAAGT TTCTTGACAA 600 ATACATGCCC CAGTTCATGA AACATCTTCA TTATAGAATA ATTGATGTGA GCACTGTTAA 660 AGAACTCTGC AGACGCTGGT ATCCAGAAGA ATATGAATTT GCACCAAAGA AGGCTGCTTC 720 TCATAGGGCA CTTCATGACA TTAGTGAAAG CATCAAAGAG CTTCAGTTTT ACCGAAATAA 780 CATCTTCAAG AAAAAAATAG ATGAAAAGAA GAGGAAAATT ATAGAAAATG GGGAAAATGA 840 GAAGACCGTG AGTTGATGCC AGTTATCATG CTGCCACTAC ATCGTTATCT GGAGGCAACT 900 TCTGGTGGTT TTTTTTTCTC ACGCTGATGG CTTGCCAGAG CACCTTCGGT TAACTTGCAT 960 CTCCAGATTG ATTACTCAAG CAGACAGCAC ACGAAATACT ATTTTTCTCC TAATATGCTG 1020 TTTCCATTAT GACACAGCAG CTCCTTTGTA AGTACCAGGT CATGTCCATC CCTTGGTACA 1080 TATATGCATT TGCTTTTAAA CCATTTCTTT TGTTTAAATA AATAAATAAG TAAATAAAGC 1140 TAGTTCTATT GAAATGCAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA 1200 AAAAAAAAAA AN 1212
(2) INFORMATION FOR SEQ ID NO: 187:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1605 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 187:
GCTTCCGGAA GTTGCTTTTG TCCAAACATC CGGGCTTCTC CTTTTTCTGT TCCGGCCGAT 60
CCCACCTCTC CTCGACCCTG GACGTCTACC TTCCGGAGGC CCACATCTTG CCCACTCCGC 120
GCGCGGGGCT AGCGCGGGTT TCAGCGACGG GAGCCCTCAA GGGACATCGC AACTACAGCG 180
GCGCCGGCGG GCGGCGCCCG AAATGGAGCT GGCCCGGAAT GGGGAGGGTT CCAAGAAAAC 240
ATCCAGGGCG GAGGCTCAGC TGTGATTGAC ATGGAGAACA TGCATGATAC CTCAGGCTCT 300
AGCTTCGAGG ATATGGGTGA GCTGCATCAG CGCCTGCGCG AGGAAGAAGT AGACGCTGAT 360
GCAGCTGATG CAGCTGCTGC TGAAGAGGAG GATGGAGAGT TCCTGGCCAT GAAGGGCTTT 420
AAGGGACAGC TGAGCCGGCA GGTGGCAGAT CAGATGTGGC AGGCTGGGAA AAGACAAGCC 480
TCCAGGGCCT TCAGCTTGTA CGCCAACATC GACATCCTCA GACCCTACTT TGATGTGGAG 540
CCTGCTCAGG TGCGAACAGG GCTCCTGGAG TCCATGATCC CTATCAAGAT GGTCAACTTC 600
CCCCAGAAAA TTGCAGGTGA ACTCTATGGA CCTCTCATGC TGGTCTTCAC TCTGGTTGCT 660
ATCCTACTCC ATGGGATGAA GACGTCTGAC ACTATTATCC GGGAGGGCAC CCTGATGGGC 720
ACAGCCATTG GCACCTGCTT CGGCTACTGG CTGGGAGTCT CATCCTTCAT TTACTTCCTT 780
GCCTACCTGT GCAACGCCCA GATCACCATG CTGCAGATGT TGGCACTGCT GGGCTATGGC 840
CTCTTTGGGC ATTGCATTGT CCTGTTCATC ACCTATAATA TCCACCTCCA CXX:CCTCTTC 900
TACCTCTTCT GGCTGTTGGT GGGTGGACTG TCCACACTGC GCATGGTAGC AGTGTTGGTG 960
TCTCGGACCG TGGGCCCCAC ACAGCGGCTG CTCCTCTGTG GCACCCTGGC TGCCCTACAC 1020
ATGCTCTTCC TGCTCTATCT GCATTTTGCC TACCACAAAG TGGTAGAGGG GATCCTGGAC 1080
ACACTGGAGG GCCCCAACAT CCCGCCCATC CAGAGGGTCC CCAGAGACAT CCCTGCCATG 1140
CTCCCTGCTG CTCGGCTTCC CACCACCGTC CTCAACGCCA CAGCCAAAGC TGTTGCGGTG 1200
ACCCTGCAGT CACACTGACC CCACCTGAAA TTCTTGGCCA GTCCTCTTTC CCGCAGCTGC 1260
AGAGAGGAGG AAGACTATTA AAGGACAGTC CTGATGACAT GTTTCGTAGA TGGGCTTTGC 1320
AGCTGCCACT GAGCTGTAGC TGCGTAAGTA CCTCCTTGAT GCNTGTCGGC ACTTCTGAAA 1380
GGCACAAGGC CAAGAACTCC TGGCCAGGAC TGCAAGGCTC TGCAGCCAAT GCAGAAAATG 1440
GGTCAGCTCC TTTGAGAACC CCTCCCCACC TACCCCTTCC TTCCTCTTTA TCTCTCCCAC 1500
ATTGTCTTGC TAAATATAGA CTTGGTAATT AAAATGTTGA TTGAAGTCTG GAAAAAAAAA 1560
AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAC TCGAG 1605
(2) INFORMATION FOR SEQ ID NO: 188:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1516 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 188: ATTCGGCATG AGGGGGTCAC GTGGTGGCTG GGCCGGCGAA ATGGCGGCTT CAGGAGAGAG 60 CGGGACTTCA GGCGGCGGAG GCAGCACCGA GGAAGCATTT ATGACCTTCT ACAGTGAGCT 120 GAAACAAATA GAGAAGAGAG ACTCGGTTCT AACTTCGAAA AATCAGATTG AAAGACTGAC 180 CCGTCCTGGT TCCTCTTACT TCAATTTGAA CCCATTTGAG GTTCTTCAGA TAGATCCTGA 240 AGTTACAGAT GAAGAAATAA AAAAGAGGTT TCGGCAGTTA TCCATCTTGG TGCATCCTGA 300 CAAAAATCAA GATGATGCTG ACAGAGCACA AAAGGCTTTT GAAGCTGTGG ACAAAGCTTA 360 CAAGTTGCTA CTCCATCAGC AGCAAAAGAA GAGCCCCCTG GATGTAATTC AGGCAGGAAA 420 AGAATACGTG GAACACACTG TGAAAGAGCG AAAAAAACAA TTAAAGAAGG AAGGAAAACC 480
TACAATTGTA GAGGAGGATG ATCCTGAGCT GTTCAAACAA GCTGTATATA AACAGACAAT 540
GAAACTCTTT GCAGAGCTGC AAATTAAAAG GAAAGAGAGA GAAGCCAAAG AGATGCATGA 600
AAGGAAACGA CAAAGGGAAG AAGAGATTGA AGCTCAAGAA AAAGCCAAAC GGGAAAGAGA 660
GTGGCAGAAA AACTTTGAGG AAAGTCGAGA TGGTCGTGTG GACAGCTGGC GAAACTTCCA 720
AGCCAATACG AAGGGGAAGA AAGAGAAGAA AAATCGGACC TTCCTGAGAC CACCGAAAGT 780
AAAAATGGAG CAACGTGAGT GACCGCCCAA GCTCACAGCC ACAGAACCTT TCCCCTGCTA 840
TCTCCCTTCC TGCTTCGAAG GACTCATTCT TTCCTCCCAC TTCCACCCCA ACATAGAGTA 900
GTATTTGCTT TTTAGTCCAT TTTGTTTTCA ATACGATTTA ATATCGATCA GAGTAATTCT 960
TTTGTACATT GAAATGAGGG GCTTGGTTTA AAAAAAGACC TTTCCCTCTC CCTGCCCCTA 1020
GAACAACCAG TATTAGAAGG TGCCACCATT GGTGCTGCCT TCTCTTCCCA CAGCCTGTAA 1080
CTCAGTGTTT TGTACTTCAC TGAATTGTGA TGGTTAGAAA CTTCGTGGAT AGTTTGTGGA 1140
AATCATCCAA TTAAACATAC TGCTTAAAAC AGTGTTGCTC TGACTTCAGA GACAAGCCTG 1200
GAAGGGGCAC CTTAGGAAGC CCCTTCGCTT CAGTTGCTCG CTTCTGCGTG TGCTCCCTTC 1260
GAAGGCCCAG ATAAGACAGG GAACACTTGT GAGCACACAG AGCAGCATCT GATGCCCTGT 1320
GGTGTTTGGC ATGTGCCCCC TCTCTACTCA CCAATCAGTG TGGCATGAGG CCCACGCCAC 1380
CCAAACCTTT CΛCTTTCCAA AGAGCTAGCC GTCCTCCACC CAGTACCATG TCCTAGCCTG 1440
TCTGCATTTG TTAGTGGTAA TATTCTTTAT GTATAATAAA TTTTTATACC GAAAAAAAAA 1500
AAAAAAAAAA ACTCGA 1516
(2) INFORMATION FOR SEQ ID NO: 189:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 681 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO 189: GCTCCCATGT TGCTCGCTCT CCGTACATCA CCCTGTCCCC TGCAGGAG G GGCTACAGGC 60 CATCTCCCTC CTGTAGGCCT CTGACTCCCC TCCACTTTTG GGCCCTCAGC TTATCTCGGG 120 CAGGGGACCA TTGCAGCATC CTCCCCTCCT CNGGACTCAA GGTGCTGAGG TATAAGCCCT 180 GGCCCCCAGA TCCCTGRTKA CACCTTCCTG GAGAAGACTC TCAAAAGTGA CTGTATATTT 240 GAGTTCACCA GCAATAACTC CCCACACTCG AAGCAGGTCC AAACCCMAGG ATCCCAGGGT 300 CCTTGCGCTC TGTGGCACTG TCTTCCCAAG ATCCTTCCTG TTGCACAATG GGAAACCTAA 360
GAGGAAAAAG ACAGGGGCCT GCTTGCCCAG CCATGCGAGG GATTCCATGC CCACCTGCCC 420
TCTGYCTGCC TCGCTGGAAT GTGGGCCCCT GCTCCCCGTC AGGTTGTGCT GTCTCTGACC 480
TATGTTTACA TCCCCGAGGG GTTTCTGCCT CCTCCCCACC CAGGTCAGGG TGTGGTCCAG 540
CAGCTTGCTG TGGCGTGCTG ACATGTGTCA CCACTCCCCC CCTTGCCCCC GGGGGGGTCA 600 TGGTCTCCTC CTGGATGCTG CTCCTTGAAT YTTTTTTYTT GAWAAACCYT TTAMAATTAA 660
AAAAAAAAAA AAAAAACTCG A 681
(2) INFORMATION FOR SEQ ID NO: 190:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 1014 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 190:
GCCTCAAGCC ACGCATATGA TAATTTTCTG GAACATTCAA ATTCAGTGTT TCTACAGCCA 60 GTTAGTCTAC AAACCATTGC AGCAGCACCA TCAAACCAGA GTCTGCCACT TTTTGTCATC 120
GCTGGATGAT TGCTGGGCAA AGGTGGCCTT TTAGAGCTCT TAAAAGCCCA CAAAAAGGCT 180
ATTCGTAGAG CCACAGTCAA CACATTTGGT TATATTGCAA AGGCCATTGG CCTCATGATG 240 TATTGGCTAC ACTTCTGAAC AACCTCAAAG TTCAAGAAAG GCAGAACAGA GTTTGTACCA 300
CTGTAGCAAT AGCTATTGTT GCAGAAACAT GTTCACCCTT TACAGTACTC CCTGCCTTAA 360
TGAATGAATA CAGAGTTCCT GAACTGAATG TTCAAAATGG AGTGTTAAAA TCGCTTTCCT 420
TCTTGTTTGA ATATATTGGT GAAATGGGAA AAGACTACAT TTATGCCGTA ACACCGTTAC 480
TTGAAGATGC TTTAATGGAT AGAGACCTTC TACACAGACA GACGGCTAGT GCAGTGGTAC 540 AGCACATGTC ACTTGCGGTT TATGGATTTG GTTGTGAAGA TTCGCTGAAT CACTTGTTGA 600
ACTATGTATG GCCCAATGTR TTTGAGACAT CTCCTCATGT AATTCAGGCA GTTATGGGAG 660
CCCTAGAGGG CCTGAGAGTT GCTATTGGAC CATGTAGAAT GTTGCAATAT TGTTTACAGG 720
GTCTGTTTCA CCCAGCCCGG AAAGTCAGAG ATGTATATTG GAAAATTTAC AACTCCATCT 780
ACATTGGTTC CCAGGACGCT CTCATAGCAC ATTACCCAAG AATCTACCAA CGATGATAAG 840 RACACCTATA TTCGTTATGA ACTTGACTAT ATCTTATAAT TTTATTGTTW ATTTKGTGKT 900
TAATCCACAS TACTTCACAC CTTAAACTTC CTTTGATTTG GTGATGTAAA CTTTTAAACA 960
TTGCAGATCA GTGTAGGACT GGTCCATAGG GGAAGAGCTA GGAANTCCAT AGGC 1014
(2 ) INFORMATION FOR SEQ ID NO : 191 :
( i ) SEQUENCE CHARACTERISTICS :
(A) LENGTH: 2779 base pairs
(B) TYPE : nucleic acid
(C) STRANDEDNESS : double
(D) TOPOLOGY : linear
(xi ) SEQUENCE DESCRIPTION: SEQ ID NO: 191 :
TCGCAGCAGG GTGTGTCCAG ATGGTCAGTC TCTGCTCCCT AGCCTCTCCT GACAGGGGAG 60
AGTTAAGCTC CCGYTCTCCA CCGTGCCGGC TGCCCAGGTG GGCTGAGGGT GACCGAGAGA 120
CCAGAACCTG CTTGCTGGAG CTTAGTGCTC AGAGCTCGGG AGGGAGGTTC CGCCGCTCCT 180
CTGCTGTCAG CGCCGGCAGC CCCTCCCGGC TTCACTTCCT CCCGCAGCCC CTGCTACTGA 240
GAAGCTCCGG GATCCCAGCA GCCGCCACGC CCTGGCCTCA GCCTGCGGGG CTCCAGTCAG 300
GCCAACACCG ACGCGCANTG GGAGGAAGAC AGGACCCTTG ACATCTCCAT CTGCACAGAG 360
GTCCTGGCTG GACCGAGCAG CCTCCTCCTC CTAGGATGAC CTCACCCTCC AGCTCTCCAG 420
TTTTCAGGTT GGAGACATTA GATGGAGGCC AAGAAGATGG CTCTGAGGCG GACAGAGGAA 480
AGCTGGATTT TGGGAGCGGG CTGCCTCCCA TGGAGTCACA GTTCCAGGGC GAGGACCGGA 540
AATTCGCCCC TCAGATAAGA GTCAACCTCA ACTACCGAAA GGGAACAGGT GCCAGTCAGC 600
CGGATCCAAA CCGATTTGAC CGAGATCGGC TCTTCAATGC GGTCTCCCGG GGTGTCCCCG 660
AGGATCTGGC TCGACTTCCA GAGTACCTGA GCAAGACCAG CAAGTACCTC ACCGACTCGG 720
AATACACAGA GGGCTCCACA GGTAAGACGT GCCTGATGAA GGCTGTGCTG AACCTTAAGG 780
ACGGGGTCAA TGCCTGCATT CTGCCACTGC TGCAGATCGA CCGGGACTCT GGCAATCCTC 840
AGCCCCTGGT AAATGCCCAG TCCACAGATG ACTATTACCG AGGCCACAGC GCTCTCCACA 900
TCGCCATTGA GAAGAGGAGW CTGCAGTGTG TGAAGCTCCT GGTGGAGAAT GGGGCCAATG 960
TGCATGCCCG GCTCTGCGGC GCTTCTTCCA GAAGGGCCAA GGGACTTGCT TTTATTTCGG 1020
TGAGCTACCC CTCTYTTTGG CCGCTTGCAC CAAGCAGTGG GATGTGCTAA GCTACCTCCT 1080
GGAGAACCCA CACCAGCCCG CCAGCCTGCA GGCACTGACT CCCAGGGCAA CACAGTCCTG 1140
CATCCCCTAG TGATGATCTC GGACAACTCA GCTGAGAACA TTGCACTGGT GACCAGCATG 1200
TATGATGGGC TCCTCCAAGC TGCGGCCCGC CTCTGCCCTA CCGTGCAGCT TGAGCACATC 1260
CGCAACCTGC AGGATCTCAC GCCTCTGAAG CTGGCCGCCA AGGAGGGCAA GATCGAGATT 1320
TTCAGGCACA TCCTGCAGCG GGAGTTTTCA GGACTGAGCC ACCTTTCCCG AAAGTTCACC 1380
GAGTGGTGCT ATGGCCCTGT CCGGGTGTCG CTGTATGACC TGGCTTCTGT GGACAGCTGT 1440
GAGGAGAACT CAGTGCTGGA GATCATTGCC TTTCATTCCA AGAGCCCGCA CCGACACCGA 1500
ATGGTCGTTT TGGAGCCCCT GAACAAACTC CTGCAGGCGA AATGGCATCT GCTCATCCCC 1560
AAGTTCTTCT TAAACTTCCT GTGTAATCTG ATCTACATGT TCATCTTCAC CGCTGTTGCC 1620
TACCATCAGC CTACCCTGAA GAAGCAGCCC GCCCCTCACC TGAAAGCGGA GGTTGGAAAC 1680
TCCATGCTGC TGACGGGCCA CATCCTTATC CTGCTAGGGG GGATCTACCT CCTCGTGGGC 1740
CAGCTGTGGT ACTTCTGGCG GCGCCACGTG TTCATCTGGA TCTCGTTCAT AGACAGCTAC 1800
TTTGAAATCC TCTTCCTGTT CCARGCCCTG CTCACAGTGG TGTCCCARGT GCTGTGTTTC 1860
CTGGSCATCG AGTGGTACCT GCCCCTGCTT GTGTCTGCGC TGGTGCTGGG CTGGCTGAAC 1920
CTGCTTTACT ATACACGTGG CTTCCAGCAC ACAGCCATCT ACAGTGTCAT GATCCAGAAG 1980
CCCTGGTGAG CCTGAGCCAG GANNTTGGCG CCCCGAAGCT CCTACAGGCC CCAATGCCAC 2040
AGAGTCAGTG CAGCCCATGG AGGGACAGGA KGACGAKGGC AACGGGGCCC AGTACAGCGC 2100
TATCCTGGAA GCCTCCTTGG AGCTCTTCAA ATTCACCATC GGCATGGGCG AGCTGGCCTT 2160
CCAGGARCAG CTGCACTTCC GCGGCATGGT GCTGCTGCTG CTGCTGGΞCT ACGTGCTGCT 2220
CACCTACATC CTGCTGCTCA ACATGCTCAT CGCCCTCATG AGCGAGACCG TCAACAGTGT 2280
CGCCACTGAC AGCTGGAGCA TCTGGAAGCT GCAGAAAGCC ATCTCTGTCC TGGAGATGGA 2340
GAATGGCTAT TGGTGGTGCA GCAAGAAGCA GCGGGCAGGT GTGATGCTGA CCGTTGGCAC 2400
TAAGCCAGAT GGCAGCCCSG ATGAGCGCTG GTGCTTCAGG GTGGAGGAGG TGAACTGGGC 2460
TTCATGGGAG CAGACGCTGC CTACGCTGTG TGAGGACCCG TCAGGGGCAG GTGTCCCTCG 2520
AACTCTCGAG AACCCTGTCC TGGCTTCCCC TCCCAAGGAG GATGAGGATG GTGCCTCTGA 2580
GGAAAACTAT GTGCCCGTCC AGCTCCTCCA GTCCAACTGA TGCCCCAGAT GCAGCAGGAG 2640
GCCAGAGGAC AGAGCAGAGG ATCTTTCCAA CCACATCTGC TGGCTCTGGG GTCCCAGTGA 2700
ATTCTGGTGG CAAATATATA TTTTCACTAA CTCAAAAAAA AAAAAAAAAA AAAAAAAAAA 2760
AAAAAAAAAA AAAAAAGGC 2779
(2) INFORMATION FOR SEQ ID NO: 192:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1923 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 192:
ACCCGCTCCG CTCCGCTCCG CTCGGCCCCG CGCCGCCCGT CAACATGATC CGCTGCGGCC 60
TGCCCTGCGA GCGCTGCCGC TGCATCCTGC CCCTGCTCCT ACTCAGCGCC ATCGCCTTCG 120
ACATCATCGC GCTGGCCGGC CGCGGCTGCT TGCAGTCTAG CGACCACGGC CAGACGTCCT 180
CGCTGTGGTG GAAATGCTCC CAAGAGGGCG GCGGCAGCGG GTCCTACGAG GAGGGCTGTC 240
AGAGCCTCAT GGAGTACGCG TGGGGTAGAG CAGCGGCTCC CATGCTCTTC TGTGGCTTCA 300
TCATCCTGGT GATCTGTTTC ATCCTCTCCT TCTTCGCCCT CTGTGGACCC CAGATGCTTG 360
TCTTCCTGAG AGTGATTGGA GGTCTCCTTG CCTTGGCTGC TGTGTTCCAG ATCATCTCCC 420
TGGTAATTTA CCCCGTGAAG TACACCCAGA CCTTCACCCT TCATGCCAAC CSTGCTGTCA 480
CTTACATCTA TAACTGGGCC TACGGCTTTG GGTGGGCAGC CACGATTATC CTGATYGGCT 540
GTGCCTTCTT CTTCTGCTGC CTGCCCAACT ACGAAGATGA CCTTCTGGGC AATGCCAAGC 600
CCAGGTACTT CTACACATCT GCCTAACTTG GGAATGAATG TGGGAGAAAA TCGCTGCTGC 660
TGAGATGGAC TCCAGAAGAA GAAACTGTTT CTCCAGGCGA CTTTGAACCC ATTTTTTGGC 720
AGTGTTCATA TTATTAAACT AGTCAAAAAT GCTAAAATAA TTTGGGAGAA AATATTTTTT 780
AAGTAGTGTT ATAGTTTCAT GTTTATCTTT TATTATGTTT TGTGAAGTTG TGTCTTTTCA 840
CTAATTACCT ATACTATGCC AATATTTCCT TATATCTATC CATAACATTT ATACTACATT 900
TGTAAGAGAA TATGCACGTG AAACTTAACA CTTTATAAGG TAAAAATGAG GTTTCCAAGA 960
TTTAATAATC TGATCAAGTT CTTGTTATTT CCAAATAGAA TGGACTCGGT CTGTTAAGGG 1020
CTAAGCAGAA GAGGAAGATA AGGTTAAAAG TTGTTAATGA CCAAACATTC TAAAAGAAAT 1080
GCAAAAAAAA AGTTTATTTT CAAGCCTTCG AACTATTTAA GGAAAGCAAA ATCATTTCCT 1140
AAATGCATAT CATTTGTGAG AATTTCTCAT TAATATCCTG AATCATTCAT TTCAGCTAAG 1200
GCTTCATGTT GACTCGATAT GTCATCTAGG AAAGTACTAT TTCATGGTCC AAACCTCTTC 1260
CCATAGTTGG TAAGGCTTTC CTTTAAGTGT GAAATATTTA GATGAAATTT TCTCTTTTAA 1320
AGTTCTTTAT AGGGTTAGCC TGTGGGAAAA TGCTATATTA ATAAATCTGT AGTCTTTTGT 1380
GTTTATATGT TCAGAACCAG AGTAGACTGG ATTGAAAGAT GGACTGGCTC TAATTTATCA 1440
TGACTGATAG ATCTGGTTAA GTTGTGTAGT AAAGCATTAG GAGGCTCATT CTTGTCACAA 1500
AAGTGCCACT AAAACAGCCT CAGGAGAATA AATGACTTGC TTTTCTAAAT CTCAGGTTTA 1560
TCTGGGCTCT ATCATATAGA CAGGCTTCTG ATAGTTTGCA ACTGTAAGCA GAAACCTACA 1620
TATAGTTAAA ATCCTGGTCT TTCTTGGTAA ACAGATTTTA AATGTCTGAT ATAAAACATG 1680
CCACAGGAGA ATTCGGCGAT TTGAGTTTCT CTGAATAGCA TATATATGAT GCATCGGATA 1740
GGTCATTATG ATTTTTTACC ATTTCGACTT ACATAATGAA AACCAATTCA TTTTAAATAT 1800
CAGATTATTA TTTTGTAAGT TGTGGAAAAA GCTAATTGTA GTTTTCATTA TGAAGTTTTC 1860
CCAATAAACC AGGTATTCTA AAAAAAAAAA AAAAAAACTN GAGGGGGGCC CCGCTACCCA 1920 ATT 1923
(2) INFORMATION FOR SEQ ID NO: 193:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2346 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 193:
AGGCTCAGGC GGACACTCTC .(AAATTACAC AGCTTTTAAC AGGTGGCAGA ATTGCGCTTC 60
AGACCCAGAT CTCGGTTCAA GTCACTCATG GTGTGATTGC GGCATTCCTT CCCGCATCTG 120
GCCCTTGCCA TCTCTCTCTC CGAGTGGACA TGGAGAGGAC GGGGGCCCAG CAGCTGGATG 180
GCTGCAGGGG ATCAAGTCTT CTCTGGGGCT GGGCACGTAN AAGAGCATGT GGCTGGTGCA 240
CGGCATCCCT GGCTCCTCAC CTGGCAGTCT GCCTGCCCTG CTAACCGGCT GTCTCTTGTT 300
CCCCTAGTGC CCTCGGCTAG CATGACCCGC CTGATGCGWT SCCGCACAGC CTCTGGTTCC 360
AGCGTCATTC TCTGGATGGC ACCCGCAGCC GCTCCCACAC CAGCGAGGCC ACCCGAAGCC 420
GCTCCCACAC CAGCGAGGCC ACCCGCAGCC GCTCGCACAC CAGCGAGGGG GCCCACCTGG 480
ACATCACCCC CAACTCGGGT GCTGCTGGGA ACAGNGCCGG GCCCAAGTCC ATGGAGGTCT 540
CCTGCTAGCC GCCCTGCCCA GCTGCCGCCC CCGGACTCTG ATCTCTGTAG TGGCCCCCTC 600
CTCCCCGGCC CCTTTTCGCC CCCTGCCTGC CATACTGCGC CTAACTCGGT ATTAATCCAA 660
AGCTTATTTT GTAAGAGTGA GCTCTGGTGG AGACAAATGA GGTCTATTAC GTGGGTGCCC 720
TCTCCAAAGG CGGGGTGGCG GTGGACCAAA GGAAGGAAGC AAGCATCTCC GCATCGCATC 780
CTCTTCCATT AACCAGTGGC CGCTTGCCAC TCTCCTCCCC TCCCTCAGAG ACACCAAACT 840
GCCAAAAACA AGACGCGTAC AGCACACACT TCACAAAGCC AAGCCTAGGC CGCCCTGAGC 900
ATCCTGGTTC AAACGGGTGC CTGGTCAGAA GGCCAGCCGC CCACTTCCCG TTTCCTCTTT 960
AACTGAGGAG AAGCTGATCC AGTTTCCGGA AACAAAATCC TTTTCTCATT T GGGAGGGG 1020
GGTAATAGTC ACATGCAGGC ACCTCTTTTA AACAGGCAAA ACAGGAAGGG GGAAAAGGTG 1080
GGATTCATGT CGAGGCTAGA GGCATTTGGA ACAACAAATC TACGTAGTTA ACTTGAAGAA 1140
ACCGATTTTT AAAGTTGGTG CATCTAGAAA GCTTTGAATG CAGAAGCAAA CAAGCTTGAT 1200
TTTTCTAGCA TCCTCTTAAT GTGCAGCAAA AGCAGGCRAC AAAATCTCCT GGCTTTACAG 1260
ACAAAAATAT TTCAGCAAAC GTTGGGCATC ATGGTTTTTC AAGCCTTTAG TTCTGCTTTC 1320
TGCCTCTCCT CCACAGCCCC AACCTCCCAC CCCTGATACA TGAGCCAGTG ATTATTCTTG 1380
TTCAGGGAGA AGATCATTTA GATTTGTTTT GCATTCCTTA GAATGGAGGG CAACATTCCA 1440
CAGCTGCCCT GCCTGTGATG AGTGTCCTTG CAGGCGCCGG AGTAGGAGCA CTGGGGTGGG 1500
GGCGGAATTG GGGTTACTCG ATGTAAGGGA TTCCTTGTTG TTGTGTTGAG ATCCAGTGCA 1560
GTTGTGATTT CTGTGGATCC CAGCTTGGTT CCAGGAATTT TCTGTGATTG GCTTAAATCC 1620
AGTTTTCAAT CTTCGACAGC TGGGCTGCAA CGTGAACTCA GTAGCTGAAC CTCTCTGACC 1680
CGGTCACGTT CTTGCATCCT CAGAACTCTT TGCTCTTGTC GGCGTGGGGG TGGGAACTCA 1740
CGTGGGGAGC GGTCGCTGAG AAAATGTAAG GATTCTGCAA TACATATTCC ATGGGACTTT 1800
CCTTCCCTCT CCTGCTTCCT CTTTTCCTGC TCCCTAACCT TTCGCCGAAT GGGGCAGCAC 1860
CACTGACGTT TCTGCGCGGC CAGTGCGGCT GCCAGGTTCC TGTACTACTG CCTTGTACTT 1920
TTCATTTTGG CTCACCGTGG ATTTTCTCAT AGGAAGTTTG GTCAGAGTGA ATTGAATATT 1980
GTAAGTCAGC CACTGGGACC CGAGGATTTC TGGGACCCCG CAGTTGGGAG GAGGAAGTAG 2040
TCCAGCCTTC CAGGTGGCGT GAGAGGCAAT GACTCGTTAC CTGCCGCCCA TCACCTTGGA 2100
GGCCTTCCCT GGCCTTGAGT AGAAAAGTCG GGGATCGGGC CAAGAGAGGC TGAGTACGGA 2160
TGGCAAACTA TTGTGCACAA GTCTTTCCAG AGGAGTTTCT TAATGAGATA TTTGTATTTA 2220
TTTCCAGACC AATAAATTTG TAACTTTGCA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA 2280
AAAAAAAAAA AAAAAAAACT CGAGGGGGGC CCGTACCCAA TTCGCCGTAT ATGATCGTAA 2340
ACAATC 2346
(2) INFORMATION FOR SEQ ID NO: 194:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 3054 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi ) SEQUENCE DESCRIPTION : SEQ ID NO : 194 : TATCTGAACC ACCCTTTATT CTACATATGA TAGGCAGCAC TGAAATATCC TAACCCCCTA 60 AGCTCMAGCT GCCCTGTGGN ACGAGCAACT GGACTATAGC AGGGCTGGGC TCTGTCTTCC 120 TGGTCATAGG CTCACTCTTT CCCCCAAATC TTCCTCTGCA GCTTTGCAGC CAAGGTGCTA 180 AAAGGAATAG GTAGGAGACC TCTTCTATCT AATCCTTAAA AGCATAATGT TGAACATTCA 240
TTCAACAGCT GATGCCCTAT AACCCCTGCC TGGATTTCTT CCTATTAGGC TATAAGAAGT 300
AGCAAGATCT TTACATAATT CAGAGTGGTT TCATTGCCTT CCTACCCTCT CTAATGGCCC 360
CTCCATTTAT TTCACTAAAG CATCACACAG TGGCACTAGC ATTATACCAA GAGTATGAGA 420
AATACAGTGC TTTATGGCTC TAACATTACT GCCTTCAGTA TCAAGGCTGC CTGGAGAAAG 480
GATGGCAGCC TCAGGGCTTC CTTATGTCCT CCACCACAAG AGCTCCTTGA TGAAGGTCAT 540
CTTTTTCCCC TATCCTGTTC TTCCCCTCCC CGCTCCTAAT GGTACGTGGG TACCCAGGCT 600
GGTTCTTGGG CTAGGTAGTG GGCACCAAGT TCATTACCTC CCTATCAGTT CTAGCATAGT 660
AAACTACGCT ACCAGTGTTA GTGGCAAGAG CTGCGTTTTC CTAGTATACC CACTGCATCC 720
TACTCCTACC TGGTCAACCC GCTGCTTCCA GGTATGGGAC CTGCTAAGTG TGGAATTACC 780
TGATAAGGGA GAGGGAAATA CAAGGAGGCC CTCTGGTGTT CCTGCCCTCA GCCAGCTGCC 840
CACAAGCCAT AAACCAATAA AACAAGAATA CTGAGTCAGT TTTTTATCTG GGTTCTCTTC 900
ATTCCCACTG CACTTGGTGC TGCTTTGGCT GACTGGGAAC ACCCCATAAC TACAGAGTCT 960
GACAGGAAGA CTCGAGACTG*TCCACTTCTA GCTCGGAACT TACTGTGTAA ATAAACTTTC 1020
AGAACTGCTA CCATCAAGTG AAAATGCCAC ATTTTGCTTT ATAATTTCTA CCCATGTTGG 1080
GAAAAACTGG CTTTTTCCCA GCCCTTTCCA GGCCATAAAA CTCAACCCCT TCGATAGCAA 1140
GTCCCATCAG CCTATTATTT TTTTAAAGAA AACTTGCACT TGTTTTTCTT TTTACAGTTA 1200
CTTCCTTCCT GCCCCAAAAT TATAAACTCT AAGTGTAAAA AAAAGTCTTA ACAACAGCTT 1260
CTTGCTTGTA AAAATATGTA TTATACATCT GTATTTTTAA ATTCTGCTCC TGAAAAATGA 1320
CTGTCCCATT CTCCACTCAC TGCATTTGGG GCCTTTCCCA TTGGTCTGCA TGTCTTTTAT 1380
CATTGCAGGC CAGTGGACAG AGGGAGAAGG GAGAACAGGG GTCGCCAACA CTTGTGTTGC 1440
TTTCTGACTG ATCCTGAACA AGAAAGAGTA ACACTGAGGC GCTCGCTCCC ATGCACAACT 1500
CTCCAAAACA CTTATCCTCC TGCAAGAGTG GGCTTTCCAG GGTCTTTACT GGGAAGCAGT 1560
TAAGCCCCCT CCTCACCCCT TCCTTTTTTC TTTCTTTACT CCTTTGGCTT CAAAGGATTT 1620
TGGAAAAGAA ACAATATGCT TTACACTCAT TTTCAATTTC TAAATTTGCA GCGGATACTG 1680
AAAAATACGG CAGCTGGCCT AAGCCTGCTG TAAAGTTGAG GGGAGAGGAA ATCTTAAGAT 1740
TACAAGATAA AAAACGAATC CCCTAAACAA AAAGAACAAT AGAACTGGTC TTCCATTTTG 1800
CCACCTTTCC TGTTCATGAC AGCTACTAAC CTGGAGACAG TAACATTTCA TTAACCAAAG 1860
AAAGTGGGTC ACCTGACCTC TGAAGAGCTC AGTACTCAGG CCACTCCAAT CACCCTACAA 1920
GATCCCAAGG AGGTCCCAGG AAGTCCAGCT CCTTAAACTG ACGCTAGNCA ATAAACCTCG 1980
GCAAGTGAGG CAAGAGAAAT GAGGAAGAAT CCATCTGTGA GGTGACAGGC AAGGATGAAA 2040
GACAAAGAAG GAAAAGAGTA TCAAAGGCAG AAAGGAGATC ATTTAGTTGG GTCTGAAAGG 2100
AAAAGTCTTT GCTATCCGAC ATGTACTGCT AGTACCTGTA AGCATTTTAG GTCCCAGAAT 2160
GGAAAAAAAA ATCAGCTATT GGTAATATAA TAATGTCCTT TCCCTGGAGT CAGTTTTTTT 2220
AAAAAGTTAA CTCTTAGTTT TTACTTGTTT AATTCTAAAA GAGAAGGGAG CTGAGGCCAT 2280
TCCCTGTAGG AGTAAAGATA AAAGGATAGG AAAAGATTCA AAGCTCTAAT AGAGTCACAG 2340
CTTTCCCAGG TATAAAACCT AAAATTAAGA AGTACAATAA GCAGAGGTGG AAAATGATCT 2400
AGTTCCTGAT AGCTACCCAC AGAGCAAGTG ATTTATAAAT TTGAAATCCA AACTACTTTC 2460
TTAATATCAC TTTGGTCTCC ATTTTTCCCA GGACAGGAAA TATGTCCCCC CCTAACTTTC 2520
TTGCTTCAAA AATTAAAATC CAGCATCCCA AGATCATTCT ACAAGTAATT TTGCACAGAC 2580
ATCTCCTCAC CCCAGTGCCT GTCTGGAGCT CACCCAAGGT CACCAAACAA CTTGGTTGTG 2640
AACCNAACTG CCTTAACCTT CTGGGGGAGG GGGATTAGCT AGACTAGGAG ACCAGAAGTG 2700
AATGGGAAAG GGTGAGGACT TCACAATGTT GGCCTGTCAG AGCTTGATTA GAAGCCAAGA 2760
CAGTGGCAGC AAAGCAAGAC TTGGCCCAGG AAAAACCTGT GGGTTGTGCT AATTTCTGTC 2820
CAGAAAATAG GGTGGACAGA AGCTTGTGGC GTGCATGGAG GAATTGGCAC CTGGTTATGT 2880
TGTTATTCTC GGACTGTGAA TTTTGGTGAT GTAAAACAGA ATATTCTGTA AACCTAATGT 2940
CTGTATAAAT AATGAGCGTT AACACAGTAA AATATTCAAT AAGAAGTCAA AAAAAAAAAA 3000
AAAAAACTCG AGGGGGGGCC CGGTACCCAA TTTNCCAAAT AGAGATNGTA TTAC 3054
(2) INFORMATION FOR SEQ ID NO: 195:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 907 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO : 195: GGCAGAGCTC GTGCCCGNAA CTTTTTCTGC TCCTGGCTGC CACCTACTGG CTGGCCGCGG 60 CCCTGGCCTG GGCCTGCACC AGCCTGCGNG CGGCCTCCCA CAGCAGCCCC CTTCCAAGCA 120 GCGTCCCCAC ACCGCGCACC TTCTGCGGCA ACGTGCTCGC CGTGCCGGGG ACCATATGGA 180 CGGAAGCCTT TGTGCTCACC TACAAGCTGG GTGAGCAGCG TGCCAGCAGC CTGTTGATCC 240 TCTTGGCTCC TGCTGGAGCA CGAGCGGCGT TTCTGCTCCC GAGTTGGGAC TGTCCAATGG 300 TGTGGCTGCT GTGGTCTGCT CCATCGCTGG CTCCTCCCTG GGTGGGACCT TGCTGGCCAA 360 GCACTGCAAA CTGCTGCCTC TGTGAGGTCG GTGCTGCGCT TCCGCCTCGG GGGCCTAGCC 420
TCTCAGACTC CCTTGGTCTT CCACCTTGGA CACCCTGGGG GCCAGCATGG ACGCTGGCAC 480
AATCTTGAGA GGGTCAGCCT TGCTGAGCCT ATGTCTGCAG CACTTCTTCG GARGCCTGGT 540
CACCACAGTC ACCTTCACTG GGAATGATGC GCTGCAGCCA GCTGGCCCCC AGGCCCTTGC 600
AGCCCACACA CTACAGCCTT CTGGCCACGC TGGAGCTGCT GGGGAAGCTG CTGCTGGGCA 660 CTYTGGSCGG AGGGCCTGGC TGATGGGTTG GGGCCACATC CCTGCTTCTT GCTCCTGCTC 720
ATCCTCTCTC CCTTTCCCGT TCTGTACCTG GACCTAGCAC CCAGCACCTT TCTCTGAGCT 780
GAGTGGCTGG AGTGGTCAAT AAAGCCACAT GTGCCTGTGG CCCAAAAAAA AAAAAAAAAA 840
AAAAAAAAAA AAAAAAACTG GAGGGCCGCC CCGGTACCCA AATCGCCGGA TATGATCGTA 900
AACAATC 907
(2) INFORMATION FOR SEQ ID NO: 196: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1290 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi ) SEQUENCE DESCRIPTION: SEQ ID NO : 196 :
GGCACGAGGA GGGACAGGGA GTGGGCAAGG GGAAGAAGCA GCTTATTTGA CTAACCAGCC 60 CCTCTGTGCT CCACCAGCGT CTTGGCTTGG TGGGAGCGCT CTCAATCAGC AGGGCCCCAG 120
KAGGGCAAGA AGAAGTGGCC CAAAGCCTGC CGCTCGGCCG CGGTCGCGGC AGCTTTGCMA 180
TCTGGAGCCA CGCCTCCTCC AGGCCATGCT CCTTGAACTT GGAAATGTCA ACCGGAGCCC 240
TTAACACCAG CCCTCCAGCA TCTAATAGAC TTGAATCTAC TCTAAACGAA TATTTAATCC 300
AACCTCAACT ACATTGTAGC TCAGTCCAAC GACTAACCCT GAAATGGCGG TGTTCCAGCC 360 TTCAGCGAGA TGGCCAAGCG GTCCCCTGGG GGCTGTGGCA GCGCGCTTAT CCTTCTCTGT 420
TGCCAACCTT GCCGTCCGAC CTCCTCCGCC CCCATGCGGT GACCCCGTCC GTGTCTGTGT 480
CTGTCCATAC GTGTGAGTCC AGCTAAAAAG ACAAAACAGA ACCCGTGCCC CCAGCTCGGA 540
AGGTGCGTGG AGAAGGCTCC GACGTCTCCG AAGTGCAGCC CTTGCGATGG CATTCCGTTG 600
TGTGCCTTAT TCCTGGAGAA TCTGTATACG GCTCGCCTAT AAGAAATATA GCCTCTTCAT 660 GCTGTATTAA AAGGACTTTT AAAAGCAAAA AAAAAAAAAA AAAAACTCGA GGGGGGGCCC 720
GGTACCCAAT TCGCCCAATA GT AGTCGTA TTACAATTCA CTGGGCCGTC STTTTAACAA 780
CGTCGTGAAC TGGGAAAACC CTGGCGTTTA CCCAACTTAA TCGCCTTGCA GCACATCCCC 840
CTTTCGCCAG CTGGCGTTAA TAGCGAAAAA NGCCCGCACC CGAATCGCCC TTCCCAACAG 900 TTTGCGCAGC CCTGAATGGC GAAATGGCAA ATTGTAAGCG TTTAATATTT TKKTTAAAAT 960 TCCNCGTTWA AWTTTTTGTT TAAATCARCT CAATTTTTTT AACCCAATAA GSCCGAAATC 1020 CGGCAAATCC CCYTTATTAA TTCCAAAAAA ATAAACCSAA AAWGGGTTTG AATTTTTTKT 1080 TTCCCCAYTT TTGGAAACAA AWTYCCCCCT TTTTAAAAAA GTTGGAACCC CCAMCCYTCC 1140 AAAGGGGAAA AAACSYTTTT YTGCGCGCNA ANGGGGCCCC CNTACTTTNA ACAYCCCCCC 1200 CCAAWCAATT TTTTTGGGGG GTCCCNAAAG GTCCCCCTAA AANCTTTTTT CGGAACCCNA 1260 AGCGGANCCC CCCATTTAAA ATTTTNGGTN 1290
(2) INFORMATION FOR SEQ ID NO: 197:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1020 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 197:
GGTCTGCCTG GATGGTCGTG TAGGTGAGTT TTACCAAGGA TTATGGTAAC AAATGAGTGA 60
GACCTCTATG GAGAAAATAT TGAAGNNCAT TAAAGAAGAC CTCATANTAG GAGAGAATGT 120
SCTTTGGAGG ATTTGTATTG AGCTTTTACA GTATTCATTT TTCAACTCAA GGCAATGGCT 180
TTCTACACCA ACTGTAATCC ATAAACGGGT CTTATGACAT CTATGAAGTA GTAGCAAGAC 240
ATGCTTAGTG TGTATTTCTC TCTTTGAGAC ACTCTAATTT CTACCAGAAA TTTCCAGAGC 300
ATTATGTAGG TAGAAAAAAA TGCAAGCAAG CTGTTAAAGA TCTTGGATCC CATTATATAG 360
TATGTATAGC TGAAATCTGT AATTCAATCA CTTTTTCTCT TTTATCCTCT AACCAAAAAA 420
TTGTTTAATT TTGCATCCCA AATGTTTTTA ATCTTTGTAT ATTTTTTAAA AAYCCTTTTC 480
TCCTCATCAT TGCCTTTTTT GTGGTTGTAA ATAGACTTAC TTGCACTTTG AAGATGAGTT 540
ACTCCTTGTC ATCTTACAAA TATGTGATAT GGTAATTTTC ATAACAGATG TCAGTTTTGA 600
ACCAAGAATT GGTGATTTGT TTATAAGAAA AAAACTGGCT TCATTTCTGT GAAATTGCTC 660
TTTGAAAATT TCTTTTTACA CGTGTAAGCC AACTGAGATA CCGTGATGGT GTTGATTTCT 720
TTCAATGATG CTTACCATCT ATTTTAGCCA CTGAGCCTTT TATTATTTGT CTATTTCTAA 780
AGTTTATTTC TCTTAACTCA TTTAATAAAT ATACTGTTTA TCTGTTTCTG AATGGGGACT 840
GAACTTTTTG GATATTGATA TTGATTTGAA AATATTTTGG AATTTTTTCT ACTTGAAATT 900
TTAGAAATCT AATKGAAAAT TCTATAATGT ACTGAAAGTA WGGTTGTGTA CAGTGAKCAC 960
TCTCTAATAA TATGATGNCT TGCCCTAAAN GAGCNGGGAC ATGTCCCACT TTCCACCACG 1020
,(2) INFORMATION FOR SEQ ID NO: 198:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 524 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 198:
AATTCCCGAA GCTGAGGGTT GTGTGCCNTC GGGCGAGCCA AGTCTTTTGA CCGGACCCTT 60 CCCGGCGCAG AAGANCTGAA GTTGATTTGA GAGCCTGTKT TTGGCCTTRA GCCGAGCTGC 120
TGCGGGCTTY GTCGCCGGCC AGGACACAAG YTACTTGCAA CGGGGCGCCG CCTGGCTTAT 180
GATGTTCCTC AACCCAGGGG CGGCCTCTGC CCTCTACTCG TGCCAGGCCC ACTTGCCAGG 240 CAGGAGCCCT CCCCAAGCCT TCAGGGCTGC TCGGAGTCAC CTGTTGGAAT GGACTAAAAG 300
GACCCTTGTG TGGGAACAGG TGCTCCAAAC ACCCTGCTGC TGGCTGCCAG GCAGGCCCTC 360
TGGAAGGGAA GGGGCAGGAC TCATCAGGAC CTCCCTGGAC CCTGCAGGGC AGGCAGTTGG 420
CCCGAGCCCA AGCATTTGGC TCTGCTTGCC CCAAGGGGAC AGGAAGCCTC TTGGGCCTCT 480
TCCCTTCCTG GACAAGGCCC CCTGCCTTTG CCTCACATAA ACTG 524
(2) INFORMATION FOR SEQ ID NO: 199: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 332 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 199:
GTGATACAAG GAAGGGTGAT CATCATCTGT CACCATGCAA TTCCTGCTCA CA∞CTTTCT 60 GTTGGTGCCA CTTCTGGCTC TTTGTGATCT CCCCATATCC CTAGGCTTCT CCCCCTCCTA 120
GAAGGGCTTC TTGATAGATT AGAAAATAAG AATGAGTGAC ATTTCCTATG TGCATATAAG 180
AAGGAGCCAC AAGACATCTC TTTTAAATAA AAGGACAGTG TCCATCCTTT TAGCTCCCGA 240
ATAGAACCTT GGTCTCATCC TCCTGGAGCT AGGSCTTAAA ACAGCTTCTG TGTTTCTSAT 300
TKGTCTCART GTTTTGCCAA GGTTTTATTC GG 332
(2) INFORMATION FOR SEQ ID NO: 200:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 376 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 200:
CCAGGGAAGC CCCARGCCTG TCCTGAATTG ACATCAGTGC TTCCCTGAAC TGCCTCCCCC 60 ACCCCTGGGC ATTATCCCAG GAAACTTATG TTTTCTAGAA GCTAAGCAGC TGCTGGGACT 120
CAGGGACTGG TGCAGGTAGG CTGAGTGGCA GCTCAGTCCT AGAAGGTCTC TGAAGATCTG 180
GACTGAGGAC CYTGCTACTC CCCAAGCCAG AGCCCATCAG CCAGGCCTGC TGTGAGCCAC 240
CTGCCTCTGG AGTGCTGAGC TCAACCAAAG GCTGGCAAGC TCTGGGCCTC ATTTAAGCGA 300
TTCTCATGAG CCGATGGGCC CTGGAGGCAG CCCATTAAAG CATCTGGCTC GTTTTTGGAA 360 AAAAAAAAAA AAAAAG 376
(2) INFORMATION FOR SEQ ID NO: 201:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1192 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 201: CCCAGTATAT TTCTATAACA TTTATTTTAG TGAACTTATA ATGTTTCTTT GTATTAAATT 60
ATTAGATTAT ATCTTTAGAT AATATTGTTA CTNAATTAGT AGGTAATATA TATTTTATTC 120
AAAAATAAAT TGTCCATCTA ATGTCTACCA ATTAATGTAC TTGTAGATGT ATCTTATCTT 180
AACTTGAGTC TTTGCTGCCC CTAATGAGGT GTGAAGGACT CTTCTCCCCT GGGGAAGTTT 240
TTCTTTTTCA GCAGGGAGGA GGGCTTTCCC AGGTAATGTG TCTAGAGTGT TGGGCAGAAR 300 AATCTGGGAC CACACCACAC CAGTTCTCTC CTTAATCCAC GTCATTTGCC TTCTATCCCA 360
GCTATCTTTC CAGTGTCCTC TCGGTCTTTC CAAGAGCAAC AAGAAAYGAA TAAATCTCTG 420
KTGAGTTGTT TATTTGTTCT TCACTTTGTT TTACACTGTA WTTTCTGAGT TTATGGGTGT 480
CTGTGAATTA AAAAGGAAAA GTRGAAATAA GTAAAACTCA GGTTGAAGCA AATATACATA 540
AATAAGATAA AGCTGACCTG TAGATATARR CAGGTTATAA RAGCTTAGAG TTGTCTAAGT 600 TGRGTGCAAA KTTTCCTCTG ATCTTTCTGA TGCCGARACA AAAAAGGCAG TCATGTTTGT 660
WATGTGATTG GAATGGAACC CGARAAGAGA GCAYGCTGTG TTCTTGGGCA CAGGAAAGCT 720 TGYGTGCACC AAGTCTKAAC CACCACCTTC ATGGGACATA GRTTATGTGC TGGAACATAT 780 TTCACACCGC CCTGGCAGTA AACACTTGTA GTGTTGTGCA GTGGAAACGG TCATCTTCCG 840 CTAAAGCACG GCGTGTTGTG CAGCGGAAAT GGTCATCTGC TGCTAAAACA CAGCTTCCAT 900 CGTAATGTAT GCTCCTTACT CAAAGAGTGT GGTCCCAAAC AGCCTTTGGG AGGTCCTCCT 960 TGATTCATGG ATGAAACCTG GAACATCTTG AGGACTGAGT TAACCATAGG TCCTTAAATA 1020 ACTCTCCACA CGTTTTTCTT AGTTTATCTC TACATGCAGG GTGTGCAGCA GCCTGTTCAA 1080 AGTCATATTT TCTGGGAAAT ATTTCCAGTG TTTATTTGCA CTTTAGCCCA CTCTGTGTAG 1140 CCTTATTTCT TCTAAACTCA CCATTAATCT GAATAATAGT CAAATTTAGG GG 1192
(2) INFORMATION FOR SEQ ID NO: 202:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 589 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO 202: ATCTTGGGCT ATCTTTGACA GGGGATTCTT GCAAGTTGAT GCTTTCTACA AGTGAATATA 60 GTCAGTCCCC AAAGATGGAG AGCTTGAGTT CTCACAGAAT TGATGAAGAT GGAGAAAACA 120 CACAGATTGA GGATACGCAA CCCATGTCTC CAGTTCTCAA TTCTAAATTT GTTCCTGCTG 180 AAAATGATAG TATCCTGATG AATCCAGCAC AGGATGCTGA AGTACAACTG AGTCAGAATG 240 ATGACAAAAC AAAGGGAGAT GATACAGACA CCMGGGATGA CATTAGTATT TTAGCCACTG 300 GTTGCAAGGG CAGAGAAGAA ACGCTAGCAG AAGATGTTTG TATTGATCTC ACTTGTGATT 360 CGGCGAGTCA GCCAGTTCCG TCACCAGCTA CTCGATCTGA GGCACTTTCT AGTGTGTTAG 420 ATCAGGAGGA AGCTATGGAA ATTAAAGAAC ACCATCCAGA GGAGGGGTCT TCAGGGTCTG 480 AGGTGGAAGA AATCCCTGAG ACACCTTGTG AAAGTCAAGC AGAGGAACTC AAAGAAGAAA 540 ATATGGAGAG TGTTCCGTTG CACCTTTCTC TGACTGAAAC TCAGTCCCA 589
(2) INFORMATION FOR SEQ ID NO: 203:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 847 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 203:
GGCACGAGCG CAAGCTCCTG GCCGCCATCA ACGCGTTCCG CCAGGTGCGG CTGAAACACC 60
GGAAGCTCCG GGAACAAGTG AACTCCATGG TGGACATCTC CAAGATGCAC ATGATCCTGT 120 ATGACCTGCA GCAGAATCTG AGCAGCTCAC ACCGGGCCCT GGAGAAACAG ATTGACACGC 180
TGGCGGGGAA GCTGGATGCC CTGACTGAGC TGCTTAGCAC TGCCCTGGGG CCGAGCAGCT 240
TCCAGAACCC AGCCAGCAGT CCAAGTAGCT GGACCCACGA GGAGGAACCA GGCTACTTTC 300
CCCAGTACTG AGTCGTGGAC ATCGTCTCTG CCACTCCTGA CCAGCCTGAA CAAAGCACCT 360
CAAGTGCAAG GACCAAAGGG GGCCTGGCTT GGATGGGTTG GCTTGCTGAT GGCTGCTGCA 420 GGGGACGCTG GCTAAAGTGG GGAGGCCTTG GCCCACCTGA GGCCCCAGGT GGGAACATGG 480
TCACCCCCAC TCTGCATACC CTCATCAAAA ACACTCTCAC TATGCTGCTA TGGACGACCT 540
CCAGCTCTCA GTTACAAGTG CAGGCGACTG GAGGCAGGAC TCTTGCGTCC CTGGGAAAGA 600
GGGTACTAGG GCCCCGGATC CAGGATTCTG GGAGGCTTCA GTTACCGCTC GCCGAGCTGA 660
AGAACTGCGT ATGAGGCTGC GGCGGGCCTG GAGGTGCCGC CCCCTGGTGG GACAACAAAG 720 AGGACACCAT TTTTCCAGAG CTGCAGAGAG CACCTGGTGG GGAGGAAGAA GTGTAACTCA 780
CCAGCCTCTG CTCTTATCTT TCTAATAAAT GTTAAAGCCA GAAAAAAAAA AAAAAAAAAA 840
AAAAAAA 847
(2) INFORMATION FOR SEQ ID NO: 204:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 852 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 204:
ACAAACATAC TCGCAGGAAG GAGTCTCATG CTGCCCGCAG CATCAGCGCA ACNCNTGGCC 60
GCCATCAACG CGTTCCGCCA GCTGCGGCTG AAACACCGGA AGCTCCGGGA ACAAGTGAAC 120
TCCATGGTGG ACATCTCCAA GATGCACATG ATCCTGTATG ACCTGCAGCA GAATCTGAGC 180 AGCTCACACC GGGCCCTGGA GAAACAGATT GACACGCTGG CGGGGAAGCT GGATCCCCTG 240
ACTGAGCTGC TTAGCACTGC CCTGGGGCCG AGGCAGCTTC CAGAACCCAG CCAGCAGTCC 300
AAGTAGCTGG ACCCACGNAG GAGGAACCAG GCTACTTTCC CCAGTACTGA GCTGGTGGAC 360
ATNCGTCTCT TGCCACTCCN TGNACCCAGC CCTGAACAAA GCACCTCAAG TGCAAGGACC 420
AAAGGGGGCC CTGGCTTGGA GTGGGTTGGC TTGCTGATGG CTGCTGGAGG GGACGCTGGC 480
) TAAAGTGGGK AGGCCTTGGC CCACCTGAGG CCCCAGGTGG GAACATGGTC ACCCCCACTC 540
TGCATACCCT CATCAAAAAC ACTCTCACTA TGCTGCTATC GACGACCTCC AGCTCTCAGT 600
TACAAGTGCA GGCGACTGGA GGCAGGACTC CTGGGTCCCT GGGAAAGAGG GTACTAGGGG 660
CCCGGATCCA GGATTCTGGG AGGCTTCAGT TACCGCTGGC CGAGCTGAAG AACTGGGTAT 720
GAGCCTGGGG CGGGGCYGGA GGTGGCGCCC CCTGGTGGGA CAACAAAGAG GACACCATTT 780 TTCCAGAGCT GCAGAGAGCA CCTGGTGGGG AGGAAGAAGT GTAACTCACC AGCCTCTGCT 840
CTTATCTTTG TA 852
(2) INFORMATION FOR SEQ ID NO: 205:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 1354 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi ) SEQUENCE DESCRIPTION : SEQ ID NO : 205 :
GATTCGGCAC GAGGCTTGCT GGAGCAGGAG AAGTCTCTRG CCGGCTGGGC ACTGGTGCTG 60
GCASGARCTG GCATTGGACT CATGGTGCTG CATGCAGAGA TGCTGTGGTT CGGGGGGTGC 120
TCGGCTGTCA ATGCCACTGG GCACCTTTCA GACACACTTT GGCTGATCCC CATCACATTC 180
CTGACCATCG GCTATGGTGA CGTGGTGCCG GGCACCATGT GGGGCAAGAT CGTYTGCCTG 240 TGCACTGGAG TCATGGGTGT CTGCTGCACA G CCTGCTGG TGGCCGTGGT GGCCCGGAAG 300
CTGGAGTTTA ACAAGGCAGA GAAGCACGTG CACAACTTCA TGATGGATAT CCAGTATACC 360
AAAGAGATCA AGGAGTCCGC TGCCCGAGTG CTACAAGAAG CCTGGATGTT CTACAAACAT 420
ACTCGCAGGA AGGAGTCTCA TGCTGCCCGC AGGCATCAGC GCAANCTCCT GGCCGCCATC 480
AACGCGTTCC GCCAGGTGCG GCTGAAACAC CGGAAGCTCC GGGAACAAGT GAACTCCATG 540 GTGGACATCT CCAAGATGCA CATGATCCTG TATGACCTGC AGCAGAATCT GAGCAGCTCA 600
CACCGGGCCC TGGAGAAACA GATTGACACG CTGGCGCGGA AGCTGGATGC CCTGACTGAG 660
CTGCTTAGCA CTGCCCTGGG GCCGAGGCAG CTTCCAGAAC CCAGCCAGCA GTCCAAGTAG 720
CTGGACCCAC GAGGAGGAAC CAGGCTACTT TCCCCAGTAC TGAGGTGGTG GACATCGTCT 780
CTGCCACTCC TGANCCCAGC CCTGAACAAA GCACCTCAAG TGCAAGGACC AAAGGGGGCC 840 CTGGCTTGGA GTGGGTTGGC TTGCTGATGG CTGCTGGAGG GGACGCTGGC TAAAGTGGGK 900
AGGCCTTGGC CCACCTGAGG CCCCAGGTGG GAACATGGTC ACCCCCACTC TGCATACCCT 960 CATCAAAAAC ACTCTCACTA TGCT CTATG GACGACCTCC AGCTCTCAGT TACAAGTGCA 1020 GGCGACTGGA GGCAGGACTC YTGGGTCCCT GGGAAAGAGG GYACTAGGGG CCCGGATCCA 1080 GGATTCTGGG AGGCTTCAGT TACCGCTGGC CGAGCTGAAG AACTGGGTAT GAGGCTGGGG 1140 CGGGGCTGGA GGTGGCGCCC CCTGGTGGGA CAACAAAGAG GACACCATTT TTCCAGAGCT 1200 GCAGAGAGCA CCTGGTGGGG AGGAAGAAGT GTAACTCACC AGCCTCTGCT CTTATCTTTG 1260 TAATAAATGT TAAAGCCAGA AAAAAATAAA AAAAAAAAAA AAAAAACTCG AGGGGGGCCC 1320 AGACCCAATC TCCCTATAGT AAGNCGCCNN ANAN 1354
(2) INFORMATION FOR SEQ ID NO: 206:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1378 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 206:
TCCCCAGGTG CACAGCCAGG GCCCTCCTGT CTCCAGGAGA ATTCACAGCT GGTGTGGGAC 60
TCAGCCCCTA GNCCATTCAA AGCCTTAATG TTGTAATCAT ATCTTACGTG TTCAAGACCT 120
GACTGGAGAA ACAAAATGTG CAATAACGYG AATTTTATCT TAGAGATCTG TGCAGCCTAT 180
TTCTGTCACA AAAGTTATAT TGTCTAATAA GAGAAGTCTT AATGGCCTCT GTGAATAATG 240
TAACTCCAGT TACACGGTGA CTTTTAATAG CATACAGTGA TTTGATGAAA GGACGTCAAA 300
CAATGTGGCG ATGTCGTGGA AAGTTATCTT TCCCGCTCTT TGCTGTGGTC ATTGTGTCTT 360
GCAGAAAGGA TGGCCCTGAT GCAGCAGCAG CGCCAGCTCT ANATAAAAAA TAATTCACAC 420
TATCAGACTA GCAAGGCACT AGAACTGGAA AAGACCACAG AAAACAAAGA ATCCAACCCT 480
TTCATCTTAC AGGTGAACAA ACTGTGATGA TGCACATGTA TGTGTTTTGT AAGCTGTGAG 540
CACCGTAACA AAATGTAAAT TTGCCATTAT TAGGAAGTGC TGGTGGCAGT GAAGAAGCAC 600
CCAGGCCACT TGACTCCCAG TCTGGTGCCC TGTCTACACC AGACAACACA GGAGCTGGGT 660
CAGATTCCCC TCAGCTGCTT AACAAAGTTC CTCGAACAGA AAGTGCTTAC AAAGCTGCCT 720
TCTCGGATAC TGAAAGGTCG AGTTTTCTGA ACTGCACTGA TTTTATTGCA GTTGAAAAAA 780
AAAAAAAGCT ATTCCAAAGA TTTCAAGCTG TTCTGAGACA TCTTCTGATC GCTTTACTTC 840
CTGAGAGGCA ATGTTTTTAC TTTATGCATA ATTCATTGTT GCCAAGGAAT AAAGTGAAGA 900
AACAGCACCT TTTAATATAT AGCTCTCTCT GGAAGAGACC TAAATTAGAA AGAGAAAACT 960
GTGACAATTT TCATATTCTC ATTCTTAAAA AACACTAATC TTAACTAACA AAAGTTCTTT 1020
TGAGAATAAG TTACACACAA TGGCCACAGC AGTTTGTCTT TAATAGTATA GTGCCTATAC 1080
TCATGTAATC GGTTACTCAC TACTGCCTTT AAAAAAAAAA ACCAGCATAT TTATTGAAAA 1140
CATGAGACAG GATTATAGTG CCTTAACCGA TATATTTTGT GACTTAAAAA ATACATTTAA 1200
AACTGCTCTT CTGCTCTAGT ACCATGCTTA GTGCAAATGA TTATTTCTAT GTACAACTGA 1260
TGCTTGTTCT TATTTTAATA AATTTATCAG AGTGAAAAAA AAAAAAAAAA AAAAAAAAAA 1320
AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAGAA NAAAANAA 1378
( 2 ) INFORMATION FOR SEQ ID NO : 207 :
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1166 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 207:
AANCCACTGC ANTTTAAACC CCCTCCCCTC CAAGAAAGTT CACAACCGGC CATGGATGAC 60
CCTCATTTTA GATGGGCCNC AATATTTAAG ATGGACTGRG GMCCCCARAG ACTGACCCTT 120
GAAAGGGGGA CTCAGAAGAA AGATCCTTGA CATTGCCMAA CATGCTGGGC TTGTCCAACA 180
CAGTGATGCG GCTCATCGAG AARCGGGCTT TCCMAGGACA AGTACTTTAT GATAGGTGGG 240
ATGCTGCTGA CCTGTGTGGT CATGTTCCTC GTGGTGCAGT ACCTGACATG AGCCAGCCAC 300
GCTCAGTGGC TGAACAGCAT TCCCACAGCC TGCAAGTGTG TGTGTGTGTG AAAGAGAGAG 360
GGGGCCCAGA GGCCGCCTTT TGAAATGTTT GCCTGTCTGA ACTGTGAAGA CACTTGGGAG 420
TGATTGTGGT CTAATTTCCA ACCTGCTCTC TTTTCTGTGA CATCTTGGAG GGGGAGCTAG 480
TGCCAMCACC ATGCGCGGTG CTTAGGAAAT GAAAGAAGTC CCGGGTCTGT CTCTCTCACT 540
CTCGCTCTCA MTGGGGGAGG GAAAGAATGG CTTTGGTGGC TTTGTTCACA CAGCTGATGC 600
GTGSCCTGGG AAGGTGTCCA CAGTGAGCCC TGTGTCCAGG ACTGTCCACN ACGGTTCACA 660
CCTTGTCACC ATCAGGCCTT TCTGGCTCCT GATAGGGTGG AGCAAAAGTG GAAAGGAAAG 720
GAAAGAGGCY TTTTCTCACA GCCATTATAT TAAATAGTAG GTCGATTCAC ATCYTCGTGC 780
TCCTGGCCAC CCTCCCCTGT GCCTCAGTGA CATGTAGATG ACTGACTGCC AATACTTGTC 840
ACCATTCCCT GGAAGCAGCT ACCTAGGGGA AACAAGATGT AGTGCTATTG CCGATAACAA 900
GTAAGATTTT CCACACTACA GCTGCGTGTT TCTCTTTTCT AAAGTGAGGC CAGTGTTATT 960
TCCCGGGAGT GTTCAGTCTT GACCCTAGTC ACTGATTTTT TCTAGTTGTT AATAGAGTGG 1020
TTGGGCTTTT AAGGTTCAGA GACTGTGGGC TTGGGCACCT GCGCCCAGGG STTTTGTGGG 1080
GGCCTTTCCC CCTTAGRAAA GTAGCTTTTA GGGGCAAAGA TTTGTTGATT TTCCCCATTA 1140
CAGTCTTCAG CTCNAGGGTT TTAAAA 1166
(2) INFORMATION FOR SEQ ID NO: 208:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 697 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi ) SEQUENCE DESCRIPTION : SEQ ID NO : 208 : TACTTCTAGG ATTATAAGGA ATTAACATTG AGATGACATT TCCATTTGAG AAGGAAAATA 60 GTTCCTTTCA GTGCCTTTTA TTTGATTCCT GGAGAGAGCA GACTCGCACS AACATTCAAC 120 CCCAGCGCTG ATATGACAGT AATCCTCAGA GGCAGAGCCC AGCACAAAAC AGCAATGCTA 180 GAAAGTTACA ATTGGAAAGT TTCCTGCCAG CTTCGGGAAT GACACTGCAA AGCTGATGCC 240 AGAAACTGCC AGRGTAATTC TCCTCATTAC TGCTCTACCC ACCCACTTTC AGCTCCCCAA 300 ATTAACTAGT GCAGTTGACT AATTCTCTTT ACCTTTATCA TTTARGGTGA RGCATTGCAC 360 AAAAACTCTC GACTTTGCCA TATAAGGGCT GTGGTTCTCT GTGGTCCCCT GGATAAGAGG 420 CATCACCATT ATCTGGAAAC ATGCAGTAAA TGCAGATTNT TCATCTTCTC CCCAGACCTC 480 CTGAGTTAGA AATTCACAAG TTCTCCAGGT GATCTCATAC ATGCTAAAGT TTGAGAACCA 540 TTGAGTAAAG TTAATGCATT AAGAAGAGAT TAGATAGGGA TGGTGGCGTA TCTTCCTACA 600 GTTTCCCTGT TAACAAGAAA GTCAGAGGTC AGTTGATCAG ACATTAGATT ATTTATTGCT 660 AAAACTAAAA AAAATTAAAA AAAACTCGAG GGGGGCC 697
(2 ) INFORMATION FOR SEQ ID NO : 209 :
( i) SEQUENCE CHARACTERISTICS :
(A) LENGTH : 932 base pairs
(B) TYPE : nucleic acid (C) STRANDEDNESS : double
(D) TOPOLOGY : linear
(xi ) SEQUENCE DESCRIPTION : SEQ ID NO : 209 : CGTGAGTCAC CTCTCTATAG TGGGCGTGGC CGAGGCCGGG GTGACCCTGC CGAAGCCTCC 60
GCTGCCAGAA ACCATGTTCA AGGTAATTAA AAGGTCCGTG GGGCCAGCCA GCCTGAGCTT 120
GCTCACCTTC AAAGTCTATG CAGCACCAAA AAAGGACTCA CCTCCCAAAA ATTCCGTGAA 180
GGTTGATGAG CTTTCACTCT ACTCAGTTCC TGAGGGTCAA TCGAAGTATG TGGAGGAGGC 240
AAGGAGCCAG CTTGAAGAAA GCATCTCACA GCTCCGACAC TATTGCGAGC CATACACAAC 300 CTGGTGTCAG GAAACGTACT CCCAAACTAA GCCCAAGATG CAAAGTTTGG TTCAATGGGG 360
GTTAGACAGC TATGACTATC TCCAAAATGC ACCTCCTGGA TTTTTTCCGA GACTTGGTGT 420
TATTGGTTTT GCTGGCCTTA TTGGACTCCT TTTGGCTAGA GCTTCAAAAA TAAAGAAGCT 480
AGTGTATCCG CCTGGTTTCA TGGGATTAGC TGCCTCCCTC TATTATCCAC AACAAGCCAT 540
CGTGTTTGCC CAGGTCAGTG GGGAGAGATT ATATGACTGG GGTTTACGAG GATATATAGT 600 CATAGAAGAT TTGTGGAAGG AGAACTTTCA AAAGCCAGGA AATGTGAAGA ATTCACCTGG 660
AACTAAGTAG AAAACTYCAT GYTCTGCCAT CTTAATCAGT TATRGGTAAA CATTGGAAAC 720
TCCATAGAAT AAATCAGTAT TTCTACAGAA AAATGGCATA GAAGTCAGTA TTGAATGTAT 780
TAAATTGGCT TTCTTCTTCA GGAAAAACTA GACCAGACCT CTGTTATCTT CTGTGAAATC 840
ATCCTACAAG CAAACTAACC TGGAATCCCT TCACCTAGAG ATAATGTACA AGCCTTAGAA 900 CTCCTCATTC TCATGTTGCT ATTTATGTAC CT 932
(2) INFORMATION FOR SEQ ID NO: 210:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 661 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 210: GTCATTCTTT AAATAAAAGC TTTCCTGTTT AAAGCTTTTC AAAGGAGCAG ACCACCTTGA 60
AGATTCCCCC TAGGGTTGAT ATGTGTCTAA TTCATTTTAT AAAAATTATT CTTGTCTTCA 120
TTTTAAAGCT TTGGCTATAT AGTCAGAAAT GTCCTAAATA ACAAACTATT TTGTATTTAA 180
TTTAGGGAAG ACTAAAGGGA AGAAAAATGA AAACTCAGTC TTTATGTAAG CTCCAAGGAT 240
ATTAGGGCTT AAAGGGCTTT TCTAGTTTTA TGAGAATTTG TACTACTGAT TTTTATATAT 300 TCCTGTTTTT GAGATGAACA GATCTCTGGG GAAATTGTTG AGTTACAATG GCATTTCACT 360
GTGATCCCTC TCAAGCTCAG ATCAGTTCTA TAACCCAATG ACAACCTGTC TCTTTGGTTT 420
ACTGTCCTGT GAAATGTCAG CTCAAGTTTC CCAGAAGTCG TGTCTTTATG ATGAGTCAGA 480
GTGCTTTTCC TCCGTCGGAC AGTTGCTGGC CCTCTTAATT TTGGTGTATC TGCTTCCAAG 540
TATCTAAACC TCCAGTCTGA TCTGTATATG CTATCCTAAC TGTTAATTGT ATTATTGATT 600 ATGTTGATTA TCTTCCTTCA AGCTTCATAC TTTTCAATTT GATAGAAATA AAGTTTTTTT 660
C 661
(2) INFORMATION FOR SEQ ID NO: 211:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 592 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 211:
GAAACTGACA TTGTTAAACA CACTAAAACA GAAGTACTTA CCTCTTGAAG ATTTAATATA 60 TAATGGTTGA CATGATACAT GTACATGAAT GGAATGACCA GATGCTTATG GTCTACATTT 120
TCCTTTATCC TGTTAGTATT ACCTTCCTTA ATCTTTGTTC CTTAACATGC TAAATTCCTC 180
TTCAGTGTTT ATTTTCTAGT GACAGAATGC TAACATTTCT TACACCCTGG CAGAAGGGAG 240 AGAAATGTGT TTTGGGGTGG GTAACTAAAT TTTTCAGTGA AATATCATAA GATGAGAATG 300
GAAAGAGGGA GACACAAAGA GTTATAACAA AAAAACAATG GTTTTTTTAG CCATTTGACT 360
GGCTCTTTAA ATAGTCTACA AGACATTCAC GTTNAACATC ACTTTTAGTG AAATAAAATG 420
TGCCATACTA GTATGTGCTT CAAAAGGGCA AATGTGCTTT AGTGCCCTAA GGCTAAATTT 480
TGGTCATTTC ACATCAGAGA TGTTGTAAGT ATTGCACTTA ATACGCACCT ATTTCTCAAT 540 AGTGNTATTT TTTTGGCTAG CATTTNCTTT ACCACTAACC TTGTTGGATA GC 592
(2) INFORMATION FOR SEQ ID NO: 212:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 938 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 212: TGGAGTGGCT TTCCAGCTGA ATGAATCCTA TGTCTCGCGT GCAGGTGGTT GGTTTTCAAT 60
GTTCTTSCTA ATTTTTTTCC TATTGGCTCT TGGGAGTTTN CTTTGTTTGC TCCTGTGTTT 120
GCCCAGCTTT AATAAAACCA GGCGCAAACA AAAACCATAG CATTCTGAAA CAATAGGGGG 180
CCCACATTGG ACCCAGTATG TCACTTTAAT GGACTTCAAG AAAAAATCTG AATCGGAAAA 240
TGACACTAGG AATCTATACT CCACACATTT TATGCCATAT AATGGTGTGT TTTCTTAATT 300 TTCTTTCTTG TGGCGAAATG TGGCTTTCAA ATTAAAATGM CCTTTTCTTC TTKGAAACTT 360
TTTGTTTKGA CTKGTATAAT TAAGGGTTTG GAAAGATTCA TAATTMTGAG AGAGGTTTGC 420
AACCAGGAGA TACAAAGAAG TCTCAGTAGT AATCTTGTTC ATGTGCTTTT ACAGCCAGCT 480
ACATTTAAGR ATGTATTAGT TACAGAAATT ATATGTCTGT GTATCTCTCT CTACTCAATA 540
AAGTACATGC CTCCACATAA TGCGCTGCTC TCCATCTCGG CAAATACTGG CCAAGTCCCT 600 TTATGACAGG CACACAGAAA CCATAGCATG GTCTGGCTTT CAGAAAATGC CTCTCATCTT 660
TCCTGGAACC TTATTTTGCT AAATGTCTGT TTTCTTGTGA TTTGTTGTAC CTCACAGCAC 720
CATTGTGACC ATGGTGATGC CTCATTTGCA TGATATGTAC CTTGTGTTTA ATGTGAAATA 780
CATTTTCATT GAAGAGTCTG ATGACTTGCT AGCGTTTTAT TTTTTCTGTA AGCTCAATGT 840
GCTGAAACCA AACCAGGCTT TTAAAAACCT GTGTAGAAGA AAACCAAAAA ATCCTGTGTG 900 GGTGTCCTTT CCCTGTCAAA CTCATTAAAA ATTCCTTT 938
(2) INFORMATION FOR SEQ ID NO: 213:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1079 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 213: AGCCTGCCGG GAGAGTGGTG GCATCTRARA GGCTGGTCGT GGACTGTGGT TGGGGGAGGT 60
GGGAGCTGTT TTAACCGTGT GCCCCCTCTC CTGTGCCKGC GTGGGCATCC CCCGGGGCAG 120
TGGAACGCGG GCGCTCCTCC AGCTTCCGAG TCCAGCCAGC CTGGGCGCGG GGCGCGCCCC 180
CGAGACACCC GAGGAGTCCG TTCCTCCCTG GTTACGTGGA CTGTGGAGCT GGTCTCTTGT 240
GGCTCAGCGC CGTGCGGAGG TTGAAGCGTA CCTGCGGAGG TCGCACCAGG GGCGTGAGGA 300 GGAGGAGGAA GGGCATGAGC CGAGCTTGAG GAATCCGTCY TCCAAACTCT ACACTCAAGG 360
RTGCMCTGCG CAACTCTGGT GGCGATGGGC TGGGGCAGAT GTCCTTGGAG TTCTACCAGA 420
AGAAGAAGTC TCGCTGGCCA TTCTCAGACG AGTGCATCCC ATGGGAAGTG TGGACGGTCA 480
AGGTGCATGT GGTAGCCCTC GCCACGGAGC AGGAGCGGCA GATCTGCCGC GAGAAGGTGG 540
GTCAGAAACT CTGCGAGAAG ATCATCAACA TCGTGGAGGT GATGAATCGG CATGAGTACT 600 TGCCCAAGAT GCCCACACAG TCGGAGGTGG ATAACGTGTT TGACACAGGC TTGCGGGACG 660
TGCAGCCCTA CCTGTACAAG ATCTCCTTCC AGATCACTGA TGCCCTGGGC ACCTCAGTCA 720
CCACCACCAT GCGCAGGCTC ATCAAAGACA CCCTTGCCCT CTGAGCGTCG CTGGATCTCT 780
GGGAGCTCCT TGATGGCTCC CAGACCTTGG CTTTTGGGAA TTGCACTTTT GGGCCΓTTGG 840
GCTCTGGAAC CTGCTCTGGG TCATTGGTGA GACTTGGAAG GGGCAGCCCC CGCTGGCTTC 900 TT∞TTTTCT GGTTGCCAGC CTCAGGTCAT CCTTTTAATC TTTGCTGACG GTTCAGTCCT 960
GCCTCTACTG TCTCTCCATA GCCCTGGTGG GGTCCCCCTT CTTTCTCCAC TGTACAGAAG 1020
AGCCACCACT GCGATGGGGA ATAAAGTTGA GAACATGAGT TTGGGCTGAA AAAAAAAAA 1079
(2) INFORMATION FOR SEQ ID NO: 214:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 3791 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 214:
TCAAGCAGGC GCTCTTGGCT CGGCGCGGCC CGCTGCAATC CGTGGAGGAA CGCGCCGCCG 60
AGCCACCATC ATGCCTGGGC ACTTACAGGA AGGCTTCGGC TGCGTGGTCA CCAACCGATT 120
CGACCAGTTA TTTGACGACG AATCGGACCC CTTCGAGGTG CTGAAGGCAG CAGAGAACAA 180 GAAAAAAGAA GCCGGCGGGG GCGGCGTTGG GGGCCCTGGG GCCAAGAGCG CATCAGGGCC 240
GCGGCCCAGA CCAACTCCAA CGCGGCAGGC AAACAGCTGC GCAAGGAGTC CCAGAAAGAC 300
CGCAAGAACC CGCTGCCCCC CAGCGTTGGC GTGGTTGACA AGAAAGAGGA GACGCAGCCG 360
CCCGTGGCGC TTTAAGAAAG AAGGAATAAG ACGAGTTGGA AGAAGACCTG ATCAACAACT 420
TCAGGGTGAA GGGAAAATAA TTGATAGAAG ACCAGAAAGG CGACCACCTC GTGAACGAAG 480 ATTCGAAAAG CCACTTGAAG AAAAGGGTGA AGGAGGCGAA TTTTCAGTTG ATAGACCGAT 540
TATTCACCGA CCTATTCGAG GTCGTGGTGG TCTTGGAAGA GGTCGAGGGG GCCGTGGACG 600
TGGAATGGGC CGAGGAGATG GATTTGATTC TCGTGGCAAA CGTGAATTTG ATAGGCATAG 660
TGGAAGTCAT AGATCTTCTT TTTCACATTA CAGTGGCCTG AAGCACGAGG ACAAACGTGG 720
AGGTAGCGGA TCTCACAACT GGGGAACTGT CAAAGACGAA TTAACTGACT TGGATCAATC 780 AAATCTCACT GAGGAAACAC CTGAAGGTGA AGAACATCAT CCAGTGGCAG ACACTGAAAA 840
TAAGGAGAAT GAAGTTGAAG AGGTAAAAGA GGAGGGTCCA AAAGAGATGA CTTTGGATGA 900
GTGGAAGGCT ATTCAAAATA AGGACCGGGC AAAAGTAGAA TTTAATATCC GAAAACCAAA 960
TGAAGGTGCT GATGGGCAGT GGAAGAAGGG ATTTGTTCTT CATAAATCAA AGAGTGAAGA 1020
GGCTCATGCT GAAGATTCGG TTATGGACCA TCATTTCCGG AAGCCAGCAA ATGATATAAC 1080
GTCTCAGCTG GAGATCAATT TTGGAGACCT TGGCCGCCCA GGACGTGGCG GCAGGGGAGG 1140
ACGAGGTGGA CGTGGGCGTG GTGGGCGCCC AAACCGTGGC AGCAGGACCG ACAAGTCAAG 1200
TGCTTCTGCT CCTGATGTGG ATGACCCAGA GGCATTCCCA GCTCTGGCTT AACTGGATGC 1260
CATAAGACAA CCCTGGTTCC TTTGTGAACC CTTCTGTTCA AAGCTTTTGC ATGCTTAAGG 1320
ATTCCAAACG ACTAAGAAAT TAAAAAAAAA AAGACTCTCA TTCATACCAT TCACACCTAA 1380
AGACTGAATT TTATCTGTTT TAAAAATGAA CTTCTCCCGC TACACAGAAG TAACAAATAT 1440
GGTAGTCAGT TTTGTATTTA GAAATGTATT GGTAGCAGGG ATGTTTTCAT AATTTTCAGA 1500
GATTATGCAT TCTTCATGAA TACTTTTGTA TTGCTGCTTG CAAATATGCA TTTCCAAACT 1560
TGAAATATAG GTGTGAACAG TCTGTACCAG TTTAAAGCTT TCACTTCATT TCTCTTTTTT 1620
AATTAAGGAT TTAGAAGTTC CCCCAATTAC AAACTGGTTT TAAATATTGG ACATACTGGT 1680
TTTAATACCT GCTTTGCATA TTCACACATG GTCAACTGGG ACATGTTAAA CTTTGATTTG 1740
TCAAATTTTA TGCTGTGTGG AATACTAACT ATATGTATTT TAACTTAGTT TTAATATTTT 1800
CATTTTTGGG GAAAAATCTT TTTTCACTTC TCATGATAGC TGTTATATAT ATATGCTAAA 1860
TCTTTATATA CAGAAATATC AGTACTTGAA CAAATTCAAA GCACATTTGG TTTATTAACC 1920
CTTGCTCCTT GCATGGCTCA TTAGGTTCAA ATTATAACTG ATTTACATTT TCAGCTATAT 1980
TTACTTTTTA AATCCTTGAG TTTCCCATTT TAAAATCTAA ACTAGACATC TTAATTGGTG 2040
AAAGTTCTTT AAACTACTTA TTGTTGGTAG GCACATCGTG TCAAGTGAAG TAGTTTTATA 2100
GGTATGGGTT TTTTCTCCCC CTTCACCAGG GTGGGTGGAA TAAGTTGATT TGGCCAATCT 2160
GTAATATTTA AACTGTTCTG TAAAATAAGT GTCTGGCCAT TTGGTATGAT TTCTCTGTGT 2220
GAAAGGTCCC AAAATCAAAA TGGTACATCC ATAATCAGCC ACCATTTAAC CCTTCCTTGT 2280
TCTAAAACAA AAACCAAAGG GCGCTGGTTG GTAGGGTGAG GTGGGGGAGT ATTTTAATTT 2340
TTGGAATTTC GGAAGCAGAC AGCTTTACTT TGTAAGGTTG GAACAGCAGC ACTATACATG 2400
AAATATAAAC CAAAAACCTT TACTGTTTCT AAATTTCCTA GATTGCTATT ATTTGGTTGT 2460
AAGTTGAGTA TTCCACAGAA AGTGGTAATT ATCTCTTCTC TCTTCCTCCA TTAGAAAATT 2520
AGGTAAATAA TGGATTCCTA TAATGGGAGC ATCACCACTT ATTAAAACAC ACATAGAATG 2580
ATGAATTAAA AAAGTTTTCT AGCATTGTCT TTTATTCTGC CACATTTATT GATAAACAGT 2640
GAAGGAATTT TTAAAAAATT TTTAAGAATT GTTTGTCACG TCATTTTTAG AAATGTTCTA 2700
CCTGTATATG GTAATGTCCA GTTTTAAAAA TATTGGACAT CTTCAATCTT AAACATTTCT 2760
ATTTAGCTGA TTGGTTCTCA CATATACTTC TAAAAGAAAC TTTTATGTTA TAAGAGTTAC 2820
TTTTTGGATA AGATTTATTA ATCTCAGTTA CCTACTATTC TGACATTTTA GGAAGGAGGT 2880
AATTGTTTTT AATGATGGAT AAACTTGTGC TGGTGTTTTG GATCTTATGA TGCTGAGCAT 2940
GTTCTGCACT GGTGCTAATG TCTAATATAA TTTTATATTT ACACACATAC GTGCTACCCA 3000
GAGATTAATT TAGTCCATAT GAACTATTGA CCCΛTTGTTC ATTGAGACAG CAACATACGC 3060
ACTCCTAAAT CAGTGTGTTT AGACTTTTCA AGTATCTAAC TCATTTCCAA ACATGTACCA 3120
TGTTTTATAA ACCTCTTGAT TTCCAGCAAC ATACTATAGA AAACACCTGC TACTCAAAAC 3180
ACAACTTCTC AGTGTCATCC ATTGCTGTCG TGAGAGACAA CATAGCAATA TCTCGTATGT 3240
TCCAAGCTTT CAAGATAGCC TGAACTTAAA AAGTTGGTGC ATTAGTTGTA TCTGATGGAT 3300
ATAAATTTGC CTCCTAGTTC ACTTTGTGTC AAGAGCTAAA ACTGTGAACC TAACTTTCTC 3360
TTATTGGTGG GTAATAACTG AAAATAAAGA TTTATTTTCA TGCTCACTTC TTAAAAGTCA 3420
TAAAAACAAT CAAATAGGRT CATGTTTATT GTCATGTGTT TCCTGGKTTC TGACCTGTGT 3480
GCACACCCCT GTGTGTTTAT AATTTTTAAA TTGAATTTTA TATGGGGTTT TTATTTGCTA 3540
AAAACCAGGC TGTTGAATCA CATTTGGGAA GGGTACTTAT CTTAATGACT AATGACTTAA 3600
TTGGGAAAGT TGAATTCTTG TAAAATACAA AATCCAAGGA CTTCTTGGGA TTTAATCTAA 3660
TTGTCACTTC NTTAGGCAGA TNCACTTTTT TGGATAATGG AAAGTTAAGC ATACCGAATG 3720
CTACTTTTGG TTGACAAACG GGCCTAATAG TCCCCGGGGA AATCCCTAAC NGGTAAGGNT 3780
CCCAAGTATG G 3791
(2) INFORMATION FOR SEQ ID NO: 215:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1334 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 215: CAGTGCTCGC TCCTGCTCGG GGCGCTGCGG CCCCGGGCGT CGCCATGACC AGTGAGCTGG 60 ACATCTTCGT GGGGAACAGA CCCTTATCGA CGAGGACGTG TATCGCCTCT GGCTCGATGG 120 TTACTCGGTG ACCGACGCGG TGGCCCTGCG GGTGCGCTCG GGAATCCTGG AGCAGACTGG 180 CGCCACGGCA GCGGTGCTNC AGAGCGACAC CATGGACCAT TACCGCACCT TCCACATGCT 240 CGAGCGGCTG CTGCATGCGC CGCCCAAGCT ACTGCACCAG YTCATCTTCC AGATTCCGCC 300 CTCCCGGCAG GCACTACTCA TCGAGAGGTA CTATGCCTTT RATGAGGCCT TTGTTCGGGA 360
GGTGCTCGGC AAGAAGCTGT CCAAAGGCAC CAAGAAAGAC CTGGATGACA TCAGCACCAA 420
AACAGGCATC ACCCTCAAGA GCTGCCGGAG ACAGTTTGAC AACTTTAAAC GGGTCTTCAA 480
GGTGGTAGAG GAAATGCGGG GCTCCCTGGT GGACAATATT CAGCAACACT TCCTCCTCTC 540
TGACCGGTTG GCCAGGGACT ATGCAGCCAT ∞TCTTCTTT GCTAACAACC GCTTTGAGAC 600
AGGGAAGAAA AAACTGCAGT ATCTGAGCTT CGGTGACTTT GCCTTCTGCG CTGAGCTCAT 660
GATCCAAAAC TGGACCCTTG GAGCCGTCGA CTCACAGATG GATGACATGG ACATGGACTT 720
AGACAAGGAA TTTCTCCAGG ACTTGAAGGA GCTCAAGGTG CTAGTGGCTG ACAAGGACCT 780
TCTGGACCTG CACAAGAGCC TGGTGTGCAC TGCTCTCCGG GGAAAGCTGG GCGTCTTCTC 840
TGAGATGGAA GCCAACTTCA AGAACCTGTC CCGGGGGCTG GTGAACGTGG CCGCCAAGCT 900
GACCCACAAT AAAGATGTCA GAGACCTGTT TGTGGACCTC GTGGAGAAGT TTGTGGAACC 960
CTGCCGCTCC GACCACTGGC CACTCAGCGA CGTGCGGTTC TTCCTGAATC AGTATTCAGC 1020
GTCTGTCCAC TCCCTCGATG GCTTCCGACA CCAGGCCTCT GGGACCGCTA CATGGGCACC 1080
CTCCGCGCCT GCCTCCTGCG CCTGTATCAT GACTGAGGTG CCTCCCAACG CTCCGCCCAC 1140
GCTGACAATA AAGTTGCTCT GAGTTTGGAG ACTGGTCCTC GCTCCGGGGA GCAAGTGGGG 1200
GGCGTGCAGA TGTGCCTGTG TCTGTCTCTG AGCACCTGGT GTCCGTGTAC AAGGATGGAT 1260
GTGTNCNGTG GCTCCTTGGG AACTGAGACA TATCTCAGGG AATGGTGTCT GTGCTCAGCC 1320
CATCCACCAG AAGA 1334
(2) INFORMATION FOR SEQ ID NO: 216:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1511 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO : 216: GTGGCGGGGA TGCTGCGAGG GGGTCTCCTG CCCCAGGCGG GCCGGCTGCC TACCCTCCAG 60 ACTGTCCGCT ATGGCTCCAA GCCTGTTACC CGCCACCGTC GTGTGATGCA CTTTCAGCGG 120 CAGAAGCTGA TGGCTGTCAC TGAATATATC CCCCCGAAAC CAGCCATCCA CCCATCATGC 180 CTGCCATCTC CTCCCAGCCC CCCACAGGAG GAGATAGGCC TCATCAGGCT TCTCCGCCGG 240 GAGATAGCAG CAGTTTTCCA GGACAACCGA ATGATAGCCG TCTCCCAGAA TGTCGCTCTG 300 AGTGCAGAGG ACAAGCTTCT TATGCGACAC CAGCTGCGGA AACACAAGAT CCTGATGAAG 360
RTCTTCCCCA ACCAGGTCCT GAAGCCCTTC CTGGAGGATT CCAAGTACCA AAATCTGCTG 420
CCCCTTTTTG TGGGGCACAA CATCCTGCTC GTCAGTGAAG AGCCCAAGGT CAAGGAGATG 480
GTACGGATCT TAAGGACTGT GCCATTCCTG CCGCTGCTAG GTGGCTGCAT TGATGACACC 540
ATCCTCAGCA GGCAGGGCTT TATCAACTAC TCCAAGCTCC CCAGCCTGCC CCTGGTGCAG 600
GGGGAGCTTG TAGGAGGCCT CACCTGCCTC ACAGCCCAGA CCCACTCCCT GCTCCAGCAC 660
CAGCCCCTCC AGCTGACCAC CCTGTTGGAC CAGTACATCA GAGAGCAACG CGAGAAGGAT 720
TCTGTCATGT CGGCCAATGG GAAGCCAGAT CCTGACACTG TTCCGGACTC GTAGCCAGCC 780
TGTTTAGCCA GCCCTGCGCA TAAATACACT CTGCGTTATT GGCTGTGCTC TCCTCAATGG 840
GACATGTGGA AGAACTTGGG GTCGGGGAGT GTCTTTGTCA CTTCGTTTTC ACTAGTAATG 900
ATATTGTCAG GTATAGGGCC ACTTGGAGAT GCAGAGGATT CCATTTCAGA TGTCAGTCAC 960
CGGCTTCGTC CTTAGTTTTC CCAACTTGGG ACGTGATAGG AGCAAAGTCT CTCCATTCTC 1020
CAGGTCCAAG GCAGAGATCC TGAAAAGATA GGGCTATTGT CCCCTGCCTC CTTCGTCACT 1080
GCCTCTTGCT GCACGGGCTC CTGAGCCACC CCCTTGGGGC ACAACCTGCC ACTGCCACAG 1140
TAGCTCAACC AAGCAGTTGT GCTGAGAATG GCACCTGGTG AGAGCCTGCT GTGTGCCAGG 1200
CTTTGTGCTG AGTGCTGTAC ATGTATTAGT TCCTTTACTG CTGACCACAT TGTACCCATT 1260
TCACAGAGAA GGAGCAGAGA AATTAAGTGG CTTGCTCAAG GTCATGCAGT TAGTAAGTGG 1320
CAGAACAGGG ACTTGAACCA AGCCCTCTGC TCTGAAGACC GCGTCCTGAA TTTCTTCACT 1380
AGAGCTTCCT CATCAGGTTA CCCAGAAGTG GGTCCCATCC ACCATCCAGG TGTGCTTGGA 1440
TGTTAGTTCT CCACCCTCGA GGTGTACGCT GTGAAAAGTT TGGGAGCACT GCTTTATAAT 1500
AAAATGAAAT A 1511
(2) INFORMATION FOR SEQ ID NO: 217:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 642 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 217: AGGCCTTACT TTTCCTCCCA CAAAGGAGTC GCAGCCACGC TAGCTCTGAC TTGCCACTGT 60 GACAAAGTTC ACGTAGCAGG TCTAGGCAAA GACTGGGCAA TTCAGCAGAG GAGACGGACC 120 TGTGAGTCTG ACCRYGAGSC GGRCCCCTTC ACCTTGGCTG GGCTGGTCCT GCTCCTTAGG 180 TTTTGTCAGG TTGTCCTTGT TTGGATCCCT CAACTAGGTC ATAAGCACTG GAGGGGGATG 240
ACCCGCCTTG GACGTGTTTC TTTAACCTCA TCCATATAAT AGGGCCGTGG GATGGTTGTA 300
GAGGTAAAGC AGGATGATCG TGTTTTAAGA CCAGAGCTTG GGACCAGGGC TCCTACACCT 360
AATTTTCTCT CCTGGTAGCT GAACAAAGGT CTAAATTAGC TTAACAAAAG AACAGGCTGC 420
CGTCAGCCAG AGTTCTGAAG GCCATGCTTT CAGTTTCCCT TGTTGACAAT TGCTCTCCAG 480 TTCCTATGAA AGCACAGAGC CTTAGGGGGC CTGGCCACAG AACACAACCA TCTTAGGCCT 540
GAGCTCTCAA CAGCAGGGGG TTGTGTGTCT GTTCTGTTTC TCTGCTTGCC GAACTTTCTC 600
AATAAACCCT ATTTCTTATT TTATATTTAC GTNGGTGCTG GG 642
(2) INFORMATION FOR SEQ ID NO: 218:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1241 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 218:
GCTCCCACTG TTCCATTTTA TGCTAATAGA TTCCATTCTA GGGCCCAGCC GTCTCTTGAC 60
TGATGGTGTT CCCTTTAACC CTTGGCATGT ATAATAGAAT TTTGGTGAAT GAAAGAACCC 120
AAATAGGCCA GATAGTCCCC CCAGGCCCTG ATATCCATAA AAGGCTTGGG AATGCATTAT 180 GTAATTGTCC TTAGTCTTTT TGTTCTTTTA GAAAAAAAAA ACAAGATGGG CTCAGATGGA 240
TGCCTACGTA AAAATGGTTC CTAGCTGTGT ACTCATAACT TTTCTTTGAA TTGAGTAGTG 300
AAAGGAAGGA GGAGGAAAGG AAATTAAATG TCCTTCTAGT ATTCTCTGGA CTCAAGTCTG 360
ACATATGRGA TAATAACCTA TATTGAAATG CCAAGAATTG TATCTGAAAC AAGRGAACAG 420
TTTGACACAT TTATCATGCC TTCATATTAC ATATTAACTG AAACCAATTA ATAAACATAT 480 GAAATATCCA TTGCACAAGG CAAAGGCACC TAAACCTTTT GTTTCTTTTT CTACATAGCA 540
GAAATTGATT TTTTTTTTAT TTTTTTAGGG GAACCTATAT AATTATGACC CAGTGATGTC 600
TTTTGGTGAC TTAAGCTTAT GAATTCAGGT TACAATTGAG TTGATTCTAG ATGGTTACTA 660
CCTTGAAAAG GATCTTGGTG CCTTATCTCA CACGAGCCAG AGCCTGCTGG GAATAAACAA 720
AGCAGATTCA TGCCAACACC AACTCGTAGC TTTAGTGGCA GATGGGAGTG GTCACAGACT 780 CCCAAAATCT GGGGCTTTGG ATTTCCACAC CATCCCACGT GTGTGTCATC TTCCTCTTTC 840
ACACTCTTGA TGATAATTTG AAAATGRTGA AATCACCTCT GAATTTGCCT ATAGCATGAG 900
CACATTCTTA TGACAACATA ACAAATAGTT CATAATGTGA ATATTAGAAA CTGTTACAGC 960
CTGCAGTTAC CATAATTTTC CATGTTTGTG GAATTGATAT TGAAATAGCA GGGCTAAGGA 1020
ATTACTGGCA AGTTTTAGCC TGTGGGTAAT ACCTTAGGGT TATTTAAATA TTTGTAATTT 1080
TATTTAAATG TTCATGAATG TTTGAAAGGA ACAAAATTAT CAGGGATGGC TCTTTGCCAT 1140
GGGTCTTATT TTCACCCTCT TTTCTGTAAG AAAAAAGAAC AATGTCTTAA TGTATTTTTA 1200
AAGTTTTTGG TATAGTTTCT AATTCCAATT TTAATAAAAG T 1241
(2) INFORMATION FOR SEQ ID NO: 219:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1080 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi ) SEQUENCE DESCRIPTION: SEQ ID NO : 219 :
TGTTTATGTG ACCTAAAACA TACACACATG CACACACACA TACATATCCA TTCATTCATT 60
CATTCAAGTG GTGTTTCCAG TGTCTGTGTG TCACTGTTTA TGCAGTTTCC ATTTCCCAGT 120
GAATTATGAG TGGAGGGCAA CTTTTCTAAC CAGATTGTCT TTTCAGAACA AAGACCKGGG 180
RATTGAGGAA GAGTTTGGAA AGAGGGAGAG GCAAGGAAAG AGAGCTTTAA ATTGAAAGGT 240
TAATTTCCTA AGAGGAACCT GGGCTGAATG ACTACAGTGT TATACCCTCC AATCTTTGCA 300
GGTGGGCATG GAACACTGCT TGTATCACTC TGTGCACGGT ATAAATCCAT ATATCCACAA 360
AAACACACAT CCATCCATCA ACATATACAT GGTTTGGGAT GAGCAGGTCA ATAGTTTTGA 420
GAGGGAGTTT GTTCCTTTTT TTTTCTCATT ATACTCTTAA ATTGTTGTCA GTTATCAAAC 480
AAACAAACAG AAAAATTGTT TGGGAAAAAC CTTGCATACG CCTTTTCTAT CMAGTGCTTT 540
AAAATATAGA CTAAATACAC ACATCCTGCC AGTTTTTTCT TACAGTGACA GTATCCTTAC 600
CTGCCATTTA ATATTAGCCT CGTATTTTTC TCACGTATAT TTACCTGTGA CTTGTATTTG 660
TTATTTAAAC AGGAAAAAAA ACATTCAAAA AAAGAAAAAT TAACTGTAGC GCTTCATTAT 720
ACTATTATAT TATTATTATT ATTGTGACAT TTTGGAATAC TGTGAAGTTT TATCTCTTGC 780
ATATACTTTA TACGGAAGTA TTACGCCTTA AAAATACGAA AATAAATTTT ACAAGGTTTC 840
TGTTTTGTGT GGAAGAGTAA TTGATGTTGC TAAGAATGAT GTTTGTTTTT TTGGGGTTTT 900
TCTTGTTTTT TTTTTAAATG TTACCAGCAC TTTTTTTCTA AGTTTCACTT TCCGAGGTAT 960
TCTACAAGTT CACACTGTTT GTGAAGTTTG AATATGAAGG AATAATTAAA AAAAAAAAAA 1020
AAACCNCGGG GGGGGCCCGG TCCCATTGGN CCCAAGGGGG CGGTTACGGG GTCACGGCCG 1080
(2) INFORMATION FOR SEQ ID NO: 220:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1258 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(Xi) SEQUENCE DESCRIPTION: SEQ ID NO: 220:
TGAATTGAGG GCTTAAAGAT AAACATATGG GRTTGGAGTT GTGTGTCCAT AGGGTTTCAC 60
TGCCTATTTG ATTTGAGTTT ATCCCTATTA ATTTTTTACA GTGAAATTTT ATTAAAGTAT 120
AATGTACATA TATTTTCAGT GGATTTTGCT CTGAAGGTTC TCCAGTGGTC TGACTACGAG 180
ATAGTGCGGC TTCAGCTGTG GGATATTGCA GGGCAGGAGC GCTTCACCTC TATCACACGA 240
TTGTATTATC GGGATGCCTC TGCCTCTCTT ATTATCTTTG ACGTTACCAA TGCCACTACC 300
TTCAGCAACA GCCAGAGGTG GAAACAGGAC CTAGACAGCA AGCTCACACT ACCCAATGGA 360
GAGCCGCTGC CCTGCCTGCT CTTGGCCAAC AAGTGTGATC TGTCCCCTTG GGCAGTGAGC 420
CGGGASCAGA TTGACCGGTT CAGTAAAGAG AACGGTTTCA CAGGTTGGAC AGAAACATCA 480
GTCAAGGAGA ACAAAAATAT TAATGAGGCT ATGAGAGTCC TCATTGAAAA GATCATGAGA 540
AATTCCACAG AAGATATCAT GTCTTTGTCC ACCCAAGGGG ACTACATCAA TCTACAAACC 600
AAGTCCTCCA GCTGGTCCTG CTGCTAGTAG TGTTTGGYTT ATTTTCCATC CCAGTTCTGG 660
GAGGTCTTTT AAGTCTCTTC CCTTTGGTTG CCCACCTGAC MATTTTATTA AGTACATTTG 720
AATTGTCTCC TGACTACTGT CCAGTAAGGA GGCCCATTGT CACTTAGAAA AGACACCTGG 780
AACCCAKGTC CATTTCTCCA TCTCCTGGAT TAGCCTTTSA CATGTTGCTG RCTCACATTA 840
GTGCCAGTTA GTGCCTTCGG TGTAAGATCT TCTCATCAGC CCTCAATTTG TGATCCGGAA 900
TTTTGTGAGA AGGATKAGAA ATCAGCACCT GCGTTTTAGA GATCATAATT CTCACCTACT 960
TCTGAGCTTA TTTTTCCATT TGATATTCAT TGATATCATG ACTTCCAATT GAGAGGAAAA 1020
TGAGATCAAA TGTCATTTCC CAAATTTCTT GTAGGCCGTT GTTTCAGATT CTTTCTGTCT 1080
TGGAATGTAA ACATCTGATT CTGGAATGCA GAAGGAGGGG TCTGGGCATC TGTGGATTTT 1140
TGGCTACTAG AAGTGTCCCA GAAGTCACTG TATTTTTGAA ACTTCTAACG TCATAATTAA 1200
GTTTCTCTTG TCTTGGGCAT CAAGANTAGT TCCAATTTTT TGGGCCGGGG CAGGGTGG 1258
(2) INFORMATION FOR SEQ ID NO: 221: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1693 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 221:
CACAATATAT GAAATAGTAC CCTCTAAAAA AGAGAAAAAA AAAATCAGGC GGTCAAACTT 60
AGAGCAACAT TGTCTTATTA AAGCATAGTT TATTTCACTA GAAAAAATTT AATATCAAGG 120
ACTATTACAT ACTTCATTAC TAGGAAGTTC TTTTTAAAAT GACACTTAAA ACAATCACTG 180
AAAACTTGAT CCACATCACA CCCTGTTTAT TTTCCTTAAA CATCTTGGAA GCCTAAGCTT 240
CTGAGAATCA TGTGGCAAGT GTGATGGGCA GTAAAATACC AGAGAAGATG TTTAGTAGCA 300
ATTAAAGGCT GTTTGCACCT TTAAGGACCA GCTGGGCTGT AGTGATTCCT GGGGCCAGAG 360
TGGCATTATG TTTTTACAAA ATAATGACAT ATGTCACATG TTTGCATGTT TGTTTGCTTG 420
TTCAATTTTT GAACAGCCAG TTGACCAATC ATAGAAAGTA TTACTTTCTT TCATATGGTT 480
TTTGGTTCAC TGGCTTAAGA GGTTTCTCAG AATATCTATG GCCACAGCAG CATACCAGTT 540
TCCATCCTAA TAGGAATGAA ATTAATTTTG TATCTACTGA TAACAGAATC TGGGTCACAT 600
GAAAAAAAAT CATTTTATCC GTCTTTTAAG TATATGTTTA AAATAATAAT TTATGTGTCT 660
GCATATTGCA GAACAGCTCT GAGAGCAACA GTTTCCCATT AACTCTTTCT GACCAATAGT 720
GCTGGCACCG TTGCTTCCTC TTTGGGAAGA GGAAAGGGTG TGTCAACATG GCTAACAATC 780
TTCAAATACC CAAATTGTGA TAGCATAAAT AAAGTATTTA TTTTATGCCT CAGTATATTA 840
TTATTTAATT TTTTAGGTAA TGCCTATCTC TTGGTCTATT AAGGAAAGAA GCAATCAGTA 900
GAGAATTCAG GATAGTTTTG TTTAAATTCT TGCAGATTAC ATGTTTTTAC AGTGGCCTGC 960
TATTGAGGAA AGGTATTCTT CYATACAACT TGTTTTAACC TTTGAGAACA TTGACAGAAA 1020
TTATGCAATG GTTTGTTGAG ATACGGACTT GATGGTGCTG TTTAATCAGT TTGCTTCCAA 1080
AGTGGCCTAC TCAAGAGGCC CTAAGACTGG TAGAAATTAA AAGGATTTCA AAAACTTTCT 1140
ATTCCTTTCT TAAACCTACC AGCAAACTAG GATTGTGATA GCAATGAATG GTATGATGAA 1200
GAAAGTTTGA CCAAATTTGT TTTTTTGTTG TTGTTGTTGT TTTGAATTTG AAATCATTCT 1260
TATTCCCTTT AAGAATCTTT ATGTATGAGT GTGAAGATGC TAGCGAACCT ATGCTCAGAT 1320
ATTCATCGTA AGTCTCCCTT CACCTGTTAC AGAGTTTCAG ATCGGTCACT GATAGTATGT 1380
ATTTCTTTAG TAAGAATGTG TTAAAATTAC AATGATCTTT TAAAAAGATG ATGCAGTTCT 1440
GTATTTATTG TGCTGTGTCT GGTCCTAAGT GGAGCCAATT AAACAAGTTT CATATGTATT 1500
TTTCCAGTGT TGAATCTCAC ACACTGTACT TTGAAAATTT CCTTCCATCC TGAATAACGA 1560
ATAGAAGAGG CCATATATAT TGCCTCCTTA TCCTTGAGAT TTCACTACCT TTATGTTAAA 1620
AGTTGTGTAT AATTGTTAAA ATCTGTGAAA GAATAAAAAG TCGATTTAAA TTAAAAAAAA 1680
AAAAAAAAAA AAA 1693
(2) INFORMATION FOR SEQ ID NO: 222:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1196 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 222:
ACGCGTGGGT CGACCCACGC GTCCGCGACN TCGCGTGGTG GGGAAGGGAG AAGGATTTGT 60
AAACCCCGGA GCGAGGTTCT GCTTACCCGA GGCCGCTGCT GTGCGGAGAC CCCCGGGTGA 120
AGCCACCGTC ATCATGTCTG ACCAGGAGGC AAAACCTTCA ACTGAGGACT TGGGGGATAA 180
GAAGGAAGGT GAATATATTA AACTCAAAGT CATTGGACAG GATAGCAGTG AGATTCACTT 240
CAAAGTGAAA ATGACAACAC ATCTCAAGAA ACTCAAAGAA TCATACTGTC AAAGACAGGG 300
TGTTCCAATG AATTCACTCA GGTTTCTCTT TGAGGGTCAG AGAATTGCTG ATAATCATAC 360
TCCAAAAGAA CTGGGAATGG AGGAAGAAGA TGTGATTGAA GTTTATCAGG AACAAACGGG 420
GGGTCATTCA ACAGTTTAGA TATTCTTTTT ATTTTTTTTC TTTTCCCTCA ATCCTTTTTT 480
ATTTTTAAAA ATACTTCTTT TGTAATGTGG TGTTCAAAAC GGAATTGAAA ACTGGCACCC 540
CATCTCTTTG AAACATCTGG TAATTTGAAT TCTAGTGCTC ATTATTCATT ATTGTTTGTT 600
TTCATTGTGC TGATTTTTGG TGATCAAGCC TCAGTCCCCT TCATATTACC CTCTCCTTTT 660
TAAAAATTAC GTGTGCACAG AGAGGTCACC TTTTTCAGGA CATTGCATTT TCAGGCTTGT 720
GGTGATAAAT AAGATCGACC AATGCAAGTG TTCATAATGA CTTTCCAATT GGCCCTGATG 780
TTCTAGCATG TGATTACTTC ACTCCTGGAC TGTGACTTTC AGTGGGAGAT GGAAGTTTTT 840
CAGAGAACTG AACTGTGGAA AAATCACCTT TCCTTAACTT GAAGCTACTT TTAAAATTTG 900
AGGGTCTGGA CCAAAAGAAG AGGAATATCA GGTTGAAGTC AAGATGACAG ATAAGGTGAG 960
AGTAATGACT AACTCCAAAG ATGGCTTCAC TGAAGAAAAG GCATTTTAAG ATTTTTTAAA 1020
AATCTTGTCA GAAGATCCCA GAAAAGTTCT AATTTTCATT AGCAATTAAT AAAGCTATAC 1080
ATCCAGAAAT GAATACAACA GAACACTGCT CTTTTTGATT TTATTTGTAC TTTTTCGCCT 1140
GGGATATGGG TTTTAAATGG ACATTGTCTG TACCAGCTTC ATTAAAATAA ACAATA 1196
(2) INFORMATION FOR SEQ ID NO: 223:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1791 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS : double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 223:
TCAGGGAGGT GGCAGGAAAG GCTTGGAACA GCTGCCGGAG TGACGGAGCG GCGGCCCCGC 60
CCGGTTGCGC TGGAGGTCGA AGCTTCCAGG TAGCGGCCCG CAGAGCCTCA CCCAGGCTCT 120
GGACATCCTG AGCCCAAGTC CCCCACACTC AGTGCAGTGA TGAGTGCGGA AGTCAAGGTG 180
ACAGGGCAGA ACCAGGAGCA ATTTCTGCTC CTAGCCAAGT CGGCCAAGGG GGCAGCGCTG 240
GCCACACTCA TCCATCAGGT GCTGGAGGCC CCTGGTGTCT ACGTGTTTGG AGAACTGCTG 300
GACATGCCCA ATGTTAGAGA GCTGGCTGAG AGTGACTTTC CCTCTACCTT CCGGCTGCTC 360
ACAGTGTTTG CTTATGGGAC ATACGCTGAC TACTTAGCTG AAGCCCGGAA TCTTCCTCCA 420
CTAACAGAGG CTCAGAAGAA TAAGCTTCGA CACCTCTCAG TTGTCACCCT GGCTGCTAAA 480
GTAAAGTGTA TCCCATATGC AGTGTTGCTG GAGGTCTTGC CCTGCGTAAT GTGCGGCAGC 540
TGGAAGACCT TGTGATTGAG GCTGTGTATG CTGACGTGCT TCGTGGCTCC CTGGACCAGC 600
GCAACCAGCG GCTCGAGGTT GACTACAGCA TCGGGCGGGA CATCCAGCGC CAGGACCTCA 660
GTGCCATTGC CCGAACCCTG CAGGAATGGT GTGTGGGCTG TRAGGTCGTG CTGTCAGGCA 720
TTCAGGAGCA GGTGAGCCGT GCCAACCAAC ACAAGGAGCA GCAGCTGGGC CTGAAGCAGC 780
AGATTGAGAG TGAGGTTGCC AACCTTAAAA AAACCATTAA AGTTACGACG GCAGCAGCAG 840
CCGCAGCCAC ATCTCAGGAC CCTGAGCAAC ACCTGACTGA GCTGAGGGAA CCAGCTCCTG 900
GCACCAACCA GCGCCASCCA GCAAGAAAGC CTCAAAGGGC AAGGGGCTCC GAGGGAGCGC 960
CAAGATTTGG TCCAAGTCGA ATTGAAAGRA CTGTCGTTTC CTCCCTGGGG ATGTGGGGTC 1020
CCAGCTGCCT GCCTGCCTCT TAGGAGTCCT CAGAGAGCCT TCTGTCCCCC TGGCCAGCTG 1080
ATAATCCTAG GTTCATGACC CTTCACCTCC CCTAACCCCA AACATAGATC ACACCTTCTC 1140
TAGGGAGGAG KCAAATCTAG GTCATGTTTT TGTTGGTACT TTCTGTTTTT TGTGACTTCA 1200
TGTCTTCCAT TGCTCCCCGC TGCCATGCTC TCTCCCTTGT TTCCTTAAGA GCTCAGCATC 1260
TGTCCCTGTT CATTACATGT CATTGAGTAG GTGGGTAGCC CTGATGGGGG TCGCTCTGTC 1320
TGGAGCATAA CCCACAGGCG TTTTTTCTCC CACCCCATCC CTGCATGCCT GATCCCCAGT 1380
TCCTATACCC TACCCCTGAC CTATTGAGCA GCCTCTGAAG AGCCATAGGG CCCCCACCTT 1440
TACTCACACC CTGAGAATTC TGGGAGCCAG TCTGCCATCC CAGGAGTCAC TGGACATGTT 1500
CATCCTAGAA TCCTGTCACA CTACAGTCAT TTCTTTTCCT CTCTCTGGCC CTTGGGTCCT 1560
GGGAATGCTG CTGCTTCAAC CCCAGAGCCT AAGAATGGCA GCCGTTTCTT AACATCTTGA 1620
GAGATGATTC TTTCTTGGCC CTGGCCATCT CGGGAAGCTT GATGGCAATC CTGGAAGGGT 1680
TTAATCTCCT TTTCTGAGTT TGGTGGGGAA GGGAAGGGTA TATAGATTGT ATTAAAAAAA 1740
AAAAGGTATA TATGCATATA TCTATATATA ATATGACGCA GAAATAAATC T 1791
(2) INFORMATION FOR SEQ ID NO: 224:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2517 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 224: ACACTAGTGG ATCCAAAGAA TTCGGCACAG CGGCACAGCA TTGTTGAGCT TTTCTGTGTG 60 TCTGGGGCCC TCAAGCGAGC TCGACTGGTC CATCCTGGGG TAGCGASGTG GTGTTTGTGA 120 AAAAGGACGA TGCCATCACC GCATAYAAGA AGTACAACAA CCGGTGTCTG GACGGGCAGC 180 CGATGAAGTG CAACCTTCAC ATGAATGGGA ATGTTATCAC CTCAGACCAG CCCATCCTGC 240 TGCGGCTGAG TGACAGCCCA TCAATGAAAA AGGAGAGCGA GCTGCCTCGC AGGGTGAACT 300 CTGCCTCCTC CTCCAACCCC CCTGCCGAAG TGGACCCTGA CACCATCCTG AAGGCACTCT 360 TCAAGTCCTC AGGGGCCTCT KTGACCACGC AGCCCACAGA WTTCAAAATC AAGCTTTGAG 420 CAGGGGAGTR AGGCAGCCAG AAGTGGGGGC AGAGGAGGGT GGCTCTGTTT CCCCAAGGCA 480 AAGCTTATGA CCAATGGGCC ATCGGACTGG AGACCCCTGA TTGTGGGAAG GGTTGCCAGG 540 GATAAAGAGC TTCCTCACTG GATGGGACCC GCCTTTCTGT GTTGTGTTCT GCCCTGTGCT 600 CTTCTCTCTA CGTTAACGTT TCCTGTAGTA TGTTTCTTCA TCTCATCGCC AAGGTAGGCT 660 TGTGTTTTTM AGTGTGTGCC TCCCCGAGCC TCAGCCCCAA GCTGATTTCT TATCTGGAAA 720 TGGTACACTG AATTCTCTGG GTGGCTTTCT TGTGGCCCCA TGGGATGCAG CGTGGGGGCT 780 GTCTGAAGGA CCCTGCTTTT TCCAGGGGCC GAGGGGCTGC CTTTCCTTTG TGTGTATTAA 840 GCTTTTCAAA CAATGGAGGG GATGGAGAGC CCTGGTGTCC TGACGCGAGC CAGGTCGGCC 900 TGAGAGCTGT GCCGCTCCTC TGTCTTGTCA GTGGAGGTGC CTGGGTGGGG AGCAGGTCTC 960 AGGCCTCTTG TCCTCTCCCC AGTGGCTCCA GGCCTCACTA GTGGCAAGGG CAGGATGAGG 1020 CTGCACCGCT GGGAAGAGTC TATCTAAGCT CTTGGCTTGG AGTCCCGTGT CGTCTCCRCC 1080
CAGAGGAAGT TCTCCAGAGT TCACCTTTCC CTTTTCCTTG AGTTGTGCTG AATGCCCCAC 1140
CCCAGCTCTC TTTCCCTTCT GGGTGTCTTT GCTGGGAGGG GGCTGTGTTG TGAGCCCTCC 1200
CGGTTCTCAC CTCGCCTGGC ACTTAACCAC ACCCTGGTTT TGTGTAGCCG CCAGCTCTCT 1260
TCTGGTTGGG CCTTTGAAAG GCTCAGCCTC CCATTGTGCA GTGCTTGGGT TTCGAGCTTA 1320
TTTGAATGGA AGAGGTCAGT TTGTTCCTGG CTCTCCATTT CTGGCCTCAG TTGTCTACAG 1380
GACAGTGGTC AGGGATGCCT GGAGGCATAT ATCCAGCTGC CACCAAGGGG CACTGTTTGT 1440
TCCCACTTAT GTGAGTGACC CCATCCATCC ATGACCAGAG GATTATTTTC CTGCCTTGGC 1500
AGAGGAGGAG GAGTCAAGGG AGCAGGGCAG CTCTACCAGG CAAGGTGTTT CCCCAGCATA 1560
GGCGCAGACA GTTGGGACGA AACTTCAGAG CCCAGGCAGT CCCTGAATGA CCAGGCCAGT 1620
GTTGTCACTG AGTGGTCCCC TGCTGGTTGG GAGTGAAGAG AATCCAGGCT GGCAGAGCTG 1680
GAGCCAGTTG GGGAGCACGG TTCTGGGAGC TCTGCAAAAT CAGTAGCAAG TGCTGGAAAA 1740
GGCACATGCC GAAGATACTC AAGAGCTCCC AAGATTTGCT TGAGGCTAGC CCAGTGAAAA 1800
AAACCAGAGA CTCATGTTTC CAGGGGTCAG TCTGTCAGGC AGGAAGGACC CAGGATTTGA 1860
ACCCAGCTTC AGTGTGCAGG CTCTGAGGCT GCCCAGGACG GGAAAGTCCA AGGAAGGGGC 1920
CTGGTGGTGC TCCACTTGCA GTTCTTTAAA GAATGCTGCT TTTTATTCTC CTAACCCTTT 1980
CAAGTGGGTG CAGACTTCTC GTTAGCAGCT GCAAGACATT CCTCCCACAC TTTTCCCTTC 2040
CTGGCCCAAG AGAGCATCCA GAAGGCAGTA GGACCTGGTT TTTCAGGTAC TGGGAGCCGG 2100
GGGCTCACTG CTTGCACTGT GCTTAGGGTA GGGATGGTAA ATATCCTCCC TGCATGGCTT 2160
TATCCTCCCT CTCATCCCAA AGCAGGTATC TTCTGGTTGT CACAGAGTTT CATTGAGTCC 2220
AGCTGCAGCC ACGTGGCCAT CTGGAGCTGG TGCTATAGGT GACCATCTGG TACATTGAGG 2280
GGACCTGTTT GCCTCCTCCA CTCTATAAGC AGTCATCTTG GGAGACCGGG AGGAGAAGGT 2340
GGTGGGCTAG TCCTGTGTCC TCCTCCACTT CCCATGCCTC TATGTTACCC ATCTGTGTCT 2400
CCTGTGCAGA AGGAGAGGAA GGGGCATTAA GAGATGAAGG GTGATTATGT ATTACTTATC 2460
CATTTCTGAA TAAACATTTG TTATTCCTAA AAAAAAAAAA AAAAAACTCG AGGGGGG 2517
(2) INFORMATION FOR SEQ ID NO: 225:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 2424 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 225:
TTGTANCTAA TCGAGGATTG ATTCTAATCA CAGAGTCTTT CAACACTTTG CACATGATGT 60
ATCACGAAGC TACAGCTTGC CATGTGACTG GAGATTTAGT AGAACTTCTG TCAATATTTC 120
TTTCGGTTTT GAAGTCTACA CGCCCTTATC TTCAGAGAAA AGATGTGAAA CAAGCATTAA 180
TCCAGTGGCA GGAGCGAATT GAATTTGCCC ATAAACT TT AACTCTTCTT AATTCCTATA 240
GTCCTCCAGA ACTTAGAAAT GCCTGTATAG ATGTCCTCAA GGAACTTGTA CTTTTGAGTC 300
CCCATGATTT TYTTCATACT CTGGTTCCCT TTCTACAACA CAACCATTGT ACTTACCATC 360
ACAGTAATAT ACCAATGTCT CTTGGACCTT ATTTCCCTTG TCRAGAAAAT ATCAAGCTAA 420
TAGGAGGGAA AAGCAATATT CGGCCTCCGC GCCCTGAACT CAATATGTGC CTCTTGCCCA 480
CAATGGTGGA AACCAGTAAG GGCAAAGATG ACGTTTATGA TCGTATGCTG CTAGACTACT 540
TCTTTTCTTA TCATCAGTTC ATCCATCTAT TATGCCGAGT TGCAATCAAC TGTGAAAAAT 600
TTACTGAAAC ATTAGTTAAG CTGAGTGTCC TAGTTGCCTA TGAAGGTTTG CCACTTCATC 660
TTGCACTGTT CCCCAAACTT TGGACTGAGC TATGCCAGAC TCAGTCTGCT ATGTCAAAAA 720
ACTGCATCAA GCTTTTGTCT GAAGATCCTG TTTTCGCAGA ATATATTAAA TGTATCCTAA 780
TGGATGAAAG AACTTTTTTA AACAACAACA TTGTCTACAC GTTCATGACA CATTTCCTTC 840
TAAAGGTTCA AAGTCAAGTG TTTTCTGAAG CAAACTGTGC CAATTTGATC AGCACTCTTA 900
TTACAAACTT GATAAGCCAG TATCAGAACC TACAGTCTGA TTTCTCCAAC CGAGTTGAAA 960
TTTCCAAAGC AAGTGCTTCT TTAAATGGGG ACCTGAGGGC ACTCGCTTTG CTCCTGTCAG 1020
TACACACTCC CAAACAGTTA AACCCAGCTC TAATTCCAAC TCTGCAAGAG CTTTTAAGCA 1080
AATGCAGGAC TTGTCTGCAA CAGAGAAACT CACTCCAAGA GCAAGAAGCC AAAGAAAGAA 1140
AAACTAAAGA TGATGAAGGA GCAACTCCCA TTAAAAGGCG GCGTGTTAGC AGTGATGAGG 1200
AGCACACTGT AGACAGCTGC ATCAGTGACA TGAAAACAGA AACCAGGGAG GTCCTCACCC 1260
CAACGAGCAC TTCTGACAAT GAGACCAGAG ACTCCTCAAT TATTGATCCA GGAACTGAGC 1320
AAGATCTTCC TTCCCCTCAA AATAGTTCTG TTAAAGAATA CCGAATGGAA GTTCCATCTT 1380
CGTTTTCAGA AGACATGTCA AATATCAGGT CACAGCATGC AGAAGAACAG TCCAACAATG 1440
GTAGATATGA CGATTGTAAA GAATTTAAAG ACCTCCACTG TTCCAAGGAT TCTACCCTAG 1500
CCGAGGAAGA ATCTGAGTTC CCTTCTACTT CTATCTCTGC AGTTCTGTCT GACTTAGCTG 1560
ACTTGAGAAG CTGTGATGGC CAAGCTTTGC CCTCCCAGGA CCCTGAGGTT GCTTTATCTC 1620
TCAGTTGTGG CCATTCCAGA GGACTCTTTA GTCATATGCA GCAACATGAC ATTTTAGATA 1680
CCCTGTCTAG GACCATTGAA TCTACAATCC ATGTCGTCAC AAGGGATATC TGGCAAAGGA 1740
AACCAAGCTG CTTCTTGACA TTAGGTGTAG CATGTCTACT TTTAAGTCCC TCACCCCCAA 1800
CCCCCATGCT GTTTCTATAA GTTTTGCTTA TTTCTTTTTG TGCTTCAGTT TGTCCAGTGC 1860
TCTCTGCTTG AATGGCAAGA TAGATTTATA GGCTTAATTC TTGGTCAGGC AGAACTCCAG 1920
ATGAAAAAAA CTTGCATCTT CAGTATACTT CCTAAAGGGC AATCAGATAA TGGATATCTT 1980
TTATGTAATT AAGAGTTCAC TTTAGTGGCT TTCATTTAAT ATGGCTGTCT GGGAAGAACA 2040
GGGTTCCCTA GCCCTGTACA ATGTAATTTA AACTTACAGC ATTTTTACTG TGTATGATAT 2100
GGTGTCCTCT GTGCCAGTTT TGTACCTTAT AGAGGCAGAT TGCCTCCGAT CGCTGTGGTT 2160
CTTATTATCA AAATTAAGTT TACTTGTATA CGGAACAACC ACAAGAAATT TGATTCTGTA 2220
AAGAATCCTC TTTAGCTGTG GCCTGGCAGT ATATAAATGG TGCTTTATTT AACAGAATAC 2280
CTGTGGAGGA AATAAAGCAC ACTTGATGTA AAAATAATTG TTTTATTTTT ATTGACATGA 2340
CTGATTGATT GCTATTCTGT GCACTNAATT AAACTGATTG TGATGACTTA AAAAAAAAAA 2400
AAAAAAAAAA AAAAAAAAAA AAAA 2424
(2) INFORMATION FOR SEQ ID NO: 226:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1080 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi ) SEQUENCE DESCRIPTION : SEQ ID NO : 226 :
ATATAGGACG GATAATCTGT TTACATTCTG TTCTTCTCGA TGCACTCACA AGCGGGTAAC 60
TAGGTGACAA GAAAACAAAG ATCTTATTCA AAAGAGGTCT TACAGCAACC CAACGTCTCA 120
TCTTCCCATA GTAAAGATGA CGGCGCCTTC AGGTAAGCTA CAGGCAACAC CACTTCCGCG 180
TTTCTCTTGC GCCCTGCTCC AAGATGGCGG ATGAAGCCAC GCGACGTGTT GTGTCTGAGA 240
TCCCGGTGCT GAAGACTAAC GCCGGACCCC GAGATCGTGA GTTGTGGGTG CAGCGACTGA 300
AGGAGGAATA TCAGTCCCTT ATCCGGTATG TGGAGAACAA CAAGAATGCT GACAACGATT 360
GGTTCCGACT GGAGTCCAAC AAGGAAGGAA CTCGGTGGTT TGGAAAATGC TGGTATATCC 420
ATGACCTCCT GAAATATCAG TTTGACATCG AGTTTGACAT TCCTATCACA TATCCTACTA 480
CTGCCCCAGA AATTGCAGTT CCTGAGCTGG ATGGAAAGAC AGCAAAGATG TACAGGGGTG 540
GCAAAATATG CCTGACGCAT CATTTCAAAC CTTTGTGGGC CAGGAATGTG CCCAAATTTG 600
GACTAGCTCA TCTCATGGCT CTGGGGCTGG GTCCATGGCT GGCAGTGGAA ATCCCTGATC 660
TGATTCAGAA GGGCGTCATC CAACACAAAG AGAAATGCAA CCAATGAAGA ATCAAGCCAC 720
TGAGGCAGGG CAGAGGGACC TTTGATAGGC TACGATACTA TTTTCCTGTG CATCACACTT 780
AACTCATCTA ACTGCTTCCC CGGACACCCT CCACCTCTAG TTGTTACTAA GTAGCTGCAG 840
TAGGCATTGC TGGGGAAGAA ACAAACACAC ACCAAACAGT ACTGCTACTT AGTTTCTAAG 900
GCTGCACAGG GAAGGGAAAG ACTGGGCTTT GGACAATCTA GAGGTAATTT ATATCCGCCC 960
CCAGGTGGAG CAACATGCGA TTCTGGAGGC ACGGGGGTAA CTGAAAGTGA GTACATATAG 1020
TCTTTCTGGT TTCTGGAGAT AACCCATCAA TAAAAGCTGC TTCCTCTGGG TAAAAAAAAG 1080
(2) INFORMATION FOR SEQ ID NO: 227:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1336 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 227:
TTGCATTCAC AATTACTGGG AGCCAGGCAG GGGCAGTTGC ATGCTGGGGG TGGCTGCATG 60
GSCTGCCASC TCTCCTGGGT TTGAAGGATG CGGTACASCT GCTTCAGCTG AGCAACGATG 120 TTATCCTTCA TGTCTGGGGT TGAGATCTGC AGGCGGACAC TGCCACTATC AAAGGATCGT 180
GTGAAATCAC CAGAAAACAT CTCGTAGATC ATCCGAGCCA CTACTGGAAT GACCTGAACC 240
AAGATGAGTT TCCTTTCCAA TGGTTTCCCA TCTGGCCATT CTTCCCCAAA GCATAAGTAG 300
ATCTCAAACG GTGGCTGCTT CTCTATCTGT CCTTTCTGGT GGGCAATGAG ATCGCTAAGG 360
AATGTTTCCA GACAAAATAG CTTGACCTTC TTTTGTCTCT CAATCAGGTT GGGAGCAACA 420 AGTGATGGGG CACATGGCCC AGACCAGTAC ACCTTGCACT GGCACAGYCT GATGGCATAA 480
ATGGCATGAC CGCTGACCTC CAGGATCAGT CCTCTGTCCA TGACGTCCAG CAGCTTGCTA 540
GTGAACAGCT TCTGCTTCTC ATTGGTAATA TGCTCAGGAC CTGCGAATTT GACCTGCTCC 600
AGNCTGACGG GACCAAAGAG CTCCTCCTGG TCAGGCATGG GACCCAGGTC CCCATAGAAG 660
AGTCGGCAGC CCTGAGGGTT GCTCACGGTC ATGGTCCTCC CCGTACTCCT TCCCACGGTA 720 CTGAAACTTG ATCTCCAGGT CAGTCATTGG GAGAGAGCTG ATCCACAGTT CTCGAGAGCT 780
ATAGAAGGRC TGTATAGGTG CCTGGGGWAC TTCCATCTCC AGGGGTTCAG TTTTGGGCCA 840
CACTGCCTCC GGSCTGCAGT TGCCCACACT GCAATTGCCC ACACTGGCTG GCGCCATGGG 900
AGAACCATTC ATGTTCAGGA AGGGGAAGGT GTCCTCGATG GGAACATGGT GCTCCCACTG 960
ATCCAGCTCA TCTTCCTCAT CTTCTTCATC CACATCATTA TCCTTCTCAT CCCAGGGAGC 1020 AGACCCTGTG GATCCTGGGT TAATGATCGA SCCCTGGGGC TGAGGGATGT CACACACTTG 1080
ATATATCTTC ACTGGGTTCA TGGGCACCTC CCTTGGTGCC ATCCATACAT CCAGGTTGAA 1140
TTCTCTGCTC TTATTGAGAG CACAGCGCAG CTGGGCCTTC CATTTAGCTG GGTCAGGGTC 1200
ATCCACCCCT TCCTGGTACT TCCCTGTCTC TACAGCCCAG GCCTTAAAAA TCGTATTTTC 1260
CTCTTCTTGT TGAGGGCTAT GCCGGGTGGC ATCTTTCCAG GGAATCTGGA AGCGTTTAGA 1320
GTCCCTGTGT AGCCAG 1336
(2) INFORMATION FOR SEQ ID NO: 228:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2043 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 228:
TCAGCTGGTC CCTTCCTTGT GTCCTGGGGG ACCTGCTGGC GGCCTCTTCC TGGGAGCCAT 60
GACCTCAGAC CCCACCCACA CTCCAGATCG AGACCCCTGC CTCCCCCCGG CAAATGTCCT 120
CCCGCTGCCT TGCAGCCTGC ACTTTGCACA TGCTCACCCC CAGCACAGTC CCACTGGCCC 180
CTCAMCTCCC CTTCCCTGAG CTCCTTCCCA AGGACTCCTG GTCACTGCCT GCTGTGCAKT 240
CAGAGGCCCA GGGTCCAGCA GCCCGGSGGG AACGGGTGCT GCCTSTTCCT CCAGTTAGCT 300
CCAGYTCAGG TCTGAGACCC GTGYTGAGTA AAGGTCTGAG CAMCGACCGT GCCCTCTGCC 360
CAGGGCTGGG TCCTGAGCAG CTGGTTTTCC TGCAGGAAGG TTGGAGCAAG CAAAGTCCTT 420
CTCTGCCCTC AGGGTCAGCT GCCCAGACTG GGGCGGATGC AGAGAGGCAG GTGGGCTGTG 480
GCTGGACTGG TCCGGAGCTG GCTTCCTTAC CAGAAAAGCC TCAGCCTTCC TCTGGAAGCA 540
TCCCCCGTTC TGGGCAAGGG GGAAGGGCTC CTTTAAGGGG TGTGCTTTCC CAGTGGGGAG 600
CAGTCTGGCC CTGCCCCCTA CTAAAGCCTC TGCTCTCAGC ACTTTCCCCC AAGTCCTTGT 660
AACTTGCTTG AAGGTCGGTT CTGGCTGCCA GCCAGTCCCT GGACAAACTC TCCTGCCCCT 720
TTTAAATTTC ACTCATTTTG TATAAACCCA GCAGGCTGGT GTTTACTTAG CCCTGTAGCT 780
TTTTTCATTT TTTCTTTCCG TCTTTCTTCT TGAGTTCACG GTTCAATATT GCCTCCTCGC 840
CCTCGTGAGG GGAGGTGCTG CTTTTCTGCC CCACCTGCCG GCTGGTTCCA GCAGCGCTGG 900
NGCCCAGCTG GGGGGCCGGG ATGGGGGCTT CTCTCTCTGG GAGGGGTGCA GGTGCCCTCC 960
CCAGGCTGGG AGGGTTCCTT CCCTAGCTCC CCATCTGCCC CCGCTGGTGA GAGTTGGGCT 1020
TCTTGGTCTT GGAACTCCCT GGCATTGGGA ACAGAGCATT TCCAGCATTT GTTGTTGTTG 1080
TTTTACTCAC CTAACCCTTA GAAAATGAAT GTTAGAAGGT GCCTGCCGAG GCGGGACAGA 1140 GTGTTTGCTC GCGCTGGAGA AGGCTCTGCT CAGCCCTCAG AGTCCCTTCC TGCCCCACCG 1200 ATACTGGCAC TTTAAAAAGG AAGCTGACCG CACAGTCTCC AGACGAATTG GCCCCCAGAA 1260 GATGGGGAGT TCTGTCCTCC CCTTCTGTGT CTGCGTGACC TCACCCAGCC TAGGAGGGAG 1320 GTGCATTCAG GGTAGATTTG CCTCTCATTC AAAGTTCTGG GGCTTTGGGY GGAAAACAGC 1380 CAGCTTTGGC GCTGTTGGGG AGACTCCTCC AGACCAGGAA CCCCAGAAGG AGACAGAGCC 1440 TGCCACATCC TCCCACGCCA GGCCCTGGGC CAGGGTGATT GGACTGAGAA TTTGGCCACA 1500 ACCAAATTGA TGCTGGCTGG AACCAGAGGC CAGAAAGCCT GGCCTTGTCC CCATGTGGGA 1560 GCCCTGTCCT CAGCCCTCTT GTCCCCTTGA GCTCAGTGAA TTCCCACCAG GTGCCCACAG 1620 CTCCTGGACT TCAAATTCTA TATATTGAGA GAGTTGGAGA GTATATCAGA GATATTTTTG 1680 GAAAGGAGTT GGTCTATGCA ATGTCAGTTT GGAATCTTCT TGAAAGTTTA ATGTTTTTAT 1740 TAGGAGATTT AAAGAAAATA AAGGTCTACA ATATCTTTAG GTTTTTTTTT TTTCCTGTTT 1800 ACCGCACAAA CTGACCACAT GGCATGTCTA TCAGCATGGA GGGTGTCCAT GTTCTCCTCT 1860 GTCTTTAGGG AGGTGATAAG GAGATGGSCG RAGGGGTGTT TTTTTCTTTG ACTCCCCTCC 1920 TTTCTAACAG AATGTTGCCA CCACTGCTTG AGTGGGCTGT GTTTGTTCCT CTGTCCCAGC 1980 TTCTGTTGTA GAAAATAACA TTGTTAGGGG AACTCAGGCT AGTGTCAGCG TCTTGGTTTG 2040 GGG 2043
(2) INFORMATION FOR SEQ ID NO: 229:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 540 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO 229: TAAAAAGAAG CGGGAGAATC TGGGCGTCGC TCTAGAGATC GATGGGCTAG AGGAGAAGCT 60 GTCCCAGTGT CGGAGAGACC TGGAGGCCGT GAACTCCAGA CTCCACAGCC GGGAGCTGAG 120 CCCAGAGGCC AGGAGGTCCC TGGAGAAGGA GAAAAACAGC CTAATCAACA AAGCCTCCAA 180 CTACGAGAAG GAACTGAAGT TTCTTCGGCA AGAGAACCGG AAGAACATGC TGCTCTCTCT 240 GGCCATCTTT ATCCTCCTGA CGCTCGTCTA TGCCTACTGG ACCATCTGAG CCTGGCACTT 300 CCCCACAACC AGCACAGGCT TCCACTTGGC CCCTTGGTCA GGATCAAGCA GGCACTTCAA 360 GCCTCAATAG GACCAAGGTG CTGGGGTGTT CCCCTCCCAA CCTAGTGTTC AAGCATGGCT 420
TCCTGGCGGC CCAGGCCTTG CCTCCCTGGC CTGCTGGGGG GTTCCGGGTC TCCAGAAGGA 480 CATGGTGCTG GTCCCTCCCT TAGCCCAAGG GAGAGGCAWT AAAGACACAA AGCTCGAAAT 540
(2) INFORMATION FOR SEQ ID NO: 230:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 448 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 230:
AATTGTGAAA TATTAGAATA TTGTTACTAT TTCACCCAAC TCAAAATCTC CATGGGAAAA 60
TACCTGTCGA TACCCACAGT ATTGTTGAAA ATAATCAGAT GCAGTATCAC AGCTGTGTCA 120
GACTCTAGTA CCAGTTGGGC AATCAAGGCA CAGCTAAAAA TTGAAAACAA AGATCTGGAC 180 AACAAAACAG CCAAAGGTGG GGGTCAAGAA GCTCTGACGT GTACCTAGCT GTAGAATGCT 240
ATGCACACGT GCCAGCTGTA GTGTGCATAT CCAGGAAAAA CTGCAGAGAG CCCCAGTCTT 300
CAMCTCTGGT TGACCATGAG CTCTGTGTAA GCAGGAAGTC AAGGCTAAGG CAGATTTAAG 360
CTCTGAAAGC ATTCCACAAC ATACACACAA ATCGTGCAAA GCATTAAGGA AATCTTGTTA 420 CTGCTAAGTG TTGCTGACCC AGGAACAA 448
(2) INFORMATION FOR SEQ ID NO: 231: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 407 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 231:
GTATGCTGCC CGAAACCAAT ATGTGTGGCT GCCTTTWACC TGACTTCTCC AACATGTAGC 60 CCCAAGAGGA GGCCTCTAGA CTRAGGGAGG GGCTGGTGAC CCAGGTGTGG TGGGGCTGCA 120
TCARACTACC AGAGAGACAG ACATTCTGGA ACTCACCCTG GGGGATCCAG TGGATCTGCC 180
TATGGTCTGG TCCACCCCAG ACCTGTGAGA TCTTCCTCAT GAGGATGCAC TTGTGCTTCT 240
GCAAGTATTG CTGCAGCTTC ATAGTGACTC CCACCAGCAC CAGCAATACA GYTAGCTACC 300
TGTGGCCTTG GATCTCAGCC AGCATGGCTG GGAGAGGGAG CARCTGGGCA TGTACCCTAA 360 ATGCTGTTAC CAGGGAAGGA CTCCCAGAGT GAAGACAAGT AGGGACT 407
(2) INFORMATION FOR SEQ ID NO: 232:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 830 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 232: GTATTTGATT TCAGGCTGCT AAATGGGCTC ATTTAGCATT CATTCCTTGA TGTAGACATT 60
AAAAAAAAAA CTGAATAGCA TTCTTTCCAG GNTAACTAAT AAAGCAGACA TGCTAAGCCT 120
ATAAATACAT CAGCACTGCA GCACACGTTT AAGGTTGCCA CGGACAAGGA TCACACAATA 180
GAGAACACTC TAGTTCGGTC TGCTCACAAG ACCCAGAACA TTGATCAGTT TTTGTTGTTG 240
GTTTATTATT TTTCTGTTAA AAAATTGTGA AAAGTTTGTT TTAGCTAGAT GATATTTTAA 300 TAGCTCCGAG TGCTTTGGAA CTATAAAGAT GTCACTACTT AACACACATA CCTTATGTTT 360
TGTTTTGTTT TGTTTTACAC TCAGTATAAA TCAGGAGAAG TTAGCCAACC ATCTAGCATT 420
TAGAATCCTC TTTTTTATTC TCTTCTAAGG ATATGGATGT TCCCATAACA GCAACAAAAC 480
AGCAACAAAA ACATTTCATA AATATCACTT GATAGACTGT AAGCACCTGC TTAACTTTGT 540
GTNCCAAATA TTTAGTGTGT ATATATATAT ATATATACAC ACACACACAC ATATATATTC 600 AACAAATAAA GCAAAATATA ACATGCATTT CACATTTTGT CTTTCCCTGT TACGATTTTA 660
ATAGCAGAAC TGTATGACAA GTTTAGGTGA TCCTAGCATA TGTTAAATTC AAATTAATGT 720
AAAACAGATT AACAACAACA AAGAAACTGT CTATTTGAGT GAAGTCATGC TTTCTATTAT 780
AATAACTTGG CTTCGGTTAT CCATCAAATC CACACTTATA CTGTTATCTG 830
(2) INFORMATION FOR SEQ ID NO: 233:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 932 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 233:
CCAGAAGAAA GACCAATCTA GAATATGGAA CTCTAATCAC TTCTAGTATT TCAACTTCCT 60
AGCAGAAATG AACTTGGCCC TAGACCTAGG GGATAAGCAA TGTTCTTTAT GTAGCCAATG 120 CTACGGAAAC AAAAGAGGTG AAAGAGACCC TTTTTTTATA CTTAATGTAC ATATATTGAC 180
TTTTTGAGCA AGAATGCCAG AAATAGCCTT CATTTCTACC CTGCAAAATA ATCCAGATCT 240
GCTTTCTAAA ATGRANTCAG TTTCTAAAGT GAAACATGCA ATATTTATGC TCTGACTGAC 300
TCCTGAATTG GARGAGGAAG RACTTCTGTT TACAGAAAAC YGTATTGTTA TATATGTCAG 360
GCTGTGTATT GTGACTATCA GCATTCTGGT GCAAATGAAC TTTTCTCCAT CATCGACTGT 420 GGAAAATTGA TACTTTTAAA GCATATTCTT CTATGAGCAC AGGTCCTCCT AGTGAAACTT 480
AATTTGACAA AGGGTGTCAT ATGCTTTCCT AACCTGAWTT GTATTAACAT TCACAGAGCC 540
TACATTTTCT CATTAGGGTT RTGATGCTCA GTATCTTTCC AAGTGCCAGG CAGRGCTTNC 600
CTTTTCTGAT CAAACATACC ATTTTTTGTA TTTCACAACT ATAGACAGTC ACTTCTGCAG 660
TCCCAATTTA AAAATGCAGA ACTGCTTTAT CCAAGAATGC TGAAAAATAC TGTTCTATCC 720 AGGTTTCCTA AACTATAAAA GCAGATTTTG CTTTTGTTTG TTAATCATAG GCATGGCCGA 780
GCATTGTGGA TTAGCCTGAG GCTTAAAATC AGATGCATGT CTGGTAAGAT GACCACTGTC 840
TCACTATCAA GAGCCTGCAG AGCCATTTTC CAGACCTGTG ATTGCCCAGA ACACATAGTC 900
CCCACGTTTC TAATTTGGAG CAAATCTAAA AG 932
(2) INFORMATION FOR SEQ ID NO: 234:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2786 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 234:
TTAGCAGGGT GAGCTGTTAA AACAGCACAC ATCTCTCATC CCCTCTTCCT TTATTCCCCC 60
CTGGGTTTCA GAAAGGAAGG ATATATGGGG ACCACCTCCC CCTTCTTTGA TCCCAGCATC 120 TCAGTCCCCC TCCCAACCCT CCATATGGCT CTCAATGGTG CTCACTTGCT TGGAAGCAGG 180
CTCCCAATAG GGAGGGGSCT GCCCTCTACA GTCTCTTTGA CTGTAAGACA GGGCTCTGTA 240
TCAGTGAGAC GATGAGAAAA GTCCCAGGCT AATGGCAGAA ATTTGCACTT TGAACATGTG 300
TGTTTTTGTG TTGTGGAACC TGAGATTCCT TATTTATTAA CAGGAAGTCT GATTTTTTTT 360
TTTTGGAGTC TTTGTTGCTA TATTTTGTGG GGCTGGGAGA GAGAGATTAG ATTATTTTGA 420 CATGGGATCC CTTCCATAAC AGGTACTTTG AAGGCAAGAC ATAGGGTTGA AGAAGCACAA 480
CCAGCCTCTC AAATCATAGC TCTCCAGTGG CTTTTAAAGA AAGCTGGTCC TCAGCACTAA 540
CAAAATCACT ACAATAGCCT AGTGCTTTTT TGGAAGCCTT TTTAGGGAAG AATGTTAGGT 600
TCATGGTAAC TAGTATGCTC TTTGAGATTT TTACAGTGTT GAAACTTAAG AATTTTGAGA 660
GGGTGAGGAG GGTTGTTCAG AATCTAAATT ACAGATAGAT GATTGTTTCT TGTGAATTTG 720
TTTCTTTTCC TTTTTTTTTC TCCCTACCAT TTCCTTACAT TTCCCTTGGG GCCCATCTCT 780
GGCTCCTTGC TTTTTGTTTC TTGCTTTGCT TTATCAGTTC ATTCCAGCTC CCTGTTAGTG 840
AAGGACACTG CTGTTAGTGA AGGAACAAAG TCTATGAGTC CTAAAATTTT AAGTCAAAGA 900
AAACTGCTCT GTTTCCCCTT TAGTAACACT TCTGAAGAGG AAAAACTTCA ATAGCCAAAG 960
TTAATAATCC TATATAATAA TTGCTTTGGC TTTCACCTAA AATTCTGGGC ATCACAATTT 1020
CCTTGGGATA GAGGTTGTCT TGGGGAATAG ATTGCTTATT GCTGTTCACT GGAGAGAAAA 1080
GGTAGTCTTT TTGTACAAGG TCATACCGCC AGAAGCCCCA AATCCTATTT TGGCTCATCT 1140
TCAGGTAAAG AGTAATTCCT ATCCTGTGTG CCTCAGAAGC TAGAATCGAA GGCTTACCCT 1200
ATTCATTGTT TATTGTCAGA AATGCATGAT GGCTCTTGGA AAGAATGACG TTTTGCTGGA 1260
AAAAAAAAAA AGAACAGTTT GTGTTTCACA AACATGGCTT ATCAATTTTT TCAAAGAATT 1320
CTTTTTTCCC AAAAAGAGGA GTAACAAAAT GTCATTTCTG AAAGAGGCTT ACTTTATACC 1380
AACTAGTGTC AGCATTTGGG ATGCCAGGGA ACAGAGAGTG AGACACCTAC AATCACCAGT 1440
CTCAAATGCG CTATTGTTTC TTTTCAGAGT GTTGCAGATT TGCCATTTCT CCATAATATG 1500
GGGATAGAAA ATGGAATAAA GATAGAAGGG ATCTAGAATA TGCTTTCCTG CCAACATGGT 1560
TTGGAGTCGA CTTTGGTATA TTGACTAGAT TTGAAAATAC AAGATTGATT AGATGAATCT 1620
ACAAAAAAGT TGTCCTCCTC TCAGGTCCCT TTTACACTTT TTGACTAACT AGCATCTATA 1680
TTCCACACTT AGCTTTTTTC TCACACTTAT CCTTTGTCTC CGTAAATTTC ATTTGCAGTG 1740
GTTAGTCATC AGATATTTTA GCCACCTACA CAAAAGCAAA CTGCATTTTT AAAAATCTTT 1800
CTGAGATGGG AGAAAATGTA TTCTCCTTTC CTATACCGCT CTCCCAACAA AAAAACAACT 1860
AGTTAGTTCT ACTAATTAGA AACTTGCTCT ACTTTTTCTT TTCTTTTAGG GGTCAAGGAC 1920
CCTCTTTATA GCTACCATTT GCCTACAATA AATTATTGCA GCAGTTTGCA ATACTAAAAT 1980
ATTTTTTATA GACTTTATAT TTTTCCTTTT GATAAAGGGA TGCTGCATAG TAGAGTTGGT 2040
GTAATTAAAC TATCTCAGCC GTTTCCCTGC TTTCCCTTCT GCTCCATATG CCTCATTGTC 2100
CTTCCAGGGA GCTCTTTTAA TCTTAAAGTT CTACATTTCA TGCTCTTAGT CAAATTCTGT 2160
TACCTTTTTA ATAACTCTTC CCACTGCATA TTTCCATCTT GAATTGGTGG TTCTAAATTC 2220
TGAAACTGTA GTTGAGATAC AGCTATTTAA TATTTCTGGG AGATGTGCAT CCCTCTTCTT 2280
KGTGGTTGCC CAAGGTTGTT TTGCGTAACT GAGACTCCTT GATATGCTTC AGAGAATTTA 2340
GGCAAACACT GGCCATGGCC GTGGGAGTAC TGGGAGTAAA ATAAAAATAT CGAGGTATAG 2400
ACTAGCATCC ACATAGAGCA CTTCAACCTC CTTTGTACCT GTTTGGGGAA AAAGTATAAT 2460 GAGTGTACTA CCAATCTAAC TAAGATTATT ATAGTCTGGT TCTTTGAAAT ACCATTTTTT 2520 TCTCCTTTTG TCTTTTTCCC ACTTTCCAAT GTACTCAAGA AAATTGAACA AATGTAATGG 2580 ATCAATTTAA AATATTTTAT TTCTTAAAAG CCTTTTTTGC CTGTTGTAAT GTGCAGGACC 2640 CTTCTCCTTT CATGGGAGAG ACAGCTAGTT ACCTGAATAT AGGTTGAAAA GGTTATGTAA 2700 AAAGAAATTA TAATAAAAGG GATACTTTGC TTTTCAAATC TTTGTTTTCT CTTATTCTAG 2760 GTAAGGCATA TTAAAAATAA ATATGT 2786
(2) INFORMATION FOR SEQ ID NO: 235:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 458 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO : 235: GGGTGCAGGA ATTCGGCACG AGAGAATGTT TCATTTTCTT TCCTATTTTA AGGATCTTCT 60 CTCTTGTTGA TGTTGAAAAC TTACCTTAGT GAAGATGTGT TTCAACATGC TGTTGTCCTT 120 TACCTGCATA ATCACAGCTA TGCATCTATT CAAAGTGATG ATCTGTGGGA TAGTTTTAAT 180 GAGGTCACAA ACCAAACACT AGATGTAAAG AGAATGATCA AAACCTGGAC CCTGCAGAAA 240 GGATTTCCTT TAGTGACTGT TCAAAAGAAA GGAAAGGAAC TTTTTATACA ACAAGAGAGA 300 TTCTTTTTAA ATATGAAGCC TGAAATTCAG CCTTCAGATA CAAGGTACAT GCCCTCTTTC 360 TTTTCATGCC ATCTCTTTTG CACTCTCAGG TGGAAATATT TTTAAGTCTT TTATAATCAT 420 AAGTTCTTGT GAAACCTAAC AAGATTATCC CTTCCTAA 458
(2) INFORMATION FOR SEQ ID NO: 236:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 591 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 236: AGGATGAAGA GGAAATTATC TCTTGGATTG CTCTCCAGGA AATCCTTCTC TATACTTTAA 60 AAGCTCTTGT TCTTTTCTAG GARTCCAATG TCCTGATTGC TGCTAACAGT CAGGGTACAA 120
TTAAGGTGCT AGAATTGGTA TGAAGGGTTA ACTCAAGTCA AATTGTACTT GATCCTGCTG 180
AAATACATCT GCAGCTGACA ATGAGAGARG AAACAGAAAA TGTCATGTGA TGTCTCTCCC 240 CAAAGTCATC ATCGGTTTTG GATTTGTTTT GAATATTTTT TCTTTTTTTC TTKTCCCTCC 300
TTTATGAGCC TTTGGGACAT TGGGAATACC CAGCCAACTC TCCACCATCA ATGTAACTCC 360
ATGGACATTG CTGCTCTTGG TGGTGTTATC TAATTTTTGT GATAGGGAAA CAAATTCTTT 420
TGAATAAAAA TAAATAACWA AACAATAAAA GTTTATTGAG CCACAGTTGA GCTTGGAAAG 480
TTTTTGTCAA ATGCNGCAAG AGATAACTCT TTTTANGAAG TAGCATATGT GAACTATAAT 540 GTAACAGTGA ATAATTTGTA AAGTTCGTAT TTCCCAACCT CTTTGGGAAT T 591
(2) INFORMATION FOR SEQ ID NO: 237:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1286 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 237: TCTTTTTAAG GTACAGCAGG GAAGAACTGG AAACTCAGAG AAAGAAACTG CCCTTCCATC 60
TACAAAAGCT GAGTTTACTT CTCCTCCTTC TTTGTTCAAG ACTGGGCTTC CACCGAGCAG 120
GAGATTACCT GGGGCAATTG ATGTTATCGG TCAGACTATA ACTATCAGCC GAGTAGAAGG 180
CAGGCGACGG GCAAATGAGA ACAGCAACAT ACAGGTCCTT TCTGAAAGAT CTGCTACTGA 240
AGTAGACAAC AATTTTAGCA AACCACCTCC GTTTTTCCCT CCAGGAGCTC CTCCCACTCA 300 CCTTCCACCT CCTCCATTTC TTCCACCTCC TCCGACTGTC AGCACTGCTC CACCTCTGAT 360
TCCACCACCG GGTTTTCCTC CTCCACCAGG CGCTCCACCT CCATCTCTTA TACCAACAAT 420
AGAAAGTGGA CATTCCTCTG GTTATGATAG TSGTTCTGCA CGTGCATTTC CATATGGCAA 480
TGCGATGAAG AACGATACAG ATACAGGGAA TATGCAGAAA GAGGTTATGA GCGTCACAGA 540
GCAAGTCGAG AAAANGAAGA ACGACATAGA GAAAGACGAC ACAGGGAGAA AGAGGAAACC 600 AGACATAAGT CTTCTCGAAG TAATAGTAGA CGTCGCCATG AAAGTCAAGA AGGAGATAGT 660
CACAGGAGAC ACAAACACAA AAAATCTAAA AGAAGCAAAG AAGGAAAAGA AGCGGGCAGT 720
GAGCCTGCCC CTGAACAGGA GAGCACCGAA GCTACACCTG CAGAATAGGC ATCGTTTTGG 780
CCTTTTGTGT ATATTAGTAC CAGAAGTAGA TACTATAAAT CTTGTTATTT TTCTGGATAA 840
TGTTTAAGAA ATTTACCTTA AATCTTGTTC TGTTTGTTAG TATGAAAAGT TAACTTTTTT 900 TCCAAAATAA AAGAGTGAAT TTTTCATGTT AAGTTAAAAA TCTTTGTCTT GTACTATTTC 960
AAAAATAAAA AGACAGCAAT GACTTTATAT CCAAGAAAGG AATGTCAATG AGTCACTTAA 1020
CAGGGAATCT AAAGAGCTGT GTTAGCTCTC TACATACACA GATTATCTGA GAAAAGGTCA 1080
AGGGTTCCAC TTGGGCCACA GTTTTTTTCT TAATCAAACA CCACTCTCTT AAGRGGCTGC 1140
ATCACAAARG GCAACCAARG GGCCCCTCTT ARGGCTTTGA GGATTAAAAC TAGTCTTTAT 1200
CCATTACTGC TGTGGACACT CTTGGCTTRG TATWTTTAGG GGGGNTCCTT ACCTTTTTTT 1260
GGTTTTCCNC ACCTTTTTGG TTGGGC 1286
(2) INFORMATION FOR SEQ ID NO: 238:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 734 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO : 238: ATGGCAGCGC AGAAGGACCA GCAGAAAGAT GCCGAGGCGG AAGGGCTGAG CGGCACGACC 60 CTGCTGCCGA AGCTGATTCC CTCCGGTGCA GGCCGGGAGT GGCTGGAGCG GCGCCGCGCG 120 ACCATCCGGC CCTGGAGCAC CTTCGTGGAC CAGCAGCGCT TCTCACGGCC CCGCAACCTG 180 GGAGAGCTGT GCCAGCGCCT CGTACGCAAC GTGGAGTACT ACCAGAGCAA CTATGTGTTC 240 GTGTTCCTGG GCCTCATCCT GTACTGTGTG GTGACGTCCC CTATGTTGCT GGTGGCTCTG 300 GCTGTCTTTT TCGGCGCCTG TTAACATTCT CTATCTGCGC ACCTTGGAGT CCAAGCTTGT 360 CCTCTTTGGC CGAAAGGTGA GCCCAGCGCA TCATATGCTC TGGCTGGAGG CATCTCCTTC 420 CCCTTCTTCT GGCTGGCTGG TGCGGGCTCG GCCGTCTTCT GGGTGCTGGG AGCCACCCTG 480 GTGGTCATCG GCTCCCACGC TGCCTTCCAC CAGATTGAGG CTGTGGACGG GGAGGAGCTG 540 CAGATGGAAC CCGTGTGAGG TCTCTTCTGG GACCTGCCGG CCTCCCGGGC CAGCTGCCCC 600 ACCCCTGCCC ATGCCTGTCC TGCACGGTCT GCTGCTCGGG CCCACAGCGC CGTCCCATCA 660 CAAGCCCGGG GAGGGATCCC GCCTTTGAAA ATAAAGCTGT TATGGGTGTC ATTCAAAAAA 720 AAAAAAAAAA AAAA 734
(2) INFORMATION FOR SEQ ID NO: 239:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 809 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 239:
CGGGGTCTTC AGGGTACCGG GCTGGTTACA GCAGCTCTAC CCCTCACGAC GCARACATGG 60
CAGCGCAGAA GGACCAGCAG AAAGATGCCG AGGCGGAAGG GCTCAGCGCC ACGACCCTGC 120 TGCCGAAGCT GATTCCCTCC GGTGCAGGCC GGGAGTGGCT GGAGCGGCGC CGCGCGACCA 180
TCCGGCCCTG GAGCACCTTC GTGGACCAGC AGCGCTTCTC ACGGCCCCGC AACCTGGGAG 240
AGCTGTGCCA GCGCCTCGTA CGCAACGTGG AGTACTACCA GAGCAACTAT GTGTTCGTGT 300
TCCTGGGCCT CATCCTGTAC TGTGTGGTGA CGTCCCCTAT GTTGCTGGTG GCTCTGGCTG 360
TCTTTTTCGG CGCCTGTTAC ATTCTCTATC TGCGCACCTT GGAGTCCAAG CTTCTGCTCT 420 TTGGCCGAGA GGTGAGCCCA GCGCATCAGT ATGCTCTGGC TGGAGGCATC TCCTTCCCCT 480
TCTTCTGGCT GGCTGGTGCG GGCTCGCCCG TCTTCTGGGT GCTGGGAGCC ACCCTGGTGG 540
TCATCGGCTC CCACGCTGCC TTCCACCAGA TTGAGGCTCT GGACGGGGAG GAGCTGCAGA 600
TGGAACCCGT GTGAGGTGTC TTCTGGGACC TGCCGGCCTC CCGGGCCAGC TGCCCCACCC 660
CTGCCCATGC CTGTCCTGCA CGGCTCTGCT GCTCGGGCCC ACAGCGCCGT CCCATCACAA 720 GCCCGGGGAG GGATCCCGCC TTTGAAAATA AAGCTGTTAT GGGTGTCATT CAGGAAAAAA 780
AAAAAAAAAA AAAAAAAAAA AAAAAAAAA 809
(2) INFORMATION FOR SEQ ID NO: 240:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 2201 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 240:
TCGACCCACG CGTCCGGCAA CATGGCGGCT GCCGTGGTGC AGCGCCCGGG CTGAGCGACA 60 GCAAGTGCAG CGGGCTCCTA CCCCGGGTGA GGGGTGGCCT CCGCGTGGGA TCGTGCCCTC 120
TTCAGCCCGC TCCTGTCCCC GACATCACGT GTATTCCGCA CGTCCCCTCC GCGCTGTGTG 180
TCTACTGAGA CGGGGAGGCG TGACAGGGCC CGGGTCCCTT CTCAGTGGTG CTCTGTGCTT 240 CAGGGCAAGC TCCCCGTCTC CGGGCGCACT TCCCTCGCCT GTGTTCGGTC CATCCTCCTT 300
TCTCCAGCCT CCTCCCCTCG CAGGCGGATG AMCCGGACGA CGGGCCAGTG CCTGGCACCC 360
CGGGGTTGCC ARGGTCCAMG GGGAACCCGA AGTCCGAGGA GCCCGARGTC CCGAACCAGG 420
ARGGGCTGCA GCGCATCAMC GGCCTCTCTC CCGGCCGTTC GGCTCTCATA GTGGCGGTGC 480
TGTGCTACAT CAATCTCCTG AACTACATGG ACCGCTTCAC CGTGGCTGGC GTCCTTCCCG 540
ACATCGAGCA GTTCTTCAAC ATCGGGGACA GTAGCTCTGG GCTCATCCAG ACCGTGTTCA 600
TCTCCAGTTA CATGGTGTTG GCACCTGTGT TTGGCTACCT GGGTGACAGG TACAATCGGA 660
AGTATCTCAT GTGCGGGGGC ATTGCCTTCT GGTCCCTGGT GACACTGGGG TCATCCTTCA 720
TCCCCGGAGA GCATTTCTGC CTGCTCCTCC TGACCCGGGG CCTGGTGGGG GTCGGGGAGG 780
CCAGTTATTC CACCATCGCG CCCACTCTCA TTGCCGACCT CTTTGTGGCC GACCAGCGGA 840
CCGGATGCTC AGCATCTTCT ACTTTGCCAT TCCGGTGGGC AGTGGTCTGG GCTACATTGC 900
AGGCTCCAAA GTGAAGGATA TGGCTGGAGA CTGGCACTGG GCTCTGAGGG TGACACCGGG 960
TCTAGGAGTG GTGGCCGTTC TGCTGCTGTT CCTGGTAGTG CGGGAGCCGC CAAGGGGAGC 1020
CGTGGAGCGC CACTCAGATT TGCCACCCCT GAACCCCACC TCGTGGTGGG CAGATCTGAG 1080
GGCTCTGGCA AGAAATCCTA GTTTCGTCCT GTCTTCCCTG GGCTTCACTG CTGTGGCCTT 1140
TGTCACGGGC TCCCTGGCTC TGTGGGCTCC GGCATTCCTG CTGCGTTCCC GCGTGGTCCT 1200
TGGGGAGACC CCACCCTGCC TTCCCGGAGA CTCCTGCTCT TCCTCTGACA GTCTCATCTT 1260
TGGACTCATC ACCTGCCTGA CCGGAGTCCT GGGTGTGGGC CTGGGTGTCG AGATCAGCCG 1320
CCGGCTCCGC CACTCCAACC CCCGGGCTGA TCCCCTGGTC TGTGCCACTG GCCTCCTGGG 1380
CTCTGCACCC TTCCTCTTCC TGTCCCTTCC CTGCGCCCGT GGTAGCATCG TGGCCACTTA 1440
TATTTTMATC TTCATTGGAG AGACCCTCCT GTCCATGAAC TGGGCCATCG TGGCCGACAT 1500
TCTGCTGTAC GTGGTGATCC CTACCCGACG CTCCACCGCC GAGGCCTTCC AGATCGTGCT 1560
GTCCCACCTG CTGGGTGATG CTGGGAGCCC CTACCTCATT GGCCTGATCT CTGACCGCCT 1620
GCGCCGGAAC TGGCCCCCCT CCTTCTTGTC CGAGTTCCGG GCTCTGCAGT TCTCGCTCAT 1680
GCTCTGCGCG TTTGTTGGGG CACTGGGCGG CGCACTTTCC TGGGCACCGC CATCTTCATT 1740
GAGGCCGACC GCCGGCGGGC ACAGCTGCAC GTGCAGGGCC TGCTGCACGA AGCAGGGTCC 1800
ACAGACGACC GGATTGTGGT GCCCCAGCGG GGCCGCTCCA CCCGCGTGCC CGTGGCCAGT 1860
GTGCTCATCT GAGARGCTGC CGCTCACCTA CCTGCACATC TGCCACAGCT GGCCCTGGGC 1920
CCACCCCACG AAGGGCCTGG GCCTAACCCC TTGGCCTGGC CCAGCTTCCA GAGGGACCCT 1980
GGGCCGTGTG CCAGCTCCCA GACACTACMT GGGTAGCTCA GGGGAGGAGG TGGGGGTCCA 2040
GGAGGGGGAT CCCTCTCCAC AGGGGCAGCC CCAAGGGCTC GGTGCTATTT GTAACGGAAT 2100
AAAATTTGTA GCCAGACCCC AGGTGCCTGC TCTCGTCTTT CTCTGGGTGG CCTCTGATCT 2160
TGCACCCCGT CTTCACCCCA GGGCTCCTGA AGACTGTGGG T 2201
(2) INFORMATION FOR SEQ ID NO: 241:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1661 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 241:
GTCCTTCCCG ACATCGAGCA GTTCTTCAAC ATCGGGGACA GTAGCTCTGG GCTCATCCAG 60
ACCGTGTTCA TCTCCAGTTA CATGGTGTTG GCACCTGTGT TTGGCTACCT GGGTGACAGG 120
TACAATCGGA AGTATCTCAT GTGCGGGGGC ATTGCCTTCT GGTCCCTGGT GACACTGGGG 180
TCATSCTTCA TCCCCGGAGA GCATTTCTGG CTGCTCCTCC TGACCCGGGG CCTGGTGGGG 240
GTCGGGGAGG CCAGTTATTC CACCATCGCG CCCACTCTCA TTGCCGACCT CTTTCTCGCC 300
GACCAGCGGA SCGGATGCTC AGCATCTTCT ACTTTGCCAT TCCGGTGGGC •'AGTGGTCTGG 360
GCTACATTGC AGGCTCCAAA GTGAAGGATA TGGCTGGAGA CTGGCACTGG GCTCTGAGGG 420
TGACACCGGG TCTAGGAGTG GTGGCCGTTC TGCTGCTGTT CCTGGTAGTG CGGGAGCCGC 480
CAAGGGGAGC CGTGGAGCGC CACTCAGATT TGCCACCCCT GAACCCCACC TCGTGGTGGG 540
CAGATYTGAG GGCTCTGGCA AGAAATCCTA GTTTCGTCCT GTCTTCCCTG GGCTTCACTG 600
CTGTGGCCTT TGTCACGGGC TCCCTGGCTC TGTGGGCTCC GGCATTCCTG CTGCGTTCCC 660
GCGTGGTCCT TGGGGAGACC CCACCCTGCC TTCCCGGAGA CTCCTGCTCT TCCTCTGACA 720
GTCTCATCTT TGGACTCATC ACCTGCCTGA CCGGAGTCCT GGGTGTGGGC CTGGGTGTCG 780
AGATCAGCCG CCGGYTCCGC CACTCCAACC CCCGGGCTGA TCCCCTGGTC TGTGCCACTG 840
GCCTCCTGGG CTCTGCACCC TTCCTCTTCC TGTCCCTTCC CTGCGCCCGT GGTAGCATCG 900
TGGCCACTTA TATTTTCATC TTCATTGGAG AGACCCTCCT GTCCATGAAC TGGGCCATCG 960
TGGCCGACAT TCTGCTGTAC GTGGTGATCC CTACCCGACG CTCCACCGCC GAGGCCTTCC 1020
AGATCGTGCT GTCCCACCTG CTGGGTGATG CTGGGAGCCC CTACCTCATT GGCCTGATCT 1080
CTGACCGCCT GCGCCGGAAC TGGCCCCCCT CCTTCTTGTC CGAGTTCCGG GCTCTGCAGT 1140
TCTCGCTCAT GCTCTGCGCG TTTGTTGGGG CACTGGGCGG CGCACTTTCC TGGGCACCGN 1200
CATCTTCATT GAGGCCGACC GCCGGCGGGC ACAGCTGCAC GTGCAGGGCC TGCTGCACGA 1260
AGCAGGGTCC ACAGACGACC GGATTGTGGT GCCCCAGCGG GGCCGCTCCA CCCGCGTGCC 1320
CGTGGCCAGT GTGCTCATCT GAGAGGCTGC CGCTCACCTA CCTGCACATC TGCCACAGCT 1380
KGCCCTGGGC CCACCCCACG AAGGGCCTGG GCCTAACCCC TTGGCCTGGC CCAGCTTCCA 1440
GAGGGACCCT GGGCCGTGTG CCAGCTCCCA GACACTACMT GGGTAGCTCA GGGGAGGAGG 1500
TGGGGGTCCA GGAGGGGGAT CCCTCTCCAC AGGGGNCACC CCAAGGGCTC GGTGCTATTT 1560
GTAACGGAAT AAAATTTGTA GCCAGACCCC AGGTGCCTGC TCTCGTCTTT CTCTGGGTGG 1620
CCTCTGATCT TGCACCCCGT CTTCACCCCA GGGCTCCTGA A 1661
(2) INFORMATION FOR SEQ ID NO: 242:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1146 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 242:
NGACAGAAAA GCAGAAGATG AGACTCTGTT CATTCACTTT TCCTAGGCCC ATCCTGTGGT 60
CATCTTTCCC CCTCCCATCA TACCTCCTCC TTCCTGGAGC CTCTGCCGGC TTGGCTCTAA 120
TGGTGGCACT TACCTGGATA TTTCAGTGGG AGGATGAAAG GCGAGACTCA CCCTACGCGG 180
TGGGACAGAT GGGGAGAGGA AAAAGGCAGA GATNGCCAGG AGAGGGGTGC AGGACAAACC 240
AGAGAGGTTG GGTCAGGGGA AAAGTGTNGG GAGAAAGTGG GGTGCAGGCC CTGCAGGCCG 300
GTTTAGCCAG CAGCTGCGGC CTCCCCGGGC CCTTGGCATC CAACTTCGCA GACAGGGTAC 360
CAGCCTCCTG GTGTGTATCA TAGGATTTGT TCACATAGTG TTATGCATGA TCTTCGTAAG 420
GTTAAGAAGC CGTGGTGGTG CACCATGACA TCCAACCCGT ATATATAAAG ATAAATATAT 480
ATATATATGT ATGTAAATTA TAGCACTGAG GGCCCTGCTG CCCTCCTGCA CCAAGCAAAA 540
CTAAGCCTTT TGCTTTGGGT ATTATGTTTC GTTTTGTTAT TTGTTTGTTT TTGTGGCTTG 600
TCTTATGTCG TGATAGCACA AGTGCCAGTC GGATTGCTCT GTATTACAGA ATAGTGTTTT 660
TAATTCATCA ATGTTCTAGT TAATGTCTAC CTCAGCACCT CCTCTTAGCC TAATTTTAGG 720
AGGTTGCCCA ATTTTGTTTC TTCAATTTTA CTCGTTACTT TTTTGTACAA ATCAATCTCT 780
TTCTCTCTTT CTCTCCTCCC CACCTCTCAC CCTTGCCCTC TCCATCTCCC TCTCCCGCCC 840
TCCCCTCCTC CCTCTGGCTC CCCGTCTCAT TTCTGTCCAC TCCATTCTCT CTCCCTCTCT 900
CCTGCCTCCT GCTGCCCCCT CCCCAGCCCA CTTSCCCGAG TTGTGCTTGC CGCTCCTTAT 960
CTGTTCTAGT TCCGAAGCAG TTTCACTCGA AGTTGTGCAG TCCTGGTTGC AGCTTTCCGC 1020
ATCTGCCTTC GTTTCGTGTA GATTGACGCG TTTCTTTCTA ATTTCAGTGT TTCTGACAAG 1080
ATTTAAAAAA AAAAAAAGGA AAAAAAAAAA AAAAAAAAAC TCGAGGGGGG GCCCGGTACC 1140
CAATTG 1146
(2) INFORMATION FOR SEQ ID NO: 243:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1350 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 243:
AACCCACGCC TGCTGCGGCA GGGCGTGGAG GGCAGAGGGC CGCGGAGGCG CAGTTGCAAA 60
CATGGCTCAG AGCAGAGACG GCGGAAACCC GTTCGCCGAG CCCAGCGAGC TTGACAACCC 120
CTTTCAGCCA CCACCAGCCT ATGAGCCTCC AGCCCCTGCC CCATTGCCTC CACCCTCAGC 180
TCCCTCCTTG CAGCCCTCGA GAAAGCTCAG CCCCACAGAA CCTAAGAACT ATGGCTCATA 240
CAGCACTCAG GCCTCAGCTG CAGCAGCCAC AGCTGAGCTG CTGAAGAAAC AGGAGGAGCT 300
CAACCGGAAG GCAGAGGAGT TGGACCGAAG GAGNCGAGAG CTGCAGCATG CTGCCCTGGG 360
RGGCACAGCT ACTCGACAGA ACAATTGGCC CCCTCTACCT TCTTTTTGTC CAGTTCAGCC 420
CTGCTTTTTC CAGGACATCT CCATGGAGAT CCCCCAAGAA TTTCAGAAGA CTGTATCCAC 480
CATGTACTAC CTCTGGATGT GCAGCACGST GGCTCTTCTC CTGAACTTCC TCGCCTCCCT 540
GGCCAGCTTC TGTGTGGAAA CCAACAATGG CGCAGGCTTT GGGCTTTCTA TCCTCTGGGT 600
CCTCCTTTTC ACTCCCTGCT CCTTTGTCTG CTGGTACCGC CCCATGTATA AGGCTTTCCG 660
GAGTGACAGT TCATTCAATT TCTTCGTTTT CTTCTTCATT TTCTTCGTCC AGGATGTGCT 720
CTTTGTCCTC CAGGCCATTG GTATCCCAGC TTGGGGATTC AGTGGCTGGA TCTCTGCTCT 780
GGTGGTGCCG AAGGCAACAC AGCAGTATCC GTGCTCATCC TGCTGGTCGC CCTGCTCTTC 840
ACTGGCATTG CTGTGCTAGG AATTGTCATG CTGAAACGGA TCCACTCCTT ATACCGCCGC 900
ACAGGTGCCA GCTTTCAGAA GGCCCAGCAA GAATTTGCTG CTGGTGTCTT CTCCAACCCT 960
GCGGTGCGAA CCGCARCTTG CCAATGCAGC CGCTGGGGCT GCTGAAAATG CCTTCCGGGC 1020
CCCGTGACCC CTGACTGGGA TGCCCTGGCC CTGCTACTTG AGGGAGCTGA CTTAGCTCCC 1080
GTCCCTAAGG TCTCTGGGAC TTGGAGAGAC ATCACTAACT GATGGCTCCT CCGTAGTGCT 1140
CCCAATCCTA TGCCCATGAC TCCTGAACCT GACAGGCGTG TGGGGAGTTC ACTGTGACCT 1200
AGTCCCCCCA TCAGGCCACA CTGCTCCCAC CTCTCACACG CCCCAACCCA GCTTCCCTCT 1260
GCTGTGCCAC GCCTCTTCCT TCGGTTATTT AAATAAAAAG AAAGTGGAAC TGGAAAAAAA 1320
AAAAAAAAAA AAAAAAAAAG GGGGGNCCNC 1350
(2) INFORMATION FOR SEQ ID NO: 244:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1529 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 244:
TCCCAGAGGC CGGGGGGTTC CAGCTCTGCC TGTAGCAGAG CCCTGAGGAG GAGGAGGAAG 60
AGGATGTGCT GAAATACGTC CGGGAGATCT TTTTCAGCTA GGGCATAAAC TCTGCACTCA 120
ACTGTCTGCC GAGAGCAGCT GGAGGACAGC TGAGCTTCCA CTGCTCCTCC TGGGCCGMCC 180
GCCTGTGGGA ATGGGGCTCT CTGTGCTCCT ACCTTTGTGC CTTCTTGGGC CTGGCAGATT 240
CACCTCAGGC CAGAAGCCCC TGGACACTCC GGGCCTTGGG GTGCCGTTCT GAGTGTGCGG 300
AAGGCAGGAC TCAAAATGAG ATCCCATTTG ACTCCCTCTG TATGTACTGT GCCCTCTCCT 360
GGCTCTTGAG GCTCTGGAGT CCCAATTGTC TGTGTTAGTC AGTGACCAGG TTCCAGGGAA 420
AATRATGTCA TGTGGTGGTC CAACTTACTC GAACCAAAGA GACAGTACTT TGCAAAGAAA 480
AGGATCACTG CCAGGTGCAC TGGAATTCCT ACAGTTTAGT CCGCATCATC TCTCCTGAAG 540
GAGGAAGCCT GTTTCAAAAA TAGTTTCCAT CATGAGTCTA TCAATGAGCT CCCACCTCTC 600
CAGCCAGCCT AGAAAGCAAA CGAGCTGCCC ACAGTTCTCT GCCCTGTCTG GGAGGTTGAG 660
GCCACAGTGT ATAGACTGGT AAGCCAGACA GGCCTCCTCC CGCAAGCTGC TACCTTGCTT 720
TCACCTGTAC CTTGGTCCCC GGGCAGCTAG CTATAAAGCA AGAGGGACAG GAGCCCAGAA 780
GAGACACTGA GGACAAGAGA TCACACCAGA GTACATGTCT CTGCCTCTGT TTTCAGTGTG 840
GCTTTGGACA GGAATATATG AATAAATCAC TGCCATACAG GTTTTCCAAT ACACAAGTGC 900
TAGAAAATAC ACACAATTCC CCAATGCGTA AGTTGTGCTA ATGTCTTTCC AAGTTCTGGG 960
TTGGGAAGTG GAGGGTGGCA GCGTTTGTTT GTGCGCAACC GTCCAGTCCT GTTCACAGCG 1020
AGGATTTGGA GTCCTCCAGG GTCTCATCAT GGGAGTGATT TGTCAGCGGA CGCCTCTGCC 1080
CTGTCTGGCT TCAGGTCCAG GGAAGCTTTG AAGCAGTCAA GCCTTGTCTT TGTACCCCAT 1140
GTGTCCTGTC TTTGTTGAGT CACTCAGAGA TCACTCCTGG ACCTCTGGGG TTGGAGTTCC 1200
AGTGATGGCT TATGGCGGCC CACTCACTAT GGTCGGCTGA GTGGAAGCTC CTTAACCATG 1260
TCCCCAGAGA CACTGAGGTG CTCGCTCTTT TAATGTCCTC GTTTGTTGCC GTAAGTTCTT 1320
TGCTAGGTTT CATTTTGGCA TTTGGCAAAT CAGCCTGGAA GTCTGGCCCC ATGACAGCAA 1380
TCACTCCCTC CCCACCCTCC TGAAGCTAGA GGAAGATTTG CTCAGATCCA TTAATTAAAG 1440
CAGGAATTGG TGTGACAATG AGCTGCATGG TTTAGGGAGT CTTTGGGAGC CTTGGAAGTC 1500 CTGAAGGACA AACAATCTTG TACTAAGAA 1529
(2) INFORMATION FOR SEQ ID NO: 245:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1537 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 245:
GTGCGAGGTC CCCGCCAGCC CCCAGCGGCC TTCCCGGCCC GGGGCGCTCC CAGAGCAAAC 60
GAGGCCCCTG AGAGCTCCAC CTAGTTCACA GGATAAAATC CCACAGCAGA ACTCGGAGTC 120
AGCAATGGCT AAGCCCCAGG TGGTTGTAGC TCCTGTATTA ATGTCTAAGC TGTCTGTGAA 180
TGCCCCTGAA TTTTACCCTT CAGGTTATTC TTCCAGTTAC ACAGAATCCT ATGAGGATGG 240
TTGTGAGGAT TATCCTACTC TATCAGAATA TGTTCAGGAT TTTTTGAATC ATCTTACAGA 300
GCAGCCTGGC AGTTTTGAAA CTGAAATTGA ACAGTTTGCA GAGACCCTGA ATGGTTGTGT 360
TACAACAGAT GATGCTTTGC AAGAACTTGT GGAACTCATC TATCAACAGG CCACATCTAT 420
CCCAAATTTC TCTTATATGG GAGCTCGCCT GTGTAATTAC CTGTCCCATC ATCTGACAAT 480
TAGCCCACAG AGTGGCAACT TCCGCCAATT GCTACTTCAA AGATGTCGGA CTGAATATGA 540
AGTTAAAGAT CAAGCTCCAA AAGGGGATGA AGTTACTCGA AAACGATTTC ATGCATTTGT 600
ACTCTTTCTG GGAGAACTTT ATCTTAACCT GGAGATCAAG GGAACAAATG GACAGGTTAC 660
AAGAGCAGAT ATTCTTCAGG TTGGTCTTCG AGAATTGCTG AATGCCCTGT TTTCTAATCC 720
TATGGATGAC AATTTAATTT GTGCAGTAAA ATTGTTAAAG TTGACAGGAT CAGTTTTGGA 780
AGATGCTTCG AAGGAAAAAG GAAAGATGGA TATGGAAGAA ATTATTCAGA GAATTGAAAA 840
CGTTCTCCTA GATGCAAACT GCAGTAGAGA TGTAAAACAG ATGCTCTTCA AGCTTGTAGA 900
ACTCCGGTCA AGTAACTGGG GCAGAGTCCA TGCAACTTCA ACATATAGAG AAGCAACACC 960
AGAAAATGAT CCTAACTACT TTATGAATGA ACCAACATTT TATACATCTG ATGGTGTTCC 1020
TTTCACTGCA GCTCATCCAG ATTACCAAGA GAAATACCAA GAATTACTTG AAAGAGAGGA 1080
CTTTTTTCCA GATTATGAAG AAAATGGAAC AGATTTATCC GGCGCTGGTG ATCCATACTT 1140
GGATGATATT GATGATGAGA TGGACCCAGA GATAGAAGAA GCTTATGAAA AGTTTTGTTT 1200
GGAATCAGAG CGTAAGCGAA AACAGTAAAG TTAAATTTCA GCATATCAGT TTTATAAAGC 1260
AGTTTAGGTA TGGTGATTTA GCAGAACACA AGAGAGCAAG AAAATGTGTC ACATCTATAC 1320
CAAATTRAGG ATGTTGAGTT ATGTTACTAA TGTATGCAAC TTTAATTTTG TTTAACACTA 1380
TCTCCCAAAA TAAACTTTAT TCCCTATAAC TTAAAATGTG TATATATATA TAATAGTTTA 1440
TTATGTACAG TTAATTCTAC TGTTTTGGCT GCAATAAAAT CGATTTTGAA ATAAAWRAAA 1500
AAAAAAAAAA AAGGGNGGCC GCTCTAGAGG ANCCAAG 1537
(2) INFORMATION FOR SEQ ID NO: 246:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 506 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO : 246: TGCAGGATTT GGCCAGGACC CSCCGCGGTG GCGGTTGCTA TCGCTTCGCA GAACCTACTC 60 AGGCAGCCAG CTGAGAAGAG TTGAGGGAAA GTGCTGCTGC TGGGTCTGCA GACGCGATGG 120 ATAACGTGCA GCCGAAAATA AAACATCGCC CCTTCTGCTT CAGTGTGAAA GGCCACGTGA 180 AGATGCTGCG GCTGGATATT ATGAACTCAC TGGTAACAAC AGTATTCATG CTCATCGTAT 240 CTGTGTTGGC ACTGATACCA GAAACCACAA CATTGACAGT TGGTGGAGGG GTGTTTGCAC 300 TTGTGACAGC AGTATGCTGT CTTGCCGACG GGGCCCTTAT TTACCGGAAG CTTCTGTTCA 360 ATCCCAGCGG TCCTTACCAG AAAAAGCCTG TGCATGAAAA AAAAGAAGTT TTGTAATTTT 420 ATATTACTTT TTAGTTTGAT ACTAAGTATT AAACATATTT CTGKATTATT CCAAAAAAAA 480 AAAAAAAAAA AAAAAAAATT TGGTGG 506
(2) INFORMATION FOR SEQ ID NO: 247:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1348 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 247: GTCTTTCTTT TNCTCTTTTG AGTTGGTGAG TGAGTGAATA GGGTAACATG GGCCTTCAGG 60 ATGACCCCTT GGAACTCTGC CGAGTTCCTT AAATCTCAGC TGGGATCCTC GACCTGGGAG 120 GCCCCTGTCA GGGCCAGCTC TCGAAAAACC TGGGAGTTGA TGCCGGAGGY TGGGAAGAAC 180
TCTGCTCGAG GGCAGGGTGC CCTGGAACAC TGGTAGTTCT GGGGCTGGGA GGGAGAGGGG 2 0
CTCCGGCTTT CTCTGAAATG AACACTGCTC TTCAGCAGTT CAAGTACTTG TTCTCAAAAC 300
ATTTTCTAAT TGATTGGTAG GTTTTCATAA GCATTGTTTC TTTAAGGCAT GGAAAGGGAA 360
GAATGCTCAA GCAAGTCATG TTTGTTTTCA GTGGGATGGG CCCGCGTTCT CACTGCTGGG 420
GGCTTCCCCT TGCATGTGGC ACCTTTGTGC AGGGCCACCA GGCAGACTCT TCCCACCTTC 480
TCCCACTGAA GCACCAAGGG GCTTGAACCG TAATTTGGCT AATCAGAGGC ATTTTTTTTG 540
TCCTAGTATC TTTCACACTT GTCCAACCGT CTTATTTTTT TAAAAGTTCT GTTGCTTGTA 600
TTAACACGAA ACTAGAGAGA AATAGTTTCT GAAGCCAGTT TATTGTGAAG ATCCCCAAGG 660
GGAGGTTCGG TAGAGAAAAA TAGTAAGCTG GTTTAGAAAC TGACGAGGGC AAACAGCCAG 720
GACGCATTGG AGAGGAATTT GCCAAAGATC TACCCTGAGA TAACGCCTGT CCAGTGTCTT 780
CACCACGTGA ATAACCAGCG CTCCAAAGTG TTTTTCTGCT TTGAAAAAAA AAATTCCACA 840
AGCTTTTAAA GGTGCATTTA AGAATCCATG TGACTTTAGA ATGGAACTGC CGGCCCTGGC 900
AACTGTCACG TGTGCTAGAA GGTTCGATGC CTCTGGAATG CATGTGATAC TCATCTCCAT 960
TTTGTTTCCT TGATTGCATT TTTCTTCTTT TAGCAGATCT GTCCCTGTGG GTGGTGTCTA 1020
AGAAGTCGGA CACCTTGGTT TTTGTGTTAG ATTGAGCTGG GCAGCTGCAA TCAGCTTCTT 1080
TATATGCAAA TTAGGCACGA CCCATCTGTG GTTCCCTGGT TGGTGGCTAA TGAAGTGAGG 1140
GGAGGGAGGG ATGTCACCCC AAAAGTAGGC CCTCCCATTG GCTTTGGCCA GGCCAGACAC 1200
TTCACATCGT TTACATGGTT CTGTGTAATT TTAAAGTTTA TGTGTATAAA GCGAAGCTGT 1260
TTCTGTGAAA CTGTATATTT TGTAAATAAA TATATTGCTA CTTTGAGAWR AAAAAAAAAA 1320
AAAAACTCGA GGGGGGCCCG GTACCCAA 13 8
(2) INFORMATION FOR SEQ ID NO: 248:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1766 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 248: GTCCCGAATC GGCAGAGCGG CACGAGCGGC CACGAGAGCA GGCGGAGTAA AGGGACTTGA 60 GCGAGCCAGT TGCCGGATTA TTCTATTTCC CCTCCCTCTC TCCCGCCCCG TATCTCTTTT 120 CACCCTTCTC CCACCCTCGC TCGCGTASCA TGGCGGAGCG TCGGCGGCCA CTCAGTCCCA 180
TTCCATCTCC TCGTCGTCCT TCGGAGCCGA GCCGTCCGCG CCCGGCGGCG GCGGGAGCCC 240
AGGAGCCTGC CCCGCCCTGG GGACGAAGAG CTGCAGCTCC TCCTGTGCGG TGCACGATCT 300
GATTTTCTGG AGAGATGTCA AGAAGACTGG GTTTGTCTTT GGCACCACGC TGATCATGCT 360
GCTTTCCCTG GCAGCTTTCA GTGTCATCAG TGTGGTTTCT TACCTCATCC TGGCTCTTCT 420
CTCTGTCACC ATCAGCTTCA GGATCTACAA GTCCGTCATC CAAGCTGTAC AGAAGTCAGA 480
AGAAGGCCAT CCATTCAAAG CCTACCTGGA CGTAGACATT ACTCTGTCCT CAGAAGCTTT 540
CCATAATTAC ATGAATGCTG CCATGGTGCA CATCAACAGG GCCCTGAAAC TCATTATTCG 600
TCTCTTTCTG GTAGAAGATC TGGTTGACTC CTTGAAGCTG GCTGTCTTCA TGTGGCTGAT 660
GACCTATGTT GGTGCTGTTT TTAACGGAAT CACCCTTCTA ATTCTTCCTG AACTGCTCAT 720
TTTCAGTGTC CCGATTGTCT ATGAGAAGTA CAAGACCCAG ATTGATCACT ATGTTGGCAT 780
CGCCCGAGAT CAGACCAAGT CAATTGTTGA AAAGATCCAA GCAAAACTCC CTGGAATCGC 840
CAAAAAAAAG GCAGAATAAG TACATGGAAA CCAGAAATGC AACAGTTACT AAAACACCAT 900
TTAATAGTTA TAACGTCGTT ACTTGTACTA TGAAGGAAAA TACTCAGTGT CAGCTTGAGC 960
CTGCATTCCA AGCTTTTTTT TTAATTTGGT GTTTTCTCCC ATCCTTTCCC TTTAACCCTC 1020
AGTATCAAGC ACAAAAATTG ATGGACTGAT AAAAGAACTA TCTTAGAACT CAGAAGAAGA 1080
AAGAATCAAA TTCATAGGAT AAGTCAATAC CTTAATGGTG GTAGAGCCTT TACCTCTAGC 1140
TTGAAAGGGG AAAGATTGGA GGTAAGAGAG AAAATGAAAG AACACCTCTG GGTCCTTCTG 1200
TCCAGTTTTC AGCACTAGTC TTACTCAGCT ATCCATTATA GTTTTGCCCT TAAGAAGTCA 1260
TCATTAACTT ATGAAAAAAT TATTTGGGGA CAGGAGTGTG ATACCTTCCT TGGTTTTTTT 1320
TTGCAGCCCT CAAATCCTAT CTTCCTGCCC CACAATGTGA GCAGCTACCC CTGATACTCC 1380
TTTTCTTTAA TGATTTAACT ATCAACTTGA TAAATAACTT ATAGGTGATA GTGATAATTC 1440
CTGATTCCAA GAATGCCATC TGATAAAAAA GAATAGAAAT GGAAAGTGGG ACTGAGAGGG 1500
AGTCAGCAGG CATGCTGCGG TGGCGGTCAC TCCCTCTGCC ACTATCCCCA GGGAAGGAAA 1560
RGCTCCGCCA TTTGGGAAAG TGGTTTCTAC GTCACTGGAC ACCGGTTCTG AGCATTAGTT 1620
TGAGAACTCG TTCCCGAATG TGCTTTCCTC CCTCTCCCCT GCCCACCTCA AGTTTAATAA 1680
ATAAGGTTGT ACTTTTCTTA CTATAAAATA AAAAAAAAAA AACTCGAGGG GGGCCCGGTA 1740
CCCAAATCGC CGGATATGAT CGTAAA 1766
(2) INFORMATION FOR SEQ ID NO: 249: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2664 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 249:
AGTGTCCTCG GAGCAGGCGG AGTAAAGGGA CTTGAGCGAG CCAGTTGCCG GATTATTCTA 60
TTTCCCCTCC CTCTCTCCCG CCCCGTATCT CTTTTCACCC TTCTCCCACC CTCGCTCGCG 120
TASCATGCCG GAGCGTCGGC GGCCACTCAG TCCCATTCCA TCTCCTCGTC GTCCTTCGGA 180
GCCGAGCCGT CCGCGCCCGG CGGCGGCGGG AGCCCAGGAG CCTGCCCCGC CCTGGGGACG 240
AAGAGCTGCA GCTCCTCCTG TGCGGTGCAC GATCTGATTT TCTCGAGAGA TGTGAAGAAG 300
ACTGGGTTTG TCTTTGGCAC CACGCTGATC ATGCTGCTTT CCCTGGCAGC TTTCAGTGTC 360
ATCAGTGTGG TTTCTTACCT CATCCTGGCT CTTCTCTCTG TCACCATCAG CTTCAGGATC 420
TACAAGTCCG TCATCCAAGC TGTACAGAAG TCAGAAGAAG GCCATCCATT CAAAGCCTAC 480
CTGGACGTAG ACATTACTCT GTCCTCAGAA GCTTTCCATA ATTACATGAA TGCTGCCATG 540
GTGCACATCA ACAGGGCCCT GAAACTCATT ATTCGTCTCT TTCTGGTAGA AGATCTGGTT 600
GACTCCTTGA AGCTGGCTGT CTTCATGTGG CTGATGACCT ATCTTGGTGC TGTTTTTAAC 660
GGAATCACCC TTCTAATTCT TGCTCAACTC CTCATTTTCA GTGTCCCGAT TGTCTATGAG 720
AAGTACAAGA CCCAGATTGA TCACTATGTT GGCATCGCCC GAGATCAGAC CAAGTCAATT 780
GTTGAAAAGA TCCAAGCAAA ACTCCCTGGA ATCGCCAAAA AAAAGGCAGA ATAAGTACAT 840
GGAAACCAGA AATGCAACAG TTACTAAAAC ACCATTTAAT AGTTATAACG TCGTTACTTG 900
TACTATGAAG GAAAATACTC AGTGTCAGCT TGAGCCTGCA TTCCAAGCTT TTTTTTTAAT 960
TTGGTGTTTT CTCCCATCCT TTCCCTTTAA CCCTCAGTAT CAAGCACAAA AATTGATGGA 1020
CTGATAAAAG AACTATCTTA GAACTCAGAA GAAGAAAGAA TCAAATTCAT AGGATAAGTC 1080
AATACCTTAA TGGTGGTAGA GCCTTTACCT GTAGCTTGAA AGGGGAAAGA TTGGAGGTAA 1140
GAGAGAAAAT GAAAGAACAC CTCTGGGTCC TTCTGTCCAG TTTTCAGCAC TAGTCTTACT 1200
CAGCTATCCA TTATAGTTTT GCCCTTAAGA AGTCATGATT AACTTATGAA AAAATTATTT 1260
GGGGACAGGA GTGTGATACC TTCCTTGGTT TTTTTTTGCA GCCCTCAAAT CCTATCTTCC 1320
TGCCCCACAA TGTGAGCAGC TACCCCTGAT ACTCCTTTTC TTTAATCATT TAACTATCAA 1380
CTTGATAAAT AACTTATAGG TGATAGTGAT AATTCCTGAT TCCAAGAATG CCATCTGATA 1440
AAAAAGAATA GAAATGGAAA GTGGGACTGA GAGGGAGTCA GCAGGCATGC TGCGGTGGCG 1500
GTCACTCCCT CTGCCACTAT CCCCAGGGAA GGAAARGCTC CGCCATTTGG GAAAGTGGTT 1560
TCTACGTCAC TGGACACCGG TTCTGAGCAT TAGTTTGAGA ACTCGTTCCC GAATGTGCTT 1620
TCCTCCCTCT CCCCTGCCCA CCTCAAGTTT AATAAATAAG GTTGTACTTT TCTTACTATA 1680 AAATAAATCT CTGTAACTGC TCTGCACTCC TCTAAACTTG TTAGAGAAAA AAATAACCTG 1740 CATGTGGGCT CCTCAGTTAT TCAGTTTTTG TGATCCTATC TCAGTCTGGG GGGGAACATT 1800 CTCAAGAGGT GAAATACAGA AAGCCTTTTT TTCTTGATCT TTTCCCGAGA TTCAAATCTC 1860 CGATTCCCAT TTGGGGGCAA GTTTTTTTCT TCACCTTCAA TATGAGAATT CAGCGAACTT 1920 GAAAGAAAAA TCATCTGTGA GTTCCTTCAG GTTCTCACTC ATAGTCATGA TCCTTCAGAG 1980 GGAATATGCA CTGGCGAGTT TAAAGTAAGG GCTATGATAT TTGATGGTCC CAAAGTACGG 2040 CAGCTGCAAA AAGTAGTGGA AGGAAATTGT CTACGTGTCT TGGAAAAATT AGTTAGGAAT 2100 TTGGATGGGT AAAAGGTACC CTTGCCTTAC TCCATCTTAT TTTCTTAGCC CCCTTTGAGT 2160 GTTTTAACTG GTTTCATGTC CTAGTAGGAA GTGCATTCTC CATCCTCATC CTCTGCCCTC 2220 CCAGGAAGTC AGTGATTGTC TTTTTGGGCT TCCCCTCCAA AGGACCTTCT GCAGTGGAAG 2280 TGCCACATCC AGTTCTTTTC TTTTGTTGCT GCTGTGTTTA GATAATTGAA GAGATCTTTG 2340 TGCCACACAG GATTTTTTTT TTTTTTAAGA AAAACCTATA GATGAAAAAT TACTAATGAA 2400 ACTGTGTGTA CGTGTCTGTG CGTGCAACAT AAAAATACAG TAGCACCTAA GGAGCTTGAA 2460 TCTTGGTTCC TGTAAAATTT CAAATTGATG TGGTATTAAT AAAAAAAAAA AAAACAMAAA 2520 AAAAAAAAAA AAAAGGGCGG CCGCTCTAGA GGATCCAAGC TTACGTACGC GTGCATGCGA 2580 CGTCCATAGC TCTTTCTATA GGGGTCCCCC AAATTCCATT CANCGGGCCG TCGGTTTTAN 2640 AAAGGTCGTC ANTGGGGGAA ANCC 2664
(2) INFORMATION FOR SEQ ID NO: 250:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 865 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 250: CGTGGGAGTG AGGTACCAGA TTCAGCCCAT TTGGCCCCGA CGCCTCTKTT CTCGGAATCC 60 GGGTGCTGCG GATTGAGGTC CCGGTTCCTA ACGGTGGGAT CGGTGTCCTC GGGATGAGAT 120 TTGGCGTTTC CTCGGGGCTT TGGTGGGATC GGTGTCCTCA GGATGAGATT TAGGGTTTCC 180 TCGGGGCTTT CGGGATCTTC ACCTAATATC CGGACTCCAA GATGGAGGAA GGCGGGAACC 240 TAGGAGGCCT GATTAARATG GTCCATCTAC TGGTCTTGTC AGGTGCCTGC GCCATGCAAA 300
TGTGGGTGAC CTTCGTCTCA GGCTTCCTGC TTTTCCGAAG CCTTCCCCGA CATACCTTCG 360
GACTAGTGCA GAGCAAACTC TTCCCCTTCT ACTTCCACAT CTCCATGGGC TGTGCCTTCA 420 TCAACCTCTG CATCTTGGCT TCACAGCATG CTTGGGCTCA GCTCACATTC TGGGAGGCCA 480
GCCAGCTTTA CCTGCTCTTC CTGAGCCTTA CGCTGGCCAC TGTCAACGCC CGCTGGCTCG 540
AACCCCGCAC CACAGCTGCC ATCTGGGCCC TGCAAACCGT GGAGAAGGAG CGAGGCCTGG 600
GTGGGGAGGT ACCAGGCAGC CACCAGGGTC CCGATCCCTA CCGCCAGCTG CGAGAGAAGG 660
ACCCCAAGTA CAGTGCTCTC CGCCAGAATT TCTTCCGCTA CCATGGGCTG TCCTCTCTTT 720 GCAATCTGGG CTGCGTCCTG AGCAATGGGC TCTGTCTCGC TGGCCTTGCC CTCGAAATAA 780
GGAGCCTCTA GCATGGGCCC TGCATGCTAA TAAATGCTTC TTCAGAAAAA AAAAAAAAAA 840
AAACTCGAGG GGGGCCCGGT ACCCA 865
(2) INFORMATION FOR SEQ ID NO: 251:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2082 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 251:
TGGGGGGGGN AATGGGTGTC TGGCTCANGG ATTGCCNAAT CTGGAAATTC TCCATAACTT 60
GCTAGCTTGT TTTTTTTTTT TTTTTTTACA CCCCCCCGCC CCACCCCCGG ACTTGCACAA 120
TGTTCAATGA TCTCAGCAGA GTTCTTCATG TGAAACGTTG ATCACCTTTG AAGCCTGCAT 180 CATTCACATA TTTTTTCTTC TTCTTCCCCT TCAGTTCATG AACTGGTGTT (^TTTTCTGT 240
GTGTGTGTGT GTTTTATTTT GTTTGGATTT TTTTTTTTAA TTTTACTTTT AGAGCTTGCT 300
GTGTTGCCCA CCTTTTTTCC AACCTCCACC CTCACTCCTT CTCAACCCAT CTCTTCCGAG 360
ATGAAAGAAA AAAAAAAGCA AAGTTTTTTT TTCTTCTCCT GAGTTCTTCA TGTGAGATTG 420
AGCTTGCAAA GGAAAAAAAA ATGTGAAATG TTATAGACTT GCAGCGTGCC GAGTTCCATC 480 C<X3TTTTTTT TTTAGCATTG TTATGCTAAA ATAGAGAAAA AAATGCTCAT GAACCTTCCA 540
CAATCAAGCC TGCATCAACC TTCTGGGTGT GACTTGTGAG TTTTGGCCTT CTGATGCCAA 600
ATCTGAGAGT TTAGTCTGCC ATTAAAAAAA CTCATTCTCA TCTCATGCAT TATTATGCTT 660
GCTACTTTCT CTTAGCAACA ATGAACTATA ACTGTTTCAA AGACTTTATG GAAAAGAGAC 720
ATTATATTAA TAAAAAAAAA AAGCCTGCAT GCTGGACATC TATGGTATAA TTATTTTTTC 780 CTTTTTTTTT CCTTTTGGCT TGGAAATGGA CGTTCGAAGA CTTATAGCAT GGCATTCATA 840
CTTTTGTTTT ATTGCCTCAT GACTTTTTTG AGTTTAGAAC AAAACAGTGC AACCGTAGAG 900
CCTTCTTCCC ATGAAATTTT GCATCTGCTC CAAAACTCCT TTGAGTTACT CAGAACTTCA 960
ACCTCCCAAT GCACTGAAGG CATTCCTTGT GCAAAGATAC CAGAATGGGT TACACATTTA 1020
ACCTCCCAAA CATTGAAGAA CTCTTRATGT TTTCTTTTTA ATAAGAATCA CGCCCCACTT 1080
TGGGGACTAA AATTGTGCTA TTGCCGAGAA GCAGTCTAAA ATTTATTTTT TAAAAAGAGA 1140
AACTGCCCCA TTATTTTTGG TTTGTTTTAT TTTTATTTTA TATTTTTTGG CTTTTGGTCA 1200
TTGTCAAATG TGGAATGCTC TGGGTTTCTA GTATATAATT TAATTCTAGT TTTTATAATC 1260
TGTTAGCCCA GTTAAAATGT ATGCTACAGA TAAAGGAATG TTATAGATAA ATTTGAAAGA 1320
GTTAGGTCTC TTTAGCTGTA GATTTTTTAA ACGATTGATG CACTAAATTG TTTACTATTG 1380
TGATGTTAAG GGGGGTAGAG TTTGCAAGGG GACTGTTTAA AAAAAGTAGC TTATACAGCA 1440
TGTGCTTGCA ACTTAAATAT AAGTTGGGTA TGTGTAGTCT TTGCTATACC ACTGACTGTA 1500
TTGAAAACCA AAGTATTAAG AGGGGAAACG CCCCTGTTTA TATCTGTAGG GGTATTTTAC 1560
ATTCAAAAAT GTATGTTTTT TTTTCTTTTC AAAATTAAAG TATTTGGGAC TGAATTGCAC 1620
TAAGATATAA CCTGCAAGCA TATAATACAA AAAAAAATTC CAAAACTGTT TAGAACGCTA 1680
ATAAAATTTA TGCAGTTATA AAAATGGCAT TACTGCACAG TTTTAAGATG ATGCAGATTT 1740
TTTTACAGTT GTATTGTGGT GCAGAACTCG ATTTTCTGTA ACTTAAAAAA AAATCCACAG 1800
TTTTAAAGGC AATAATCAGT AAATGTTATT TTCAGGGACT GACATCCTGT CTTTAAAAAG 1860
AAATGAAAAG TAAATCTTAC CACAATAAAT ATAAAAAAAT CTTGTCAGTT ACTTTTCTTT 1920
TACATATTTT GCTGTGCAAA ATTGTTTTAT ATCTTGAGTT ACTAACTAAC CACGCGTGTT 1980
GTTCCTATGT GCTTTTCTTT CATTTTCAAT TCTGGTTATA TCAAGAAAAG AATAATCTAC 2040
AATAATAAAC GGCATTTTTT TTTGAAAAAA AAAAAAAAAA AA 2082
(2) INFORMATION FOR SEQ ID NO: 252:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1482 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 252: CAGGCAGGCT GGCCCCGGGG ACTTCTCTCT GGCCCTGCTC CCTCCGAGCG CTCCGCCGTT 60 GCCCGCCTGG CCCCTACGGA GTCCTTAGCC AGGATGGAGG CTGTTGTGAA CTTGTACCAA 120
GAGGTGATGA AGCACGCAGA TCCCCGGATC CAGGGCTACC CTCTGATGGG GTCCCCCTTG 180
CTAATGACCT CCATTCTCCT GACCTACGTG TACTTCGTTC TCTCACTTGG GCCTCGCATC 240
ATCGCTAATC GGAAGCCCTT CCAGCTCCGT GGCTTCATGA TTGTCTACAA CTTCTCACTG 300
GTGGCACTCT CCCTCTACAT TGTCTATGAG TTCCTGATGT CGGGCTGGCT GAGCACCTAT 360
ACCTGGCGCT GTGACCCTGT GGACTATTCC AACAGCCCTG AGGCACTTAG GATCGTTCGG 420
GTGGCCTGGC TCTTCCTCTT CTCCAAGTTC ATTGAGCTGA TGGACACAGT GATCTTTATT 480
CTCCGAAAGA AAGACGGGCA GGTGACCTTC CTACATGTCT TCCATCACTC TGTGCTTCCC 540
TGGAGCTGGT GGTGGGGGGT AAAGATTGCC CCGGGAGGAA TGGGCTCTTT CCATGCCATG 600
ATAAACTCTT CCGTGCATGT CATAATGTAC CTGTACTACG GATTATCTGC CTTTGGCCCT 660
GTGGCACAAC CCTACCTTTG GTGGAAAAAG CACATGACAG CCATTCAGCT GATCCAGTTT 720
GTCCTGGTCT CACTGCACAT CTCCCAGTAC TACTTTATGT CCAGCTGTAA CTACCAGTAC 780
CCAGTCATTA TTCACCTCAT CTGGATGTAT GGCACCATCT TCTTCATGCT GTTCTCCAAC 840
TTCTGGTATC ACTCTTATAC CAAGGGCAAG CGGCTGCCCC GTGCACTTCA GCAAAATGGA 900
GCTCCAGCTA TTGCCAAGCT CAAGGCCAAC TGAGAAGCAT GGCCTAGATA GGCGCCCACC 960
TAAGTGCCTC AGGACTGCAC CTTAGGGCAG TGTCCGTCAG TGCCCTCTCC ACCTACACCT 1020
GTGACCAAGG CTTATGTGGT CAGGACTGAG CAGGGGACTG GCCCTCCCCT CCCCACAGCT 1080
GCTCTACAGC GACCACGGCT TTGGTTCCTC ACCCACTTCC CCCGGGCAGC TCCAGGGATG 1140
TGGCCTCATT GCTGTCTGCC ACTCCAGAGC TGGGGGCTAA AAGGGCTGTA CAGTTATTTC 1200
CCCCTCCCTG CCTTAAAACT TGGGAGAGGA GCACTCAGGG CTGGCCCCAC AAAGGGTCTC 1260
GTGGCCTTTT TCCTCACACA GAAGAGGTCA GCAATAATGT CACTGTGGAC CCAGTCTCAC 1320
TCCTCCACCC CACACACTGA AGCAGTAGCT TCTGGGCCAA AGGTCAGGGT GGGCGGGGGC 1380
CTGGGAATAC AGCCTGTGGA GGCTGCTTAC TCAACTTGTG TCTTAATTAA AAGTGACAGA 1440
GGAAACCAAA AAAAAAAAAA AAAAACTCGA GGGGGGCCCG TA 1482
(2) INFORMATION FOR SEQ ID NO: 253:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 834 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 253: GGCACGAGCG CCGTTGCCCG CCTGGCCCCT ACGGAGTCCT TAGCCAGGAT GGAGGCTGTT 60
GTGAACTTGT ACCAAGAGGT GATGAAGCAC GCAGATCCCC GGATCCAGGG CTACCCTCTG 120
ATCGGGTCCC CCTTCCTAAT GACCTCCATT CTCCTGACCT ACGTGTACTT CGTTCTCTCA 180
CTTCGGCCTC GCATCATGGC TAATCGGAAG CCCTTCCAGC TCCGTGGCTT CATGATTGTC 240
TACAACTTCT CACTGGTGGC ACTCTCCCTC TACATTGTCT ATGAGTTCCT GATGTCGGGC 300 TGGCTGAGCA CCTATACCTG GCGCTGTGAC CCTCAGGACT GCACCTTAGG GCAGTGTCCG 360
TCAGTGCCCT CTCCAMCTAC ACCTGTGACC AAGGCTTATG TGGTCAGGAC TGAGCAGGGG 420
ACTGGCCCTC CCCTCCCCAC AGCTGCTCTA CAGGGACCAC GGCTTTGGTT CCTCACCCAC 480
TTCCCCCGGG CAGCTCCAGG GATGTGGCCT CATTGCTGTC TGCCACTCCA GAGCTGGGGG 540
CTAAAAGGGC TGTACAGTTA TTTCCCCCTC CCTGCCTTAA AACTTGGGAG AGGAGCACTC 600 AGGGCTGGCC CCACAAAGGG TCTCGTGGCC TTTTTCCTCA CACAGAAGAG GTCAGCAATA 660
ATGTCACTGT GGACCCAGTC TCACTCCTCC ACCCCACACA CTGAAGCAGT AGCTTCTGGG 720
CCAAAGGTCA GGGTGGGCGG GGGCCTGGGA ATACAGCCTG TGGAGGCTGC TTACTCAACT 780
TGTGTCTTAA TTAAAAGTGA CAGAGGAAAC CACGAAAAAA AAAAAAAAAA AAAA 834
(2) INFORMATION FOR SEQ ID NO: 254:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1508 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 254:
TTGAACTTTT AAAATTTTAG ATCAGCAAAC TCTAAGATCC TAGAATGGAA GCTGTTCCTC 60
ATTTCTCCAT GCTCACCCTC CCAGGTCAGC GAGATGGTCA AGAAGCTGCA CGCGCCAACA 120 CCACCAACGT TCGGAGTGGA CCTCATCAAT GAGCTTGTGG AGAACTTTGG CAGATGTCCC 180
AAGTGGTCTG GTCGGCAAGC CTTTGTCTTT GTCTGCCAGA CTGTCATTGA GGATGACTGC 240
CTTCCCATGG ACCAGTTTGC TGTGCATCTC ATGCCGCATC TGCTAACCTT AGCAAATGAC 300
AGGGTTCCTA ACGTGCGAGT GCTGCTTGCA AAGACATTAA GACAAACTCT ACTAGAAAAA 360
GACTATTTCT TGGCCTCTGC CAGCTGCCAC CAGGAGGCTG TGGAGCAGAC CATCATGGCT 420 CTTCAGATGG ACCGTGACAG CGATGTCAAG TATTTTGCAA GCATCCACCC TGCCAGTACC 480
AAAATCTCCG AAGATGCCAT GAGCACAGCG TCCTGAACCT ACTAGAAGGC TTCAATCTCG 540
GTGTCTTTCC TGCTTCCATG AGAGCCGAGG TTCAGTGGGC ATTCGCCACG CATGTGACCT 600
GGGATAGCTT TCGGGGGAGG AGAGACCTTC CTCTCCTGCG GACTTCATTG CAGGTGCAAG 660
TTCCCTACAC CCAATACCAG GGATTTCAAG AGTCAAGAGA AAGTACAGTA AACACTATTA 720
TCTTATCTTG ACTTTAAGGG GAAATAATTT CTCAGAGGAT TATAATTGTC ACCGAAGCCT 780
TAAATCCTTC TGTCTTCCTG ACTGAATCAA ACTTCAATTG GCAGAGCATT TTCCTTATGG 840
AAGGGATGAG ATTCCCAGAG ACCTGCATTG CTTTCTCCTC GTTTTATTTA ACAATCGACA 900
AATGAAATTC TTACAGCCTG AAGGCAGACG TGTGCCCAGA TGTGAAAGAG ACCTTCAGTA 960
TCAGCCCTAA CTCTTCTCTC CCAGGAAGGA CTTGCTGGGC TCTCTGGCCA GCTGTCCAGC 1020
CCAGCCCTGT GTGTGAATCG TTTGTGACGT GTGCAAATGG GAAAGGAGGG GTTTTTACAT 1080
CTCCTAAAGG ACCTGATCCC AACACAAGTA GGATTCACTT AAACTCTTAA GCGCAGCATA 1140
TTGCTGTACA CATTTACAGA ATGGTTGCTG AGTGTCTGTG TCTGATTTTT TCATGCTGGT 1200
CATGACCTGA AGGAAATTTA TTAGACGTAT AATGTATGTC TGGTGTTTTT AACTTGATCA 1260
TGATCAGCTC TGAGGTGCAA CTTCTTCACA TACTGTACAT ACCTGTGACC ACTCTTGGGA 1320
GTGCTGCAGT CTTTAATCAT GCTCTTTAAA CTGTTGTGGC ACAAGTTCTC TTGTCCAAAT 1380
AAAATTTATT AATAAGATCT ATAGAGAGAG ATATATACAC TTTTGATTGT TTTCTAGATG 1440
TCTACCAATA AATGCAATTT GTGACCTGTA TTAAAAAAAA NTAAAAAAAC TCGAGGGGGG 1500
CCCGGTAC 1508
(2) INFORMATION FOR SEQ ID NO: 255:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2514 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO; 255: GAGAGACTCA CACTTCTTTT CCATTATCAC TGACGATGTA GTCGACATAG CAGGGGAAGA 60 GCACCTACCT GTGTTGGTGA GGTTTGTTGA TGAATCTCAT AACCTAAGAG AGGAATTTAT 120 AGGCTTCCTC CCTTATGAAG CCGATGCAGA AATTTTGGCT GTCAAATTTC ACACTATGAT 180 AACTGAGAAG TGGGGATTAA ATATGGAGTA TTGTCGTGGC CAGGCTTACA TTGWCTCTAG 240 TGGATTTTCT TCCAAAATGA AAGTTGTTGC TTCTAGACTT TTAGAGAAAT ATCCCCAAGC 300 TATCTACACA CTCTGCTCTT CCTGTGCCTT AAATATGTGG TTGGCAAAAT CAGTACCTGT 360 TATGGGAGTA TCTGTTGCAT TAGGAACAAT TGAGGAAGTT TGTTCTTTTT TCCATCGATC 420 ACCACAACTG CTTTTAGAAC TTGACAACGT AATTTCTGTT CTTTTTCAGA ACAGTAAAGA 480
AAGGGGTAAA GAACTGAAGG AAATCTGCCA TTCTCAGTGG ACAGGCAGGC ATGATGCTTT 540
TCAAATTTTA GTGGAACTCC TGCAAGCACT TGTTTTATGT TTAGATGGTA TAAATAGTGA 600
CACAAATATT AGATGGAATA ACTATATAGC TGGCCGAGCA TTTGTACTCT GCAGTGCAGT 660
GTCAGATTTT GATTTCATTG TTACTATTGT TGTTCTTAAA AATGTCCTAT CTTTTACAAG 720
AGCCTTTGGG AAAAACCTCC AGGGGCAAAC CTCTGATGTC TTCTTTGCGG CCGGTAGCTT 780
GACTGCAGTA CTGCATTCAC TCAACGAAGT GATTGGAAAA TATTGAAGTT TATCATGAAT 840
TTTGGTTTGA GGAAGCCACA AATTTGGCAA CCAAACTTGA TATTCAAATG AAACTCCCTG 900
GGAAATTCCG CAGAGCTCAC CAGGGTAACT TCGAATCTCA GCTAACCTCT GAGAGTTACT 960
ATAAAGAAAC CCTAAGTGTC CCAACAGTGG AGCACATTAT TCAGGAACTT AAAGATATAT 1020
TCTCAGAACA GCACCTCAAA GCTCTTAAAT GCTTATCTCT GGTACCCTCA GTCATGGGAC 1080
AACTCAAATT CAATACGTCG GAGGAACACC ATGCTGACAT GTATAGAAGT GACTTACCCA 1140
ATCCTGACAC GCTGTCAGCT GAGCTTCATT GTTGGAGAAT CAAATGGAAA CACAGGGGGA 1200
AAGATATAGA GCTTCCGTCC ACCATCTATG AAGCCCTCCA CCTGCCTGAC ATCAAGTTTT 1260
TTCCTAATGT GTATGCATTG CTGAAGGTCC TGTGTATTCT TCCTCTGATC AAGGTTGAGA 1320
ATGAGCGGTA TGAAAATGGA CGAAAGCGTC TTAAAGCATA TTTGAGGAAC ACTTTGACAG 1380
ACCAAAGGTC AAGTAACTTG GCTTTGCTTA ACATAAATTT TGATATAAAA CACGACCTGG 1440
ATTTAATGGT GGACACATAT ATTAAACTCT ATACAAGTAA GTCAGAGCTT CCTACAGATA 1500
ATTCCGAAAC TGTGGAAAAT ACCTAAGAGA CTTTTAAAAA TAGGCTTTCT TATATTTGAT 1560
ATTTGGAAGA AAAAGCCGTA AGTGTATGTA GACCACTTAA TCACTAAATA TCTTTGCCTA 1620
TAGGACTCCA TTGAATACAT TAGCCATTGA TAATCTACCT GTTTAAATGG CCCCTGTTTG 1680
AACTCTCAAG CTTTGAAGAC CTACCTGTTC TTCCAGAAGA GAACGTTGAA AGTGCCATGT 1740
TTCCTTTTGC GTGATCTCTG TTGATGGCAC TCTGGAATTG TTTCAGTTAA GTCATTTTAG 1800
ACATAGCATT TATTATCACT GTGGATCTCT ACTTGTTGGG TGTTATGAAT TCTTTGAAGA 1860
AATATATTTT GAAGAGGTGT GGGAGGAAGG AATACATTTT ATAAAATGTT GTAGTGAAGC 1920
CCACAATTGA CCTTTGACTA ATAGGAGTTT TAAGTATGTT AAAAATCTAT ACTGGACAGT 1980
TACAAGAAAT TACCGGAGAA AAGCTTGTGA GCTCACCAAA CAAGGATTTC AGTGTAGATT 2040
TTGTCTTTCT TGAACTTAAA GAAACAAATC ACAAAGTTTG AATGGAAAAG CCTGCTGTTG 2100
TTCCACATCT CGTTCCTGTT TACATTCCTT TCTGGAGCCT ACATCTTCCT AAGCTTTTTA 2160
GCAGGTATAT GTTGAACACT TCTGTTTCAT GGTTGAGACA GAATCAGAGG CCATGGATAC 2220
TCACAACTGA TTTGTCTGTT TTTTTTCTCT GTCTTTTTCC ATGACTCTTA TATACTGCCT 2280
CATCTTGATT TATAAGCAAA ACCTGGAAAA CCTACAAAAT AAGTGTTGTG GTTTATCTAG 2340
AAAAATATCG AAAATATTGC TGTTATTTTT GGTGAAGAAA ATCAATTTTG TATAGTTTAT 2400
TTCAATCTAA ATAAAATGTG AATTTTCTTT AAAGCTTAGG CACATTATTT TTTGTGGGGT 2460
CAAAACATTC TTGTGTAAAT TCTCTTAAAC ATTTGATAAA CAGCTTCACA ATTC 2514
(2) INFORMATION FOR SEQ ID NO: 256:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2357 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 256:
CTGCCTTATG AAGCCGATGC ACiAAATTTTC GCTGTGAAAT TTCACACTAT GATAACTGAG 60
AAGTGGGGAT TAAATATGGA GTATTGTCGT GGCCAGGCTT ACATTGTCTC TAGTGGATTT 120
TCTTCCAAAA TGAAAGTTGT TGCTTCTAGA CTTTTAGAGA AATATCCCCA AGCTATCTAC 180
ACACTCTGCT CTTCCTGTGC CTTAAATATG TGGTTGGCAA AATCAGTACC TGTTATGGGA 240
GTATCTGTTG CATTAGGAAC AATTGAGGAA GTTTGTTCTT TTTTCCATCG ATCACCACAA 300
CTGCTTTTAG AACTTGACAA CGTAATTYCT GTTCTTTTTC AGAACAGTAA AGAAAGGGGT 360
AAAGAACTGA AGGAAATCTC CCATTCTCAG TGGACAGGCA GGCATGATGC TTTTGAAATT 420
TTAGTGGAAC TCCTGCAAGC ACTTGTTTTA TGTTTAGATG GTATAAATAG TGACACAAAT 480
ATTAGATGCA ATAACTATAT AGCTGGCCGA GCATTTGTAC TCTGCAGTGC AGTGTCAGAT 540
TTTGATTTCA TTGTTACTAT TGTTGTTCTT AAAAATGTCC TATCTTTTAC AAGAGCCTTT 600
GGGAAAAACC TCCAGGGGCA AACCTCTGAT GTCTTCTTTG CGGCCGGTAG CTTCACTGCA 660
GTACTGCATT CACTCAACGA AGTGANTGGA AAATATTGAA GTTTATCATG AATTTTGGTT 720
TGAGGAAGCC ACMATTTCG CAACCAAACT TGATATTCAA ATGAAACTCC CTGGGAAATT 780
CCGCAGAGCT CACCAGGGTA ACTTGGAATC TCAGCTAACC TCTGAGAGTT ACTATAAAGA 840
AACCCTAAGT GTCCCAACAG TGGAGCACAT TATTCAGGAA CTTAAAGATA TATTCTCAGA 900
ACAGCACCTC AAAGCTCTTA AATGCTTATC TCTGGTACCC TCAGTCATCC GACAACTCAA 960
ATTCAATACG TCGGAGGAAC ACCATGCTGA CATGTATAGA AGTGACTTAC CCAATCCTGA 1020
CACGCTGTCA GCTGAGCTTC ATTGTTGGAG AATCAAATGG AAACACAGGG GGAAAGATAT 1080
AGAGCTTCCG TCCACCATCT ATGAAGCCCT CCACCTGCCT GACATCAAGT TTTTTCCTAA 1140
TCTCTATGCA TTGCTGAAGG TCCTGTGTAT TCTTCCTGTG ATGAAGGTTG AGAATGAGCG 1200
GTATGAAAAT GGACGAAAGC GTCTTAAAGC ATATTTGAGG AACACTTTGA CAGACCAAAG 1260
GTCAAGTAAC TTGGCTTTGC TTAACATAAA TTTTGATATA AAACACGACC TGGATTTAAT 1320
GGTGGACACA TATATTAAAC TCTATACAAG TAAGTCAGAG CTTCCTACAG ATAATTCCGA 1380
AACTGTGGAA AATACCTAAG AGACTTTTAA AAATAGGCTT TCTTATATTT GATATTTGGA 1440
AGAAAAAGCC GTAAGTGTAT GTAGACCACT TAATCACTAA ATATCTTTGC CTATAGGACT 1500
CCATTGAATA CATTAGCCAT TGATAATCTA CCTGTTTAAA TGGCCCCTGT TTGAACTCTC 1560
AAGCTTTGAA GACCTACCTG TTCTTCCAGA AGAGAACGTT GAAAGTGCCA TGTTTCCTTT 1620
TGCGTGATCT CTGTTGATGG CACTCTGGAA TTGTTTCAGT TAAGTCATTT TAGACATAGC 1680
ATTTATTATC ACTGTGGATC TCTACTTGTT GGGTGTTATG AATTCTTTGA AGAAATATAT 1740
TTTGAAGAGG TGTGGGAGGA AGGAATACAT TTTATAAAAT GTTGTAGTGA AGCCCACAAT 1800
TGACCTTTGA CTAATAGGAG TTTTAAGTAT GTTAAAAATC TATACTGGAC AGTTACAAGA 1860
AATTACCGGA GAAAAGCTTG TGAGCTCACC AAACAAGGAT TTCAGTGTAG ATTTTGTCTT 1920
TCTTGAACTT AAAGAAACAA ATGACAAAGT TTGAATCGAA AAGCCTGCTG TTGTTCCACA 1980
TCTCGTTGCT GTTTACATTC CTTTGTCGAG CCTACATCTT CCTAAGCTTT TTAGCAGGTA 2040
TATGTTGAAC ACTTCTGTTT CATGGTTGAG ACAGAATCAG AGGCCATGGA TACTGACAAC 2100
TGATTTGTCT GTTTTTTTTC TCTGTCTTTT TCCATGACTC TTATATACTG CCTCATCTTG 2160
ATTTATAAGC AAAACCTGGA AAACCTACAA AATAAGTGTT GTGGTTTATC TAGAAAAATA 2220
TGGAAAATAT TGCTGTTATT TTTGGTGAAG AAAATCAATT TTGTATAGTT TATTTCAATC 2280
TAAATAAAAT GTGAATTTTC TTTAAAGCTT AGGCACATTA TTTTTTGTCG GGTCAAAACA 2340
TTCTTGTGTA AATTCTC 2357
(2) INFORMATION FOR SEQ ID NO: 257:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 689 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 257:
ACTTTCTGGT GCAAAAAGAT GTTCAAGCCT TATTTTATAC TTGCCTGCCC CTTTCTCTTT 60
CATTTATTGG AGTGAGCTGC AGCTCTAAGA AGACCTGTTC TTTTGAATGG AGAGTAGCAT 120
CAGGAACCAG GATGTGGGTG CGAGGCGTGC TCCTGGCTGT TGCAGATTGC TGCACCCGGG 180
AGCTCTTAGT GGACAGAGCT AGAGGATATG TGCACGTACT TCCATCTCTC TCTCTGTCTC 240
CGATTTTAGC CCAGCACCAC AGGGTACGTT CCAGTTTTTC TCTCTTTCCA TAGCTGTAAG 300
GCCCTTTCTG GGAATGGTTC TCATTCTCCT TAATCTATTA TTGGGTCAGT TTTCCTGCAT 360
GTCCCCAGCC TCCCATCACT GCCACCCACT CCCCACAGAG ATGCCCTGCT CATCCGACTG 420 GGGCTTTGAC TCCCACACTG TGTACCCCTC TTGTGTGGAC GCCCTGCTGC CAAAACCTTC 480
AGCAAACAGC TTTCCAAATC GAAGTTGTCA CTGTCARGGS CTTTACAATC AGCAACAGCA 540
AAATCTACAT GCTGCTGAGG GTCCTCCCTC ATTAAGATCC AATAAATATC TAAGTACATA 600
AAAACAGCAA TAGAAGAAAC GTAATGCTTT ATTCTCAAAT ATCNATGTCT ACATAGAAAA 660
GCCAAAATTA TTAAGAATAG TAAGGAATT 689
(2) INFORMATION FOR SEQ ID NO: 258: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2377 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 258:
TCGACCCACG CGTCCGCCGA TGTGATGATT CCTGCGTATT CCAAGAACCG GGCCTATGCC 60 ATCTTCTTCA TAGTCTTCAC TGTGATAGGG GACGCCCCCG GCGCTGTGCT ATCCTGTGCC 120
GGCCACCCTT GCGTTGGTTT TGCTGCTGTA CTGGTGGCGC CCCTGACCGT GGCTGTCTCC 180
TCTTGAAGGA AGCCTGTTTC TGATGAACCT GCTGACAGCC ATCATCTACA GTCAGTTCCG 240
GGGCTACCTG ATGAAATCTC TCCAGACCTC GCTGTTTCGG AGGCGGGTGG GAACCCGGCT 300
GCCTTTGAAG TCCTATCCTC CATGGTGGGG GAGGGAGGAG CCTTCCCTCA GGCAGTTGGG 360 GTGAAGCCCC AGAACTTGCT GCAGGTGCTT CAGAAGGTCC AGCTGGACAG CTCCCACAGA 420
CAGGCCATGA TGGAGAAGGT GCGTTCCTAT GGCAGTGTTC TGCTCTCAGC TGAGGAGTTT 480
CAGAAGCTCT TCAACGAGCT TGACAGAAGT GTGGTTAAAG AGCACCCGCC GAGGCCCGAG 540
TACCAGTCTC CGTTTCTGCA GAGCGNCCCA GTTCCTCTTC GGCCACTNAC TACTTTGACT 600
ACCTGGGGAA CCTCATCGCC CTGGCAAACC TGGTGTCCAT TTGCGTGTTC CTGGTGCTGG 660 ATGCAGATGT TGCTGCCTGC TGAGCGTGAT GACTTCATCC TGGGGGGTCT CAACTGCGTC 720
TTCATTGTGT ACTACCTCTT GGAGATGCTG GCTCAAGGTC TTTTGCCCTG GGGCCTGCGA 780
RGGTACYKKT CCTAACCCCA RCAAMGTGTT TTGAACGGGC TCCTCAMCGT TTGTCCTGGC 840
TGGWWKKGSM GATCTCAACT CTGGCTGTCT ACCGATTGCC ACACCCAGGC TGGAGGCCGG 900
ANATGGTGGG CCTCCTGTCG CTGTGGGACA TGACCCGCAT ACTGAACATG CTCATCGTGT 960
TCCGCTTCCT GCGTATCATC CCCAGCATGA AGCCGATGGC CGTGGTGGCC AGTACCGTCC 1020
TGGGCCTGGT GCAAAACATG CGTGCGTTTG GCGGGATCCT GGTGGTGGTC TACTACGTAT 1080
TTGCCATCAT TGGGATCAAC TTGTTTAGAG GCGTCATTGT GGCTCTTCCT GGAAACAGCA 1140
GCCTGGCCCC TGCCAATAGG TCGGCGCCCT GTGGGAGCTT CGAGCAGCTG GAGTACTGGG 1200
CCAACAACTT CGATGACTTT GCGGCTGCCC TGGTCACTCT GTGGAACTTG ATGGTGGTGA 1260
ACAACTGGCA GGTGTTTCTG GATGCATATC GGCGCTACTA AGGCCCGTGG TCCAAGATCT 1320
ATTTTGTATT GTGGTGGCTG GTGTCGTCTG TCATCTGGGT CAACCTGTTT CTGGCCCTGA 1380
TTCTGGAGAA CTTCCTTCAC AAGTGGGACC CCCGCAGCCA CCTGCAGCCC CTTGCTGGGA 1440
CCCCAGAGGC CACCTACCAG ATGACTGTGG AGCTCCTGTT CAGGGATATT CTGGAGGAGC 1500
CCGGGGAGGA TGAGCTCACA GAGAGGCTGA GCCAGCACCC GCACCTGTGG CTGTGCAGCT 1560
GACGTCCGGG TCTGCCATCC CAGCAGGGGC GGCAGGAGAG AGAGGCTGGC ATAACACAGG 1620
TGCCCATCAT GGAAGAGGCG GCCATGCTGT GGCCAGCCAG GCAGGAAGAG ACCTTTCCTC 1680
TGACGGACCA CTAAGCTGGG GACAGGAACC AAGTCCTTTG CGTGTGGCCC AACAACCATT 1740
TACAGAACAG CTGCTGGTGC TTCAGGGAGG CGCCGTGCCC TC∞CTTTCT TTTATAGCTG 1800
CTTCAGTGAG AATTCCCTTG TCGACTCCAC AGGGACCTTT CAGACAAAAA TGCAAGAAGC 1860
AGCGGCCTCC CCTGTCCCCT GCAGCTTCCG TCGTCCCTTT GCTGCCGGCA GCCCTTGGGG 1920
ACCACAGGCC TGACCAGGGC CTCCACAGGT TAACCGTCAG ACTTCCGGGG CATTCAGCTG 1980
GGAATGATAC TAATACCTCC GATTTTAGCC CAGCACCACA GGGTACGTTC CAGTTTTTAT 2040
TTCTTTCCAT AGCTGTAAGG CCCTTTCTGG GAATGGTTAT CATTCTCCTT AATCTATTAT 2100
TGGGTCAGTT TTCCTCCATG TCCCCAGCCT CCCATCACTG CCACCCACTC CCCACAGAGA 2160
TGCCCTGCTC ATCCGACTGG GGCTTTGACT CCCACACTGT GTACCCCTCT TGTGTGGACG 2220
CCCTCCTCCC AAAACCTTCA GCAAACAGCT TTCCAAATGG AAGTTCTCAC TGTCAGGGCC 2280
TTTACAATCA GCAACAGCAA AATCTACATG CTGCTGAGGG TCCTGCCTCA TTAAGATGCA 2340
ATAAATATGT AAGTACATAA AAAAAAAAAA AAAAAAA 2377
(2) INFORMATION FOR SEQ ID NO: 259:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1193 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 259:
TCTGNTCGCC GTCGCCCCGC CCCTGGCCTT TGCCCGGTCG GGCGGGACTT CCTGTCTCGT 60
ATTTCCAAGG ACTCCAAAGC GAGGCCGGGG ACTGAAGGTG TGGGTGTCGA GCCCTCTGGC 120
AGAGGGTTAA CCTGGGTCAA ATGCACGGAT TCTCACCTCG TACAGTTACG CTCTCCCGCG 180
GCACGTCCGC GAGGMYTTGA AGTCCTGAGC GCTCAAGTTT GTCCGTAGTC GAGAGAAGGC 240
CATGGAGGTG CCGCCACCGG CACCGCGGAG CTTTCTCTGT AGAGCATTGT GCCTATTTCC 300
CCGAGTCTTT GCTGCCGAAG CTGTGACTGC CGATTCGGAA GTCCTTGAGG AGCGTCAGAA 360
GCGGCTTCCC TACGTCCCAG AGCCCTATTA CCCGGAATCT GGATGGGACC GCCTCCGGGA 420
GCTGTTTGGC AAAGACACAG TGAACACTAG TCTGAATGTA TACCGAAATA AAGATGCCTT 480
AAGCCATTTT GTAATTGCAG GAGCTGTCAC GGGAAGTCTT TTTAGGATAA ACGTAGGCCT 540
GCGTGGCTGG TGGCTGGTGG CATAATTGGA GCCTTGCTGG GCACTCCTGT AGGAGGCCTG 600
CTGATGGCAT TTCAGAAGTA CTCTGGTGAG ACTGTTCAGG AAAGAAAACA GAAGGATCGA 660
AAGGCACTCC ATGAGCTAAA ACTGGAAGAG TGGAAAGGCA GACTACAAGT TACTGAGCAC 720
CTCCCTGAGA AAATTGAAAG TAGTTTACAG GAAGATGAAC CTGAGAATGA TGCTAAGAAA 780
ATTGAAGCAC TGCTAAACCT TCCTAGAAAC CCTTCAGTAA TAGATAAACA AGACAAGGAC 840
TGAAAGTGCT CTGAACTTGA AACTCACTGG AGAGCTGAAG GGAGCTGCCA TGTCCGATCA 900
ATGCCAACAG ACAGGCCACT CTTTGGTCAG CCTGCTGACA AATTTAAGTG CTGGTACCTG 960
TGGTGGCAGT GGCTTGCTCT TGTCTTTTTC TTTTCTTTTT AACTAAGAAT GGGGCTGTTG 1020
TACTCTCACT TTACTTATCC TTAAATTTAA ATACATACTT ATCTTTGTAT TAATCTATCA 1080
ATATATGCAT ACATGAATAT ATCCACCCAC CTAGATTTTA AGCAGTAAAT AAAACATTTC 1140
GCAAAAGATT AAAGTTGAAT TTTACAGTTA AAAAAAAAAA AAAAAAAAAA AAA 1193
(2) INFORMATION FOR SEQ ID NO: 260:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1262 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 260: GAAAAACCCA AAGATGCAGA CAATCTCTTT GAACATGAAT TGGGGGCTCT CAATATGGCT 60
GCATTACTAC GAAAAGAAGA AAGAGCAAGT CTTCTTAGTA ATCTTGGCCC ATGTTGTAAG 120
GCGTTGTGCT TCAGACGGGA TTCTGCAATT CGAAAGCAGC TTGTTAAAAA TGAGAAGGGC 180
ACCATAAAAC AAGCTTACAC GAGTSCTCCA ATGGTAGACA ATGAATTACT TCGATTGAGT 240
CTTCGGTTAT TTAAGCGGAA GACTACTTGC CATGCTCCAG GACATGAAAA GACTGAAGAT 300
AATAAACTTT CACAGTCCAG TATCCAACAG GAACTGTCTG TGTCTTAAGA CCGAAGTTCA 360
ATATGGTATT TTTGGTACTG TCTTCCTTCA GCAGTGCATA TTCTTTTGCA AAGTTCTTTG 420
GTTTGACAAG CATTAGTGAC AAAGGCAGAA AAGATTTATC AGCCATGCTA AAAGAGTGAA 480
GAATTTTGAT CTTTAGAGAC ACTAGTTTTG GCCAACTTAA GATTTTACGT TAATTTTTAC 540
ATAGTATTTG ACACTCATGC AAAATAATGT GAAAACATCT AGATTTAGTA GTTTATTCTG 600
CGCCTTTTGT TAAAACTGAA GATTTTGGAA AATGGTTGTC ACTGCTCTTC CAGCCTATGA 660
ATATTTTTGT GAAATGGAAC CATGGATTTA TGTCTGGATC ATCCATACAG AACCAACAAT 720
TTTATTCAAA AACAATGTGT TCATCAAAGT AATTGCTCAC ATTGTGCAGT ACTATGTTGT 780
ACAGACCACG TGAAAGGGAA TGCTGGTCTA GCTGGCGTGG TATGTTTATA GGCGAATTTC 840
AGCAGAAGGA AGCCAAAATA GTTTTTTCCT TTTGAAAGTT TTTTAAAAAT TATTTCATGG 900
GTCTTTTTTT TAATTAATAT GTGTGCATTG TTACAATGTA TGTTGGATGT CTTTTGACCC 960
TAAATGCTTT TTTTGTTATC AGAGATTGTG TACTATTTTT ATTTTTAATA AATCTATCTT 1020
CCCTTTCCTT GTTTTAGATT TACTTTGCTC TTCGTTAATC TTATTCCTGA TGATCTAGAA 1080
CATTAGTCAT CAACATTACA TGTTTCATGC TTCAGATATT TTACTGCTTG TCTCCTTATT 1140
GTTGGACAGC TTTAAACAGA GTTGATGGTA CTTCAAATAT AGCTCATTGA TACTTAAGGG 1200
CANCTTCCTT GGGATGTGGG CTTTTTGGAA GGAAAAAAAT TNCCCCAAAG GCAAATCCCA 1260
GT 1262
(2) INFORMATION FOR SEQ ID NO: 261:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1179 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 261: GGCAAACTTT CCCCCAANGC TTCGAAACTT GCAAGCCGAA ACCTTGAATC GTTAAAAGTT 60 GGGTTGCGNC GGCGCCCTGG CCCGAAGAAG CGCAATTGGC GTTCCGCGAA CGTTGGCCCT 120 CAACGGCTCG GCAGCCAGCC ATGTCCTGCA CCCAGGACAG CGGCCCTGGG CTACAAGGAC 180
CTGGACCTCA TCTTCCTGCG CCGACCTGCG CGGGGAAGGG GAGTTTCAGA CTGTGAAGGA 240
CGTCGTGCTG GACTGCCTGT TGGACTTCTT ACCCGAGGGG GTGAACAAAG AGAAGATCAC 300
ACCACTCACG CTCAAGGAAG CTTATGTGCA GAAAATGGTT AAAGTGTCCA ATGACTCTGA 360
CCGATGGAGT CTTATATCCC TGTCAAACAA CAGTGGCAAA AATGTGGAAC TGAAATTTGT 420
GGATTCCCTC CGGAGGCAGT TTGAATTCAG TGTAGATTCT TTTCAAATCA AATTAGACTC 480
TCΓTCTGCTC TTTTATGAAT GTTCAGAGAA CCCAATGACT GAGACATTTC ACCCCACAAT 540
AATCGGGGAG AGCGTCTATG GCGATTTCCA GGAAGCCTTT GATCACCTTT GTAACAAGAT 600
CATTGCCACC AGGAACCCAG AGGAAATCCG AGGGGGAGGC CTGCTTAAGT ACTGCAACCT 660
CTTGGTGAGG GGCTTTAGGC CCGCCTCTGA TGAAATCAAG ACCCTTCAAA GGTATATGTG 720
TTCCAGGTTT TTCATCGACT TCTCAGACAT TGGAGAGCAG CAGAGAAAAC TGGAGTCCTA 780
TTTGCAGAAC CACTTTGTGG GATTGGAAGA CCGCAAGTAT GAGTATCTCA TCACCCTTCA 840
TGGAGTGGTA AATGAGAGCA CAGTGTGCCT GATGGGACAT GAAAGAAGAC AGACTTTAAA 900
CCTTATCACC ATGCTGGCTA TCCGGGTGTT AGCTGACCAA AATGTCATTC CTAATGTGGC 960
TAATGTCACT TGCTATTACC AGCCAGCCCC CTATGTAGCA GATGCCAACT TTAGCAATTA 1020
CTACATTGCA CAGGTTCAGC CAGTATTCAC GTGCCAGCAA CAGACCTACT CCACTTGGCT 1080
ACCCTGCAAT TAAGAATCAT TTAAAAATGT CCTGTGGGGA AGCCATTTCA GACAAGACAG 1140
GAGAGAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAGAGC 1179
(2) INFORMATION FOR SEQ ID NO: 262:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1162 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO : 262: GGCAAACTTT CCCCCAANGC TTCGAAACTT GCAAGCCGAA ACCTTGAATC GTTAAAAGTT 60 GGGTTGCGNC GGCGCCCTGG CCCGAAGAAG CGCAATTGGC GTTCCGCGAA CGTTGGCCCT 120 CAACGGCTCG GCAGCCAGCC ATGTCCTGCA CCCAGGACAG CGGCCCTGGG CTACAAGGAC 180 CTGGACCTCA TCTTCCTGCG CCGACCTGCG CGGGGAAGGG GAGTTTCAGA CTGTGAAGGA 240 CGTCGTGCTG GACTGCCTGT TGGACTTCTT ACCCGAGGGG GTGAACAAAG AGAAGATCAC 300 ACCACTCACG CTCAAGGAAG CTTATGTGCA GAAAATGGTT AAAGTGTCCA ATGACTCTGA 360
CCGATGGAGT CTTATATCCC TGTCAAACAA CAGTGGCAAA AATGTGGAAC TGAAATTTGT 420
GGATTCCCTC CGGAGGCAGT TTGAATTCAG TGTAGATTCT TTTCAAATCA AATTAGACTC 480
TCTTCTGCTC TTTTATGAAT GTTCAGAGAA CCCAATGACT GAGACATTTC ACCCCACAAT 540
AATCGGGGAG AGCGTCTATG GCGATTTCCA GGAAGCCTTT GATCACCTTT GTAACAAGAT 600
CATTGCCACC AGGAACCCAG AGGAAATCCG AGGGGGAGGC CTGCTTAAGT ACTGCAACCT 660
CTTGGTGAGG GGCTTTAGGC CCGCCTCTGA TGAAATCAAG ACCCTTCAAA GGTATATGTG 720
TTCCAGGTTT TTCATCGACT TCTCAGACAT TGGAGAGCAG CAGAGAAAAC TGGAGTCCTA 780
TTTGCAGAAC CACTTTGTGG GATTGGAAGA CCGCAAGTAT GAGTATCTCA TCACCCTTCA 840
TGGAGTGGTA AATGAGAGCA CAGTGTGCCT GATGGGACAT GAAAGAAGAC AGACTTTAAA 900
CCTTATCACC ATGCTGGCTA TCCGGGTGTT AGCTGACCAA AATGTCATTC CTAATGTGGC 960
TAATGTCACT TGCTATTACC AGCCAGCCCC CTATGTAGCA GATGCCAACT TTAGCAATTA 1020
CTACATTGCA CAGGTTCAGC CAGTATTCAC GTGCCAGCAA CAGACCTACT CCACTTGGCT 1080
ACCCTGCAAT TAAGAATCAT TTAAAAATGT CCTGTGGGGA AGCCATTTCA GACAAGACAG 1140
GAGAGAAAAA NAANGAAAAG AG 1162
(2) INFORMATION FOR SEQ ID NO: 263:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 735 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 263: CGGGCTGGGT ATTTGCCTCG CACCATCGCG CCCAAGGGCA AAGTGGGCAC GAGAGGGAAG 60 AAGCAGATAT TTGAAGAGAA CAGAGAGACT CTGAAGTTCT ACCTCCGGAT CATACTGGGG 120 GCCAATGCCA TTTACTGCCT TGTGACGTTG GTCTTCTTTT ACTCATCTGC CTCATTTTCG 180 GCCTGGTTGG CCTTGGGCTT TAGTCTGGCA GTGTATGGGG CCAGCTACCA CTCTATGAGC 240 TCGATCGCAC GAGCAGCGTT CTTCTGAGGA TGGGGCCCTG ATGGATGGTG GCACGAGCTC 300 AACATGGAGC AGGGCATGCC AGAGCACCTT AAGGATGTGA TCCTACTGAC AGCCATCGTG 360 CAGCTGCTCA GCTGCTTCTC TCTCTATGTC TGGTCCTTCT GGCTTCTGGC TCCAGGCCGG 420 GCCCTTTACC TCCTCTGGGT GAATGTGCTG GGCCCCTGGT TCACTGCAGA CAGTGGCACC 480 CCAGCACCAG AGCACAATGA GAAACGGCAG CGCCGACAGG AGCGGCGGCA GATGAAGCGG 540 TTATAGCCAT TGACATTGTC GCCACAGGCC ACTGGCCCTG GGTGGCTCTG TCAGGGTGCA 600
CAGCCCCTCA TGCCTGGAGC AATGAGGGTC TAGTCCAGGG GCCAAAAGCA GTCTGAGGTA 660 TTCGGTATAC TTATACTCTA TAGGGTCGTT GAATAAATGG CTTAGAATGT GAAAAAAAAA 720 AAAAAAAAAA ATTTT 735
(2) INFORMATION FOR SEQ ID NO: 264:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 783 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi ) SEQUENCE DESCRIPTION : SEQ ID NO : 264 :
AAGTGCATGA GCTGCCGATG TCGTCCTTAG TGATTGCGGT TTCGGTCGCT CTCCCGTGTT 60
TCCCGGGCTG GGTATTTGCC TCGCACCATG GCGCCCAAGG GCAAAGTGGG CACGAGAGGG 120 AAGAAGCAGA TATTTGAAGA GAACAGAGAG ACTCTGAAGT TCTACCTGCG GATCATACTG 180
GGGGCCAATG CCATTTACTG CCTTGTGACG TTGGTCTTCT TTTACTCATC TGCCTCATTT 240
TGCGCCTCGT TGCCCTGGGC TTTAGTCTGG CAGTGTATCG GGCCAGCTAC CACTCTATGA 300
GCTCGATGGC ACGAGCAGCG TTCTCTCAGG ATGGGGCCCT GATGGATGGT GGCATGGACC 360
TCAACATGGA GCAGGGCATG GCAGAGTGAG TGTCCCCCAC CGCCAGCCCA GGCACCTTAA 420 GGATGTCATC CTACTGACAG CCATCGTGCA GGTCCTCAGC TGCTTCTCTC TCTATGTCTG 480
GTCCTTCTGG CTTCTGGCTC CAGGCCGGGC CCTTTACCTC CTGTGGGTGA ATGTCCTGGG 540
CCCCTGGTTC ACTGCAGACA GTGGCACCCC AGCACCAGAG CACAATGAGA AACGGCAGCG 600
CCGACAGGAG CGGCGGCAGA TGAAGCGGTT ATAGCCATTG ACGATTTKGC SACNRGCCAC 660
TGGCCCTGGG TGGCTCTGTC AGGGTGCACA GCCCCTCATG CCTGGAGCAA TGAGGGTCTA 720 GTCCAGGGGC CAAAAGCAGT CTGAGGTATT GGGTATACTT ATACTCTATA GGGTCGTTGA 780
ATA 783
(2) INFORMATION FOR SEQ ID NO: 265:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 1638 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 265:
GGCACGAGGC GGCGGCAGCG GTGGCGGCGG CGCCCCCCGG CGGGAGCCGT NCCCTTTCCC 60
GTCGGGGAGC GCGGGGYCGG GGYCCAGGGG ANCCCGGGMC ACGGAGAGCG GGAAGAGGAT 120
GGATTGCCCG GCCCTCCCCC CCGGATGGAA GAAGGAGGAA GTGATCCGAA AATCTGGGCT 180
AAGTGCTGGC AAGAGCGATG TCTACTACTT CAGTCCAAGT GGTAAGAAGT TCAGAAGCAA 240
GCCTCAGTTG GCAAGGTACC TGGGAAATAC TCTTCATCTC AGCAGTTTTG ACTTCAGAAC 300
TGGAAAGATG ATCCCTAGTA AATTACAGAA GAACAAACAG AGACTGCGAA ACGATCCTCT 360
CAATCAAAAT AAGGGTAAAC CAGACTTGAA TACAACATTG CCAATTAGAC AAACAGCATC 420
AATTTTCAAA CAACCGGTAA CCAAAGTCAC AAATCATCCT AGTAATAAAG TGAAATCAGA 480
CCCACAACGA ATGAATGAAC AGCCACGTCA GCTTTTCTGG GAGAAGAGGC TACAAGGACT 540
TAGTGCATCA GATGTAACAG AACAAATTAT AAAAACCATG GAACTACCCA AAGGTCTTCA 600
AGGAGTTGGT CCAGGTAGCA ATGATGAGAC CCTTTTATCT GCTGTTGCCA GTGCTTTGCA 660
CACAAGCTCT GCGCCAATCA CAGGGCAAGT CTCCGCTGCT GTCGAAAAGA ACCCTGCTGT 720
TTGGCTTAAC ACATCTCAAC CCCTCTGCAA AGCTTTTATT GTCACAGATG AAGACATCAG 780
GAAACAGGAA GAGCGAGTAC AGCAAGTACG CAAGAAATTG GAAGAAGCAC TGATGGCAGA 840
CATCTTGTCG CGAGCTGCTG ATACAGAAGA GATGGATATT GAAATGGACA GTGGAGATGA 900
AGCCTAAGAA TATGATCAGG TAACTTTCGA CCGACTTTCC CCAAGAGAAA ATTCCTAGAA 960
ATTGAACAAA AATGTTTCCA CTGGCTTTTG CCTGTAAGAA AAAAAATGTA CCCGAGCACA 1020
TAGAGCTTTT TAATAGCACT AACCAATGCC TTTTTAGATG TATTTTTGAT GTATATATCT 1080
ATTATTCAAA AAATCATGTT TATTTTCAGT CCTAGGACTT AAAATTAGTC TTTTGTAATA 1140
TCAAGCAGGA CCCTAAGATG AAGCTGAGCT TTTGATGCCA GGTGCAATCT ACTCGAAATG 1200
TAGCACTTAC GTAAAACATT TGTTTCCCCC ACAGTTTTAA TAAGAACAGA TCAGGAATTC 1260
TAAATAAATT TCCCAGTTAA AGATTATTGT GACTTCACTG TATATAAACA TATTTTTATA 1320
CTTTATTGAA AGGGGACACC TGTACATTCT TCCATCRTCA CTGTAAAGAC AAATAAATGA 1380
TTATATTCAC AGACTGATTG GAATTCTTTC TGTTGAAAAG CACACACAAT AAAGAACCCC 1440
TCGTTAGCCT TCCTCTGATT TACATTCAAC TCTGATCCCG GGGCCTTAGG TTTGACATGG 1500
GAGGTGGGAG GAAGATAGCG CATATATTTG CAGTATGAAC TATTGCCTCT GGGACGTTGT 1560
GAGGAATTGT GCTTTCACCA GAATTTCTAA GGATTTCTGG CTTAAATATC ACCTAGCCTG 1620
TGGTAATTTT TTTTCCCT 1638
(2) INFORMATION FOR SEQ ID NO: 266:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1455 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 266:
CGTGCGTACT GCCATGCAGG TACCGGGTCC GGAATTCCCA GGGTCGACCC ACGCGTCCGC 60
TCAGTTGGCA AGGTACCTGG GAAATACTGT TGATCTCAGC AGTTTTGACT TCAGAACTGG 120
AAAGATGATG CCTAGTAAAT TACAGAAGAA CAAACAGAGA CTGCGAAACG ATCCTCTCAA 180
TCAAAATAAG GGTAAACCAG ACTTGAATAC AACATTGCCA ATTAGACAAA CAGCATCAAT 240
TTTCAAACAA CCGGTAACCA AAGTCACAAA TCATCCTAGT AATAAAGTGA AATCAGACCC 300
ACAACGAATG AATGAACAGC CACGTCAGCT TTTCTGGGAG AAGAGGCTAC AAGGACTTAG 360
TGCATCAGAT GTAACAGAAC AAATTATAAA AACCATGGAA CTACCCAAAG GTCTTCAAGG 420
AGTTGGTCCA GGTAGCAATG ATGAGACCCT TTTATCTGCT GTTCCCAGTG CTTTGCACAC 480
AAGCTCTGCG CCAATCACAG GGCAAGTCTC CGCTGCTGTG GAAAAGAACC CTCCTCTTTG 540
GCTTAACACA TCTCAACCCC TCTGCAAAGC TTTTATTGTC ACAGATGAAG ACATCAGGAA 600
ACAGGAAGAG CGAGTACAGC AAGTACGCAA GAAATTGGAA GAAGCACTGA TGGCAGACAT 660
CTTGTCGCGA GCTGCTGATA CAGAAGAGAT GGATATTGAA ATGGACAGTG GAGATGAAGC 720
CTAAGAATAT GATCAGGTAA CTTTCGACCG ACTTTCCCCA AGAGAAAATT CCTAGAAATT 780
GAACAAAAAT GTTTCCACTG GCTTTTGCCT GTAAGAAAAA AAATGTACCC GAGCACATAG 840
AGCTTTTTAA TAGCACTAAC CAATGCCTTT TTAGATGTAT TTTTGATGTA TATATCTATT 900
ATTCAAAAAA TCATGTTTAT TTTCAGTCCT AGGACTTAAA ATTAGTCTTT TGTAATATCA 960
AGCAGGACCC TAAGATGAAG CTGAGCTTTT GATGCCAGGT GCAATCTACT GGAAATGTAG 1020
CACTTACGTA AAACATTTGT TTCCCCCACA GTTTTAATAA GAACAGATCA GGAATTCTAA 1080
ATAAATTTCC CAGTTAAAGA TTATTGTGAC TTCACTGTAT ATAAACATAT TTTTATACTT 1140
TATTGAAAGG GGACACCTGT ACATTCTTCC ATCRTCACTG TAAAGACAAA TAAATGATTA 1200
TATTCACAGA CTGATTGGAA TTCTTTCTGT TGAAAAGCAC ACACAATAAA GAACCCCTCG 1260
TTAGCCTTCC TCTCATTTAC ATTCAACTCT GATCCCGGGG CCTTAGGTTT GACATGGGAG 1320
GTGGGAGGAA GATAGCGCAT ATATTTGCAG TATGAACTAT TGCCTCTGGG ACGTTGTGAG 1380
GAATTGTGCT TTCACCAGAA TTTCTAAGGA TTTCTGGCTT AAATATCACC TAGCCTGTGG 1440
TAATTTTTTT TCCCT 1455
(2) INFORMATION FOR SEQ ID NO: 267:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1086 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 267: CGCCTGCAGT ACCGGTCCGG AATTCCCGGG TCGACCCACG CGTCGCTCAC CCAGGAGAAG 60 CTGCCTCTCT ACATCAGCCT GGGCTGCAGC GCGCTGCCGC CGCGGGGCCG GCAGCTGAAC 120 TATGTGCTCT TCAGGGCGGG CACCGTGTTG CATTCATCTT TGTACCCCCA GCATCTAGCA 180 GTGTTGGCAT GTAGTAGGCA CTCAAGAAAT GTGTGTTGAA TGAACGATGC CTGTGACAAG 240 CAAGCGGACT TTATTCTTTC CTGACCCTTG CTCCTATGAC ACACCTCCTC CTGACTGCCA 300 CTGTCACTCC TTCAGAGCAG AACTCCTCTA GGGAACCTGG ATGGGAAACA GCCATGGCCA 360 AGGACATCCT GGGTGAAGCA GGGCTACACT TTGATGAACT GAACAAGCTG AGGGTGTTGG 420 ACCCAGAGGT TACCCAGCAG ACCATAGAGC TGAAGGAAGA GTGCAAAGAC TTTGTGGACA 480 AAATTCGCCA GTTTCAGAAA ATAGTTGGTG GTTTAATTGA GCTTGTTGAT CAACTTGCAA 540 AAGAAGCAGA AAATGAAAAG ATGAAGGCCA TCGGTGCTCG GAACTTGCTC AAATCTATAG 600 CAAAGCAGAG AGAAGCTCAA CAGCAGCAAC TTCAAGCCCT AATAGCAGAA AAGAAAATGC 660 AGCTAGAAAG GTATCGGGTT GAATATGAAG CTTTGTGTAA AGTAGAAGCA GAACAAAATG 720 AATTTATTGA CCAATTTATT TTTCAGAAAT GAACTGAAAA TTTCGCTTTT ATAGTAGGAA 780 GGCAAAACAA AAAAAAGCCT CTCAAAACCA AAAAAACCTC TGTAGCATTC CAGCGGCTTG 840 ACCAATGACC TATGTCACAA GAGGTGGCGT GTAAGGAATG CAGCCCCCTG AAGACAGCAC 900 TACAAGTCTG GGGGAGCCAG TTTTAACATC AGTGCACAGC TGCTGCTGGT GGCCCTGCAG 960 TGTACGTTCT CACCTCTTAT GCTTAGTTGG AACTAAGCAG TTTGTAAACT TTCATCCTTT 1020 TTTTTGTAAA TTCACAAAGC TTTGGAAGGA GARGCAATAA ATTTTTGKTT TCNAAATGGC 1080 TTGATG 1086
(2) INFORMATION FOR SEQ ID NO: 268:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1003 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 268: GGCACGGGAG CAGCCGGGCT GGTCCTGCTG CGAGCCGGCG GCCCGGAGTG GGGCGGCGGA 60
GCAAACATGA ACGTTGGAGT TGCCCACAGT GAAGTGAATC CAAATACCCG TGTCATGAAC 120
AGCCGGGGTA TGTGGCTGAC ATATGCATTG GGAGTTGGCT TGCTTCATAT TGTCTTACTC 180
AGCATTCCCT TCTTCAGTGT TCCTGTTGCT TCGACTTTAA CAAATATTAT ACATAATCTG 240
GGGATGTACG TATTTTTGCA TGCAGTGAAA GGAACACCTT TCGAAACTCC TGACCAGGGT 300 AAAAGCAAGG CTCCTAACTC ATTGGGAACA ACTGGACTAT GGAGTACAGT TTACATCTTC 360
ACGGAAGTTT TTCACAATTT CTCCAATAAT TCTATATTTT CTGGCAAGTT TCTATACGAA 420
GTATGATCCA ACTCACTTCA TCCTAAACAC AGCTTCTCTC CTCAGTGTAC TAATTCCCAA 480
AAT CCACAA CTACATGGTG TTCGGATCTT TGGAATTAAT AAGTATTGAA ATGTTTTGAA 540
ACTCAAAAAA AATTTTACAG CTACTGAATT TCTTATAAGG AAGGAGTGGT TAGTAAACTG 600 CACTGTTTCT CTGATAATGT GAAATGAGAA GTATTTACAT TGGAGGGCCA ATGGCTGGTC 660
CTTCAAGTGC TGTTTTGAAG TGCAGATTTC CATTAAATGA TGCCTCTCTT TAATACACCT 720
GGTACATTTC TGAAGAGGGG CTTTATAAGC AGGCTGGGCA GGCCCAGCTT ATAAGTTAAA 780
GGGCATCACA GTCAGGGTGT AGTAGATAAA TTCAAGGAAA TAAGAGATTT GTAAGAAACT 840
AGGACCAGCT TAACTTATAA TGAATGGGCA TTCTGTTAAG AAAAGAACAT TTCCAGTCAT 900 TCAGCTGTGG TTATTTAAAG CAGACTTACA TGTAAACCGG AATCCTCTCT ATACAAGTTT 960
ATTAAAGATT ATTTTTATTA CCGTAAAAAA AAAAAAAAAA AAA 1003
(2) INFORMATION FOR SEQ ID NO: 269:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 1234 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi ) SEQUENCE DESCRIPTION: SEQ ID NO : 269 :
ATCAGCATCT ACAAGTAGCA TATTTTGGAT GGTGTTTGTG TGCTACTTCA AAGTAACTAG 60
GAAAAAATAA TCCTCGCAAC ACAGGTACCT TCTCATGTCA GAATTGGGGG TGTTAGGTTG 120
CCAGTTCTAT CAGTGTTGAT TCATTTCATT ACTTCCTACA GAGCAAACAT GAACGTTGGA 180
GTTCCCCACA GTGAAGTGAA TCCAAATACC CGTCTCATGA ACAGCCGGGG TATGTGGCTG 240 ACATATGCAT TGGGAGTTGG CTTGCTTCAT ATTCTCTTAC TCAGCATTCC CTTCTTCAGT 300
GTTCCTCTTG CTTGGACTTT AACAAATATT ATACATAATC TGGGGATGTA CGTATTTTTG 360
CATGCAGTGA AAGGAACACC TTTCGAAACT CCTGACCAGG GTAAAGCAAG GCTCCTAACT 420
CATTGGGAAC AACTGGACTA TGGAGTACAG TTTACATCTT CACGGAAGTT TTTCACAATT 480
TCTCCAATAA TTCTATATTT TCTGGCAAGT TTCTATACGA AGTATGATCC AACTCACTTC 540
ATCCTAAACA CAGCTTCTCT CCTGAGTGTA CTAATTCCCA AAATCCCACA ACTACATGGT 600
GTTCGGATCT TTGGAATTAA TAAGTATTGA AATGTTTTGA AACTGAAAAA AAATTTTACA 660
GCTACTGAAT TTCTTATAAG GAAGGAGTGG TTAGTAAACT GCACTGTTTC TSTGATAATG 720
TGAAATGAGA AGTATTTACA TTGGAGGGCC AATGGCTGGT CCTTCAAGTG CTGTTTTGAA 780
GTGCAGATTT CCATTAAATC ATGCCTCTGT TTAATACACC TGGTACATTT CTGAAGAGGG 840
GCTTTATAAG CARGCTGGGC AGGCCCAGCT TATAAGTTAA AGGGCATCAC AGTGAGGGTG 900
TAGTAGATAA ATTCAAGGAA ATAAGAGATT TGTAAGAAAC TAGGACCAGC TTAACTTATA 960
ATGAATGGGC ATTGTGTTAA GAAAAGAAGA TTTCCAGTCA TTCAGCTGTG GTTATTTAAA 1020
GCAGACTTAC ATGTAAACCG GAATCCTCTC TATACAAGTT TATTAAAGAT TATTTTTATT 1080
ACCRTACATA TTTCKCTTGT TTTATGTAAG YGGATGTATA TCCTCTTGTT TTATACAAGC 1140
CAGTTCCCAC TTATGAGGGT ACΓTTTTTGG TTTTGCTGGG CTTAATATTG TGTATTGGTC 1200
AATGAGGCCA TTTTTACANT TATTAACGTT ACAG 1234
(2) INFORMATION FOR SEQ ID NO: 270:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 574 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: ; 270: NGAGGTGCGT TCTGAGCCGT CTCTCCTGCG CCAAGATGCT TCAAAGTATT ATTAAAAACA 60 TATGGATCCC CATGAAGCCC TACTACACCA AAGTTTACCA GGAGATTTCG ATAGGAATGG 120 GGCTGATGGG CTTCATCGTT TATAAAATCC GGGCTCCTGA TAAAAGAAGT AAGGCTTTGA 180 AAGCTTCAGC GCCTGCTCCT GGTCATCACT AACCAGATTT ACTTGGAGTA CATGTGAAAG 240 AAAACGTCAG TCTGCCTGTA AATTTCAGCA AGCCGTGTTA GATGGGGAGC GTGGAACGTC 300 ACTGTACACT TGTATAAGTA CCGTTTACTT CATCGCATGA ATAAATGCAT CTGTGAGATG 360 CACTGCTACC TGGTACTGCT TTCAGTGTGT TCCCCCTCAG CCCTCCGGCG TGTCAGGCAT 420
ACTCTGAGTA GATAATTTGT CATGCAGCGC ATGCAATCAG AATCTCACTG AGCCACCCAT 480
CATTCTGAAA TAATTACCTC AGTTGTACAG GACTTGGTGA TCAGCATCCA GGCACTCACT 540 TGTATTCTAC TGCTCAATAA ACGTTTATTA AACT 574
(2) INFORMATION FOR SEQ ID NO: 271:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1731 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 271: GCTGCAAGGT GCGCCTCGTG CCGCTGCAGA TCCAGCTCAC TACCCTGGGA AATCTTACAC 60
CTTCAAGCAC TCTCTTTTTC TGCTGTGATA TGCAGGAAAG GTTCAGACCA GCCATCAAGT 120
ATTTTGGGGA TATTATTAGC GTGGGACAGA GATTGTTGCA AGGGGCCCGG ATTTTAGGAA 180
TTCCTGTTAT TGTAACAGAA CAATACCCTA AAGGTCTTGG GAGCACGGTT CAAGAAATTG 240
ATTTAACAGG TGTAAAACTG GTACTTCCAA AGACCAAGTT TTCAATGGTA TTACCAGAAG 300 TAGAAGCGGC ATTAGCAGAG ATTCCCGGAG TCAGGAGTGT TGTATTATTT GGAGTAGAAA 360
CTCATGTGTG CATCCAACAA ACTGCCCTGG AGCTAGTTGG CCGAGGAGTC GAGGTTCACA 420
TTCTTGCTGA TGCCACCTCA TCAAGAAGCA TGATGGACAG GATGTTTGCC CTCGAGCGTC 480
TCGCTCRARC CNGGGATCAT AGTGACCACG AGTGNAGGCT GTTCTGCTTC AGCTGGTAGC 540
TGATAAGCAC CATCCAAAAT TCAAGGAAAT TCAGAATCTA ATTAAGGCGA GTGCTCCAGA 600 GTCGGGTCTG CTTTCCAAAG TATAGGACAT TTGAAGAACT GGTATGCTAC TCACTGGTGA 660
AGGACACTCA GGTGAAGGAC TGTAAGCCCA CACAAGCTCT TCTTATCTCT ACTAGAATTA 720
AAATGTTAAG TCAAAAACGG CTCCTTTTTT GCGCCTCCTA GTGAACTTAA CCAGCTAGAC 780
CATTTGAGTA CCAGCATTTA GTTACAAACG TCAAAGGCTT CCGGTGCTGC TTACCTTCCT 840
TTTTTGTTAA TGTGCTTTTA TTTATTAAAA AAAATTACAA TGAAGATGCC TGTTTTGTCT 900 CTACTGTGTA CTCTGATCGT ATCTTTCCAA AGTGCAGACT CTTGTGAAGT TTTCTTAAAT 960
TGTTCACTTT AAAGAAAATG ACGTACCAAC AATGATTTGG CTTTTATATT ACTGTAAGAT 1020
GTTATAATGT TAATGTGGAT GTAGTGCTTT TACTTTACAG ATTGATTGGA ATAAGATTAT 1080
TGCATATGAA TTTACCCACA GGACTCTGAA TCATGTTACC CACTCCCCTG ACAATCTTCT 1140
CCACTTAGTG AGTTGCATTC ATCTATCCGT ACCAAATGAT GTTGAATAAT TACATATCTT 1200 TCTKGACTAT ACTGATTTCT TATTTTCGTC ACTATTACTA AATCTCTGTT AATATTCTCT 1260
CTTTTAACTG AAAAGGGATG GGATAGAAGG GTTTGCAATG CCATATTATT GGTGGAGGGC 1320 TGTTTTAACA TCTTTGAAGT ATGGCTTGCT GAATATCTTT ACCAACATCT TCAATATATA 1380 TTCTAGTGTC CACAAGATTT AGCAAAAAGA TAAAGCTTGG GTGGAATATC ATTTTAAAAT 1440 GTTCATGTTC TGTTCTATAT TTTCTTCACC TACTCTCCAA ATATTGTAAT GCAAAAAGTC 1500 TCAGTAATGA TTTGGTAGTA TTAATTTTGT GGTCATTGTT TCTCTTCGAT AAATTTATTT 1560 TCATTAAATA CTTRTTAGAG GGTTTTGAAA TCTTTTTCAA ATATGTGAAA TGTGAAACTG 1620 CTGTCTTTTA TATTAAAGTA ATTAAAGAAA ATGTATTGTG ATTGAAATTA TTTTGNCCTC 1680 CACAAGATGG CTCTATGAGT ATTCTTCCAG GGATTCTAAT ATTTATTTAA G 1731
(2) INFORMATION FOR SEQ ID NO: 272:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1320 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 272:
CTGCTTAGGA AGAGAAGGTC AGAGTTCGCG GGGGCAGAGG CATTCTTGCC GCTGGCCCAG 60
TCACTATGTA GTGGAGGGGC AGACACCCTC CCGCAAATTC TGGAAGGTTC TTAGTCTCGA 120
CTAGGGCAGT AGCCCAGGAC TCCTAGTCGC CGGCTTCAGG TCACTGCCGG CTGAACGGAG 180
CTGCCGTCGC CATGTTTGGC TGCTTGGTGG CGGGGAGGCT GGTGCAAACA GCTGCACAGC 240
AAGTGGCAGA GGATAAATTT GTTTTTGACT TACCTGATTA TGAAAGTATC AACCATGTTG 300
TGGTTTTTAT GCTGGGAACA ATCCCATTTC CTGAGGGAAT GGGAGGATCT GTCTACTTTT 360
CTTATCCTGA TTCAAATGGA ATGCCAGTAT GGMAACTCCT AGGATTTGTC ACGAATGGGA 420
AGCCAAGTCC CATCTTCAAA ATTTCAGGTC TTAAATCTGG AGAAGGAAGC CAACATCCTT 480
TTGGAGCCAT GAATATTGTC CGAACTCCAT CTCTTGCTCA GATTGGAATT TCAGTGGAAT 540
TATTAGACAG TATGGCTCAG CAGACTCCTG TAGGTAATGC TGCTGTATCC TCAGTTGACT 600
CATTCACTCA GTTCACACAA AAGATGTTGG ACAATTTCTA CAATTTTGCT TCATCATTTG 660
CTGTCTCTCA GGCCCAGATG ACACCAAGCC CATCTGAAAT GTTCATTCCG GCAAATGTGG 720
TTCTGCAAAT GGTATGAGGC ATNTTCTGTC TCCAATATTA AGGCTTTTTA TAACTGAATA 780
TCTATTTTGT CTATGAATAT ATTCCTTTTT TGACATTTAA ACATATTCTT TTATTGTGAA 840
CATCAGCACT GCATGCCATT AAAGTATGTA CTATAGAGAT CTGATGAGAA ACAGTTCTTA 900
CCCTAAATAT TTTGTTATAT TGTCGCCATT ATGAATTTAT AAAGACAGGA AAATATAGTT 960 GCCTATGTTT TAGGGACCAC TATTAAAGCT TATAAATATT TGTGTATTTT CATTTAGAAG 1020 TACCATCTAT GAGAGTAGTT TATACTGCAC TCTGTACATG AATGGCTAAT GAATCTATTT 1080 TCCAACTTTC CCGTGTTTTA TAGATATTTC TTTTCACTTT GAGTATCCTA GAGATGGGAG 1140 GATCCCTAGG AAGAGTTTGT TGAGAAGTGG TACCATGGTG TAGCATGGGA GAGCATTGGG 1200 AATCCACTAG GTTTGAATTT GGCATAATCG TAGCTATCTG ACCCTGAGCA AATTTCTCTC 1260 ATCTGCTCAT CTGANGAATG AGGAAATAGG AGTGAATTTC ATNTTTCCTA GGTCCNTCTA 1320
(2) INFORMATION FOR SEQ ID NO: 273:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 515 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO : 273: CCCTGGAGAG GGGCTGCTGT GCCAGCTTGG GGAGGGTCTC GGATGGGGCT GCCCCTGATG 60 GCCCTGATGT GGAGTACCTT GCCAGCATCT GCTGGGGTGA ACTTTATTTT AGCCCTTCCC 120 TTGTTGYTCT TATCAAGAAC AGAGGAGGGG TGGGCAGGTC AGTGATGTCA GCACTCAGTA 180 TTCCCAGCAC AGCGGCTCTG GAAGAGGCAT GAGGCATTTC TTTCAGGAAA TCRTCATTAT 240 TCAGCCAGAA GGCATTCATT AAGTAAGTCC TCACTTTGTG CCCAGCTCTG TGTTATAGGC 300 CCTTGGCGAG ACTCAGGAGG GGCARAGGAC GCTAGKTTKT AGWTAACACG GAACCTCARA 360 GGWTATATGG TCCAAGAAGA CCCGGGGGCG GTGAAAACCC TGTGGACTAA TGCTCACGGG 420 AGCCCGAGGT CACACTTTGA CTTTGCTACC ATGGGCTGTG TCTANGNACG TATATATGCT 480 GCGTAATTAT TACAGAGGCA GTCCATGTCC ATTGT 515
(2) INFORMATION FOR SEQ ID NO: 274:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2995 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 274: TGACACCCAT AAGGAATTCA TGAAGAAAGT AGAAGAAAAG CGAGTGGACG TTAACTCAGC 60
AGTAGCCATG GGAGAAGTCA TCCTGGCTGT CTGCCACCCC GATTGCATCA CAACCATCAA 120
ACACTGGATC ACCATCATCC GAGCTCGCTT CGAGGAGGTC CTGACATGGG CTAAGCAGCA 180
CCAGCAGCGT CTTGAAACGG CCTTGTCAGA ACTGGTGGCT AATGCTGAGC TCCTGGAAGA 240
ACTTCTCGCA TGGATCCAGT GGGCTGAGAC CACCCTCATT CAGCGGGATC AGGAGCCAAT 300
CCCGCAGAAC ATTGACCGAG TTAAAGCCCT TATCGCTGAG CATCAGACAT TTATGGAGGA 360
GATGACTCGC AAACAGCCTG ACGTGGACCG GGTCACCAAG ACATACAAAA GGAAAAACAT 420
AGAGCCTACT CACGCGCCTT TCATAGAGAA ATCCCGCAGC GGAGGCAGGA AATCCCTAAG 480
TCAGCCAACC CCTCCTCCCA TGCCAATCCT TTCACAGTCT GAAGCAAAAA ACCCACGGAT 540
CAACCAGCTT TCTGCCCGCT GGCAGCAGGT GTGGCTGTTA GCACTGGAGC GGCAAAGGAA 600
ACTGAATGAT GCCTTGGATC GGCTGGAGGA GTTGAAAGAA TTTGCCAACT TTGACTTTGA 660
TGTCTGGAGG AAAAAGTATA TGCGTTGGAT GAATCACAAA AAGTCTCGAG TCATGGATTT 720
CTTCCGGCGC ATTGATAAGG ACCAGGATGG GAAGATAACA CGTCAGGAGT TTATCGATGG 780
CATTTTAGCA TCCAAGTTCC CCACCACCAA GTTAGAGATG ACTGCTGTGG CTGACATTTT 840
CGACCGAGAT GGGGATGGTT ACATTGATTA TTATGAATTT GTGGCTGCTC TTCATCCCAA 900
CAAGGATGCG TATCGACCAA CAACCGATGC AGATAAAATC GAAGATGAGG TTACAAGACA 960
AGTGGCTCAG TGCAAATGTG CAAAAAGGTT TCAGGTGGAG CAGATCGGAG AGAATAAATA 1020
CCGGTTCTTC CTCGGCAATC AGTTTGGGGA TTCTCAGCAG TTGCGGCTGG TCCGTATTCT 1080
GCGCAACCGT GATGGTTCGC GTTGGTGGAG GATGGATGGC CTTGGATGAA TTTTTAGTGA 1140
AAAATGATCC CTGCCGAGCA CGAGGTAGAA CTAACATTGA ACTTAGAGAG AAATTCATCC 1200
TACCAGAGGG AGCATCCCAG GGAATGACCC CCTTCCGCTC ACGGGGTCGA AGGTCCAAAC 1260
CATCTTCCCG GGCAGCTTCC CCTACTCGTT CCAGCTCCAG TGCTAGTCAG AGTAACCACA 1320
GCTGTACATC CATGCCATCT TCTCCAGCCA CCCCAGCCAG TGGAACCAAG GTTATCCCAT 1380
CATCAGGTAG CAAGTTGAAA CGACCAACAC CAACTTTTCA TTCTAGTCGG ACATCCCTTG 1440
CTGGTGATAC CAGCAATTAG TTCTTCCCCG GCCTCCACAG GTGCCAAAAC TAATCGGGCA 1500
GACCCTAAAA AGTCTGCCAG TCGCCCTGGG AGTCGGGCTG GGAGTCGAGC CGGGAGTCGA 1560
GCCAGCAGCC GGCGAGGAAG TGACGCTTCT GACTTTGACC TCTTAGAGAC GCATTGCTTG 1620
TTCCGACACT TCAGAAAGCA GCGCTGCAGG GGGCCAAGGC AACTCCAGGA GAGGGCTAAA 1680
CAAACCTTCC AAAATCCCAA CCATGTCTAA GAAGACCACC ACTGCCTCCC CCAGGACTCC 1740
AGGTCCCAAG CGATAACACT GTCTAAGCAC CCCCAAGCCA CTATCCACTT TGAATCCTGC 1800
TCCATACATT GGGTGTATAT TTATTCTGAA CGGGAGAAGT TATATTGTTA AAAGTGTAAA 1860
AGAATAATTG TCTTATGAAG CTGCCTTATT TTTTTTCTTT TTGTAAGTTA CTATTTTCAT 1920
GTCAATATTT ATCTAGATAA AATTTGCCTC CTGCTAACCC TCTAATGCAT GGGGCCCAGA 1980
AATCAAATAT TTGAGAAAAA CAAGTCAAAA GCTCAAGATA CAAATCTGTA TTAAAAAAAA 2040
AAAAGCCTAT TAATAGCGTT TCTGCGCGGT GCAGGGTTGT AAACCTGCTT TATCTTTTAG 2100
GATTATTCCT AAATGCATCT TCTTTATAAA CTTGACTTGC TATCTCAGCA AGATAAATTA 2160
TATTAAAAAA ATAAGAATCC TGCAGTGTTT AAGGAACTCT TTTTTTGTAA ATCACGGACA 2220
CCTCAATTAG CAAGAACTGA GGGGAGGGCT TTTTCCATTG TTTAATCTTT TGTGATTTTT 2280
AGCTAAAGAG AGCGAACCTC ATCTAAGTAA CATTTGCACA TGGATACAGC AAAAGGAGTT 2340
CATTGCAATA CTGTCTTTGG ATATTGTTTC AGTACTGGGT GTTTAAAGGA CAAATAGCTG 2400
CTAGAATTCA GGGGTAAATG TAAGTGTTCA GAAAACGTCA GAACATTTGG GGTTTTAAAC 2460
TCATTTGTTG CTCCCTATCC AGCCTAGACA CCAGTAACTC TTGTGTTCAC CAGGACCCAG 2520
ACCCTTGGCA AGGGATAGGC TCGTTGGTGA CATTGTGAAT TTCAGATTTG TTTTATCCAC 2580
TTTTTTTGCT ATTTATTTAA ATGGTCGATC AACTTCCCAC AAACTGAGGA ATGAATTCCA 2640
CGAGCCTGTT CTGAAAATGT GGACGTAAGA CAAACACGTG CTCGTCCTTT AATGGAGTTC 2700
ACCAGCACAC TTGTTAACCA GTCCTGTTTG CTTTCGTCTT TTTTTGTGCG TAATAAAGTC 2760
AACTGACCAA GTGACCATGA AAAGGGGCTG TCTGGGGCTC CTGTTTTTTA GCTGCTGTTC 2820
TTCAGCTCCG ACCATGTTGC TGTGTGATTA TCTCAATTGG TTTTAATTGA GGCAGAAACT 2880
GAAGCTCTAC CAATGAACTG TTTAGAAACA AGACACACTT TTGTATTAAA ATTGCTTGCA 2940
GTAACAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAACTCG AGGGGGGCCC GGTAC 2995
(2) INFORMATION FOR SEQ ID NO: 275:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1990 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 275: GGGACCCGCG CGSCTCCCGG GGATGGTGAG CAAGGCGCTG CTGCNWCGTG TCTGCCGTCA 60 ACCGCAGAGG ATGAAGCTGC TGCTGGGCAT CGCCTTGCTG GCCTACGTCG CCTCTCTTTG 120 GGGCAACTTC GTTAATATGA GGTCTATCCA GGAAAATGGT GAACTAAAAA TTGAAAGCAA 180 GATTCAAGAG ATGGTTGAAC CACTAAGAGA GAAAATCAGA GATTTAGAAA AAAGCTTTAC 240 CCAGAAATAC CCACCAGTAA AGTTTTTATC AGAAAAGGAT CGGAAAAGAA TTTTGAWTAA 300
CAGGAGGCGC AGKGTTCGTG GGCTCCCATC TKAACTGACA AACTCATGAT GGACGGCCAC 360
GAGGTGACCG TGGTGGACAA TTTCTTCACG GGCAGGAAGA GAAACGTGGA GCACTGGATC 420
GGACATGAGA ACTTCGAGTT GATTAACCAC GACCTGTGGG AGCCCCTCTA CATCGAGGTT 480
GACCAGATAT ACCATCTGGC ATCTCCAGCC TCCCCTCCAA ACTACATGTA TAATCCTATC 540
AAGACATTAA AGACCAATAC GATTGGGACA TTAAACATGT TGGGGCTGGC AAAACGAGTC 600
GGTGCCCGTC TGCTCCTGGC CTCCACATCG GAGGTGTATG GAGATCCTGA AGTCCACCCT 660
CAAAGTGAGG ATTACTGGGG CCACGTGAAT CCAATAGGAC CTCGGGCCTG CTACGATGAA 720
GGCAAACGTG TTGCAGAGAC CATGTGCTAT GCCTACATGA AGCAGGAAGG CGTGGAAGTG 780
CGAGTGGCCA GAATCTTCAA CACCTTTGGG CCACGCATGC ACATGAACGA TGGGCGAGTA 840
GTCAGCAACT TCATCCTGCA GGCGCTCCAG GGGGAGCCAC TCACGGTATA CGGATCCGGG 900
TCTCAGACAA GGGCGTTCCA GTACGTCAGC GATCTAGTGA ATGGCCTCGT GGCTCTCATG 960
AACAGCAACG TCAGCAGCCC GGTCAACCTC GGGAACCCAG AAGAACACAC AATCCTAGAA 1020
TTTGCTCAGT TAATTAAAAA CCTTGTTGGT AGCGGAAGTG AAATTCAGTT TCTCTCCCAA 1080
GCCCAGGATG ACCCACAGAA AAGAAAACCA GACATCAAAA AAGCAAAGCT GATGCTGGGG 1140
TGGGAGCCCG TGGTCCCGCT GGAGGAAGGT TTAAACAAAG CAATTCACTA CTTCCGTAAA 1200
GAACTCGAGT ACCAGGCAAA TAATCAGTAC ATCCCCAAAC CAAAGCCTGC CAGAATAAAG 1260
AAAGGACGGA CTCGCCACAG CTGAACTCCT CACTTTTAGG ACACAAGACT ACCATTGTAC 1320
ACTTGATGGG ATGTATTTTT GGCTTTTTTT TGTTGTCGTT TAAAGAAAGA CTTTAACAGG 1380
TGTCATGAAG AACAAACTGG AATTTCATTC TGAAGCTTGC TTTAATGAAA TGGATGTGCC 1440
TAAAAGCTCC CCTCAAAAAA CTGCAGATTT TGCCTTGCAC TTTTTGAATC TCTCTTTTTA 1500
TGTAAAATAG CGTAGATGCA TCTCTGCGTA TTTTCAAGTT TTTTTATCTT GCTGTGAGAG 1560
CATATGTTGT GACTGTCGTT GACAGTTTTA TTTACTGGTT TCTTTGTGAA GCTGAAAAGG 1620
AACATTAAGC GGGACAAAAA ATGCCGATTT TATTTATAAA AGTGGGTACT TAATAAATGA 1680
GTCGTTATAC TATGCATAAA GAAAAAYCCT AGCAGTATTG TCAGGTGGTG GTGCGCCGGC 1740
ATTGATTTTA GGGCAGATAA AAGAATTCTG TGTGAGAGCT TTATGTTTCT CTTTTAATTC 1800
AGAGTTTTTC CAAGGTCTAC TTTTGAGTTC CAAACTTGAC TTTGAAATAT TCCTGTTGGT 1860
CATGATCAAG GATATTTGAA ATCACTACTG TGTTTTGCTG CGTATCTGGG GCGGGGGCAG 1920
GTTGGGGGGC ACAAAGTTAA CATATTCTTG CTTAACCATG CTTAAATATG CTATTTTAAT 1980
AAAATATTGA 1990
(2) INFORMATION FOR SEQ ID NO: 276:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2436 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 276:
AACTTCGCTT AGCTCTCCAG GCTNAAACGG GTGAGNCCTT AAAAACAGAA GAGAACAAGA 60
TTTAAAGTCC GTTGCATTGA AAATAACAAA CAATATCAAT GTTTTAATCA AGGATCTCTT 120
CCACATTCCT CCTTCTTATA AGAGCACAGT AACACTATGC TGGAAACCTG TACAAAAGGT 180
TGAGATTGGG CAAAAGAGAG CCAGTGAAGA TACAACTTCA GGTTCACCAC CCAAGAAATC 240
TTCAGCAGGA CCAAAAAGAG ATGCCAGGCA GATTTATAAC CCTCCCAGTG GGAAATATAG 300
CAGCAATTTG GGCAACTTTA ATTATGAGCA GAGAGGAGCC TTCAGGGGAA GTAGAGGTGG 360
CCGAGGTTGG GGCACACGAG GAAATCGTAG TCGGGGAAGA CTCTACTGAA TAAGACATCA 420
GCATTCTTCA GCATTGTCAT GAGCTTAATA TACTTAAATT CTACTACTCA TTGGATTGCC 480
GGGGATCTCC CTTTAAACAG ACTGCTGCCT TCAGCTAAAA ACTTAATGTT CTTTATACCT 540
TTGTATGTAT GACCTACTTT TGTAACAGAC CATGGTTGTG TCCAAGGTAA AACCACAGTG 600
ATATTTTTGG ATGCTTTGTC TGCAATCTTG ACTTGTTTTT GCAGTATCAT TATTCAGACT 660
TCAAATTGTG AATCTTTTAA ACATCTTGAT AATTTGTTGT TGAGAGCTGT TCATTCTAAA 720
ATGTAATGAA ATTCAGTCTA GTTCTGCTCA TAAAGATCAT CAGTTTTGAA AGGTTACTGA 780
TTTTCCTCTT CCCTCTTAGT TTTTTACCCA ATATATGGAG AAGAGTAATG GTCAATCTTA 840
ACATTTTGTT TTAATTGTTT AATAAAGCTG CTGGGCAGTG GTGCAGCATT CCTACCTAGT 900
GTCATAAAAG CAAAATACTT ACATAGCTTT CTTAAAATAT AGGAATGACA TTACATTTTT 960
AGGAGAAAGT AAGTTGCTTT GCACCGCCTA CTTAATTCTT TTCCATATAT TGTCATACAA 1020
ACTTTTGAAT ATGGAATCTT ACTATTTGAA TAGAAATGTG TATGTATAAT ATACATACAT 1080
ACATAAGCAT ATATGTCTGT GTGTGTGTGT ATATATATAT ATATGCATGC TGTGAAACTT 1140
GACTACACAA CATAAATCAC TTTTTAAATT CCAGGAACGG GTAGTCTGAC ACGGTCATTA 1200
TCCTTTTGAG GCTGAATCCG TTATTAACTT GTTATTTAGG TTTTTACTCC CAGTAGCAAG 1260
GGATTCTAAG TTAGTTGCAC TTACATGATT ATTGTTATTT AAAACTAAGA ATAAAGGCTG 1320
CATTTTCAAA GATAAATTGG AATTGCTGTT GGTGAAATAA CAACCAAAAT ACTGAATCTG 1380
ATGTACATAC AGGTTTCTAC AGGAAGAGAT GGTATAATTT ACAATTTGGA GATTTAATAA 1440
CCAGGGCTAC CCAGAAAAAG TGACTTGATA ACATGGTACC AATAAGTAAG GGATGCTCTC 1500
TCGGTTTGCT TTTGCCACTT TCAAGATTTT AACTTCTCAG GTTATTAATC AAAATTATTG 1560
TATAAGTTAG CCAATAGAAT TTTTAGGTTA AAACAACAGA TGGGGGGTTT GTGGAGTGTT 1620
TAATGTCATG GGCATTTTTA GTAGCATAGA CCCTTTGTTC TGCATTTCAA TGTTTCGTAT 1680
ATTTTTGTTT CACAGTTAAT CTTCCCTCCC CAAGTTTGCT ATTCAAATCA ACTGCCTGAA 1740
TCACATTTCT AGTAGTCTGA TGTATTTTTC TGAGGAATAG TTTGTGATTC CAATGCAGCT 1800
GTCTTCATTA CCATTACCTC TACACTGCAG AAGAAGCAAA ACTCCTTTAT TAGAATTACT 1860
GCACATGTCT ATGGGGAAAA TAGTTCTGAA AGGCTAGAAT GATACAACTC AGCAAAAGTT 1920
GGTCAGCTTG GCTATGGAGT GGTGGCAATA ATCTCTAAAC ATTCCAAAAG ACCATGAGCT 1980
GAACCTAAAC TCCCTTGGAA TCTGAACAAA GGAATATAAA ATTGCCATTT GAAAACTGAC 2040
CAGCTAATCT GGACCTCAGA GATAGATCAG CCAGTGGCCC AAAGCCATTT CAAGTACAGA 2100
AATTATAGAG ACTACAGCTA AATAAATTTG AACATTAAAT ATAATTTTAC CACTTTTTGT 2160
CTTTATAAGC ATATTTGTAA ACTCAGAACT GAGCAGAAGT GACTTTACTT TCTCAAGTTT 2220
GATACTGAGT TGACTCTTCC CTTATCCCTC ACCCTTCCCC TTCCCTTTCC TAAGCCAATA 2280
GTGCACAACT TAGGTTATTT TTGCTTCCGA ATTTGAATCA AAAACTTAAT GCCATGGATT 2340
TTTTTCTTTT GCAAGACACC TGTTTATCAT CTTGTTTAAA TGTAAATGTC CCCTTATGCT 2400
TTTGAAATAA ATTTCCTTTT GTAATTTTAA AAAAAA 2436
(2) INFORMATION FOR SEQ ID NO: 277:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 782 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO : 277: GCCACTGACT TCTCCCACCC TTCTGTCTCC CCCATAATAG TTTATTTGGT TGGTCTGGAC 60 TCACTTGTGG CCTTTRATTA AATTCCTAAG GGGCCTGAAG AAGACATTTC TACTGCAGAG 120 GGTTAGAGGC ACTTGAGCAA GGCCCCCACA TCCCAACTCT GGGAGTTGTG GTGGGAGGAG 180 GCACTTCTGG GGGATAGGAC CAGACAAGAT AACAGGAGCT CACATGGNAA GCAGAAGCTG 240 TGACAAGTTT AGTAGTCCCA AAATGGGTTA TATCCCTTCC CCCTTTACAT CAGAATCTTG 300 TGAAATGGGA AAACAACAGA AGGAGGGGAT CAAAGATAGC TGATCTCACA TCCTTCCCAG 360 GCAGGGCARA GGTGGGAGTC AAACCCGGGT GACAGGTGGG TGGAGAGCCC TGTTTGAGGT 420
TCTCGCTCAT CCCTCTCTGG TATTAGTTTT TCCCCTGGGA GCAGGAAGCC CTAGGAAGAG 480
GGGACTGCAG GGTCCCCRGG GGATCTTTCC TCCCTGCCCT GCATGAGGCA GAGGCAAGCT 540
GCCTGCCAAC CCCCTCCCTC AAGGAATGGC CTTGCCCAGG AATGCCCACC ACACATACCC 600
TCTTCTTTTT TTCTAGTCAA ACTCTTCTTT ATTCCTTGGC TTGCCTCCCT CCTTCCTCCC 660 CTCTCAACCT TTACTTCTCA TTTCTATTTC ATGGAATTTG GGATTCAAGT TAAACTACAA 720
CAGTGCCGCC AACACCAAGT CTTCCAGGAA AAAAATACAA AGAAATTTAA CAAAAAAAAA 780
AA 782
(2) INFORMATION FOR SEQ ID NO: 278:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 961 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 278:
GAGTTCCGGC TGGAGACCCG TGCTCTGGGC CGGCGCCTTC ACCATGGCCT CGGCAGAGCT 60
GGACTACACC ATCGAGATCC CGGATCAGCC CTGCTGGAGC CAGAAGAACA GCCCCAGCCC 120
AGGTGGGAAG GAGGCAGAAA CTCGGCAGCC TGTGGTGATT CTYTTCGGCT GGGGTGGCTG 180 GAAGGACAAG AACCTTGCCA AGTACAGTGC CATCTACCAC AAAAGGGGCT GCATCGTAAT 240
CCGATACACA GCCCCGTGGC ACATGGTCTT CTTCTCCGAG TCACTGGGTA TCCCTTCACT 300
TCGTGTTTTC GCCCAGAAGC TGCTCGAGCT GCTCTTTGAT TATGAGATTG AGAAGGAGCC 360
CCTGCTCTTC CATGTCTTCA GCAACGGTGG CGTCATGCTG TACCGCTACG TGCTCGAGCT 420
CCTGCAGACC CGTCGCTTCT GCCGCCTGCG TGTGGTGGGC ACCATCTTTG ACAGCGCTCC 480 TGGTGACAGC AACCTGGTAG GGGCTCTGCG GGCCCTGGCA GCCATCCTGG AGCGCCGGGC 540
CGCCATGCTG CGCCTGTTGC TGCTGGTGGC CTTTGCCCTG GTGGTCGTCC TGTTCCACGT 600
CCTGCTTGCT CCCATCACAG CCCTCTTCCA CACCCACTTC TATGACAGGC TACAGGACGC 660
GGGCTCTCGC TGGCCCGAGC TCTACCTCTA YTCGAGGGCT GACGAAGTAG TCCTGGCCAG 720
AGACATAGAA CGCATGGTGG AGGCACGCCT GGCACGCCGG GTCCTGGCGC GTTCTGTGGA 780 TTTCGTGTCA TCTGCACACG TCAGCCACCT CCGTGACTAC CCTACTTACT ACACAAGCCT 840
CTGTGTCGAC TTCATGCGCA ACTGCGTCCG CTGCTGAGGC CATTGCTCCA TCTCAMCTCT 900
GCTCCAGAAA TAAATCCCTG ACAMCTCCCC ACAAAAAAAA AAAAAAAAAA ACTCGAGGGG 960
961
(2) INFORMATION FOR SEQ ID NO: 279:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1228 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 279:
CGCGCTTTCC AGTTCGGTCT CCTGGTGTAC GGCCAACGCC AAGTAGGGGA TTGCGTTCCC 60
TCCAGTCGCA GCCCTATCAG ATTTGGATAT GTCCTTCATA TTTGATTGGA TTTACAGTGG 120
TTTCAGCAGT GTGCTACAGT TTTTAGGATT ATATAAGAAA ACTGGTAAAC TGGTATTTCT 180
TGGATTGGAT AATGCAGGAA AAACAACATT GCTACACATG CTAAAAGATC ACAGACTTGG 240
ACAACATGTC CCAACATTAC ATCCCACTTC CGAAGAACTG ACCATTGCTG GCATGACGTT 300
TACAACTTTT GATCTGGGTG GACATGTTCA AGCTCGAAGA GTGTGGAAAA ACTACCTTCC 360
TGCTATCAAT GGCATTGTAT TTCTGGTGGA TTGTGCAGAC CACGAAAGGC TGTTAGAGTC 420
AAAAGAAGAA CTTGATTCAC TAATGACAGA TCAAACCATT GCTAATGTGC CTATACTGAT 480
TCTTGGGAAT AAGATCGACA GACCTGAAGC CATCAGTGAA GAGAGGTTGC GAGAGATGTT 540
TGGTTTATAT GGTCAGACAA CAGGAAAGGG GAGTATATCT CTGAAAGAAC TGAATGCCCG 600
ACCCTTAGAA GTTTTCATGT GTACTCTCCT CAAAAGACAA GGTTACGGAG AAGGCTTCCG 660
CTGGATGGCA CAGTACATTC ATTAACACAA ACTCACATTG GTTCCAGGTC TCAACGTTCA 720
GGCTTACTCA GAGATTTGAT TGCTCAACAT GCATAACTTG AATTCAATAG ACTTTTGCTG 780
GTTATAAAAC AGATGTTTTT TAGATTATTA ATATTAAATC AACTTAATTT GAATGAGAAT 840
TGAAAACTGA TTCAAGTAAG TTTGAGTATC ACAATCTTAG CTTTCTAATT CCATAAAAGT 900
ACTTGGTTTT TACAGTTTAT AATCTGACAT CACCCCAGCG CCATTTCTAA AGAGCAACTT 960
TCCAGCAGTA CATTTGAAGC ACTTTTTAAC AACATGAAAC TATAAACCAT ATTTAAAAGC 1020
TCATCATGTT AAATTTTTTA TCTACTTTTC TGGAACTAGT TTTTAAATTT TAGATTATAT 1080
GTGCACCTAT CKTAAGTGTA CAGTTAATAA TTAGCTTATT CAATGATTGC ATCATCCCTT 1140
ACAGTTTTCA ATAACTTTTT TTCTTATGCA AACGTCATGC AATAAAACAA ACTCTAATGT 1200
TTGGCAAAAA AAAAAAAAAA AAANTCGA 1228
(2) INFORMATION FOR SEQ ID NO: 280:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1327 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 280:
TCTCGGGTCT CGGGACAGGT GAGCACCCTG ATGAAGGCCA CGGTCCTCAT GCGGCACCTG 60
GGCGGGTGCA GGAGATCGTG GGCGCCCTCC GCAAGGGCGS CGGAGACCGG TTACAGGTGA 120
TTTCTGATTT TRACATGACC TTGAGCAGGT TTGCATATAA TGGAAAGCGA TGCCCTTCTT 180
CTTACAATAT TCTGGATAAT AGCAAGATCA TCAGTGAGGA GTGTCGGAAA GAGCTCACAG 240
CGCTCCTTCA CCACTATTAC CCAATTGAGA TCGACCCACA CCGGACCGTC AAGGAGAAGC 300
TACCTCATAT GGTGGAATGG TGGACCAAAG CGCACAATCT CCTATGTCAG CAGAAGATTC 360
AGAAGTTTCA GATAGCCCAG GTGGTTAGAG AGTCCAATGC AATGCTCAGG GAGGGATATA 420
AGACCTTCTT CAACACACTC TACCATAACA ACATTCCCCT TTTCATCTTT TCTGCGGGCA 480
TTGGTGATAT CCTGGAAGAA ATTATCCGAC AGATGAAAGT GTTCCACCCC AACATCCACA 540
TCGTGTCTAA CTACATGGAT TTTAATGAAG ATGGTTTTCT CCAGGGATTT AAGGGCCAGC 600
TGATACACAC ATACAACAAG AACAGCTCTG TGTGTGAGAA CTGTGGTTAC TTCCAGCAAC 660
TTGAGGGCAA AACCAATGTC ATCCTGCTCG GAGACTCTAT CGGGGACCTC ACCATGGCCG 720
ATGGGGTTCC TGGTGTGCAG AACATTCTCA AAATTGGCTT CCTGAATGAC AAGGTGGAGG 780
AGCGGCGCGA NCGCTAACAT GGACTCCTAT GACATCGTGC TGGAGAAGGA CGAGACTCTG 840
GATGTCGTCA ACGGGCTACT GCAGCACATC CTGTGCCNAG GCGGTCCAGC TGGAGATGCA 900
AGGCCCCTCA AGGCGCAGGC TCCNAAGKCC SCTGCAGGCC GTGGTGAGGA GGGGCGCCTC 960
CCCAGAGTCT GCTCCCCCGT GAACACAGAG CAGAGCCAGG GTGGCCAGCA GTCGCTGGGT 1020
CCTTCCGCGC CCCTCCGTCC TCCTTTCCCT GAGCACCTTC ATCACCAGAG GCTTGAAGGA 1080
ACCCCGCCAT GTGGCAGGGC ACAGGCACTG TTCCTGCTGA ACCTTGGACC ACAGCATGTC 1140
AGTGCTCTAG GGATTGTCTA CTCCAGGGAT TTTCTTCAAA ATTTTTAAAC ATGGGAAGTT 1200
CAAACAAATA TAATGTGTGA AACAGATCAA AATTTTTAAA ATGAAAAAAA AGCTGCTCTG 1260
ATTCAGGGGA TGTGGGTCGG GGTAGAACCT GGACCTCTTG GCCTGGGGGC ACATGGGATG 1320
CTTCTAG 1327
(2) INFORMATION FOR SEQ ID NO: 281:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 799 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 281: TCACCCTGCC TACAGCGTGG AGCTCAGATC ACTGCGCCCT CCACGGTCAC TGTGAGCAGG 60
TGGTATTCAC AGCCTGCATG ACCCTCACGG CCAGCCCTGG GGTGTTCCCC GTCACTGTGT 120
GGCTTTGGCT GAAGCCTAAT TCCACAGCTC CTTGTTTTTT GAGAGAGACT GAGAGAACCA 180
TAATCCTTGC CTGCTGAACC CAGCCTCGGC CTGGATGCTC TGTGAATACA TTATCTTGCG 240
ATGTTGGGTT ATTCCAGCCA AAGACATTTC AAGTGCCTGT AACTGATTTC TACATATTTA 300 TAAAAATCTA TTCAGAAATT GGTCCAATAA TGCACGTGCT TTGCCCTGGG TACAGCCAGA 360
GCCCTTCAAC CCCACCTTGG ACTTGAGGAC CTACCTGATG GGACGTTTCC ACGTGTCTCT 420
AGAGAAGGAT TCCTGGATCT AGCTGGTCAC GACGATGTTT TCACCAAGGT CACAGGAGCA 480
TTGCGTCGCT GATGGGGTTG AAGTTTGGTT TGGTTCTTGT TTCAGCCCAA TATGTAGAGA 540
ACATTTGAAA CAGTCTCCAC CTTTGATACG GTATTGCATT TCCAAAGCCA CCAATCCATT 600 TTGTGGATTT TATGTGTCTG TGGCTTAATA ATCATAGTAA CAACAATAAT ACCTTTTTCT 660
CCATTTTGCT TGCAGGAAAC ATACCTTAAG TTTTTTTTGT TTTGTTTTTC TTTTTTTGTT 720
TTTTGTTTTC CTTTATGAAG AAAAAATAAA ATAGTCACAT TTTTAATACY AAAAAATGGA 780
CAAAAAAAGT CGAGGGGGG 799
(2) INFORMATION FOR SEQ ID NO: 282:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2196 base pairs (B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 282:
AAAGACTCTA ACATCCATGA GCTTGAACAT GAGCAAGAGC CTACTTCTCC CKSCCAGATG 60
GCTGAGCCCT TCCGTACCTT CCGAGATGGA TGGGTCTCCT ACTACAACCA GCCTGTGTTT 120 CTGGCTGGCA TGGGTCTTGC TTTCCTTTAT ATGACTGTCC TGGGCTTTGA CTGCATCACC 180
ACAGGGTACG CCTACACTCA GGGACTGAGT GCTTCCATCC TCAGTATTTT GATGGGAGCA 240
TCAGCTATAA CTGGAATAAT GGGAACTGTA GCTTTTACTT GGCTACGTCG AAAATGTGGT 300
TTGGTTCGGA CAGGTCTGAT CTCAGGATTC GCACAGCTTT CCTGTTTGAT CTTGTGTGTG 360
ATCTCTGTAT TCATGCCTGG AAGCCCCCTG GACTTGTCCG TTTCTCCTTT TGAAGATATC 420
CGATCAAGGT TCATTCAAGG AGAGTCAATT ACACCTACCA AGATACCTGA AATTACAACT 480
GAAATATACA TCTCTAATGG GTCTAATTCT GCTAATATTC TCCCGGAGAC AAGTCCTGAA 540
TCTGTGCCCA TAATCTCTGT CAGTCTGCTG TTTGCAGGCG TCATTGCTGC TAGAATGGGT 600
CTTTGGTCCT TTGATTTAAC TGTGACACAG TTGCTGCAAG AAAATGTAAT TGAATCTGAA 660
AGAGGCATTA TAAATGGTGT ACAGAACTCC ATGAACTATC TTCTTGATCT TCTGCATTTC 720
ATCATGGTCA TCCTGGCTCC AAATCCTGAA GCTTTTGGCT TGCTCGTATT GATTTCAGTC 780
TCCTTTGTGG CAATGGGCCA CATTATGTAT TTCCGATTTG CCCAAAATAC TCTCGGAAAC 840
AAGCTCTTTG CTTGCGGTCC TGATGCAAAA GAAGTTAGGA AGGAAAATCA AGCAAATACA 900
TCTGTTGTTT GAGACAGTTT AACTGTTGCT ATCCTGTTAC TAGATTATAT AGAGCACATG 960
TGCTTATTTT GTACTGCAGA ATTCCAATAA ATGGCTGGGT GTTTTCCTCT GTTTTTACCA 1020
CAGCTGTGCC TTGAGAACTA AAAGCTGTTT AGGAAACCTA AGTCAGCAGA AATTAACTGA 1080
TTAATTTCCC TTATGTTGAG GCATGGAAAA AAAATTGGAA AAGAAAAACT CAGTTTAAAT 1140
ACGGAGACTA TAATGATAAC ACTGAATTCC CCTATTTCTC ATGAGTAGAT ACAATCTTAC 1200
GTAAAAGAGT GGTTAGTCAC GTGAATTCAG TTATCATTTG ACAGATTCTT ATCTGTACTA 1260
GAATTCAGAT ATGTCAGTTT TCTGCAAAAC TCACTCTTGT TCAAGACTAG CTAATTTATT 1320
TTTTTGCATC TTAGTTATTT TTAAAAACAA ATTCTTCAAG TATGAAGACT AAATTTTGAT 1380
AACTAATATT ATCCTTATTG ATCCTATTGA TCTTAAGGTA TTTACATGTA TGTGGAAAAA 1440
CAAAACACTT AACTAGAATT CTCTAATAAG GTTTATGGTT TAGCTTAAAG AGCACCTTTG 1500
TATTTTTATT ATCAGATGGG GCAACATATT GTATGAAGCA TATGTAGCAC TTCACAGCAT 1560
GGTTATCATG TAAGCTGCAG GTAGAAGCAA AGCTGTAAAG TAGATTTATC ACACAATGAC 1620
TGCATACAGA CTTCAAATAT GTCAATAGTT TGGTCATAGA ACCTAGAAGC CAAAAGCCAC 1680
ACAGAAGGGC AAGAATCCCA ATTTAACTCA TGTTATCATC ATTAGTGATC TGTGTTGTAG 1740
AACATGAGGG TGTAAGCCTT CAGCCTGGCA AGTTACATGT AGAAAGCCCA CACTTGTGAA 1800
GGTTTTGTTT TACAAATCAC TTGATTTAAC ACACTCAGGT AGAATATTTT TATTTTTACT 1860
GTTTTATACC CAGAAGTTAT TTCTACATTC TTCTACAGCA AGAATATTCA TAAAAGTATC 1920
CCTTTCAAAT GCCTTTGAGA AGAATAGAAG AAAAAAAGTT TGTATATATT TTAAAAAATT 1980
GTTTTAAAAG TCAGTTTGCA ACATGTCTCT ACCAAGATGG TACTTTGCCT TAACCGTTTA 2040
TATGCACTTT CATGGAGACT GCAATACGTT GCTATGAGCA CTTTCTTTAT CCTTGGAGTT 2100
TAATCCTTTG CTTCATCTTT CTACAGTATG ACATAATCAT TTGCTATGTT GTAAAATCTT 2160 TGTAAAAAAT TTCTATATAA AATATTTGAA ACTTAA 2196
(2) INFORMATION FOR SEQ ID NO: 283:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1185 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 283:
GCAGTTAAGG CTTCTGATAA GGAAAGAGAG TCTGAACAGA GCACACACAT CTGGAGCTCC 60
AGGAGTGGGG GATCCAGGAT CAGATTCCAT CTTGAATTTC TGCTAAAATA CTTTGTACTC 120
ATAATCGATC TCAACAAAGA TCTGTATTTC ATCTGTGGCT CCATCTTCCC TCTGGGTCAA 180
GTAGATGTTA AGCTCGACCT TGGCACGCCT CTTAACATGA AGAGATCTAG CTAGACAGAC 240
AGACTCCCCC ATTTATGGGA ACAAGAATTC AATTTATTCT CTATTTATAA AACATTTTTT 300
TAAAGTGCCT TGGGTATAAA AATCTAAATG TCTGCGGTCT GATCAGTCAG GAGCACGTAA 360
CTATCACTCT TCGCATCCTT TCGTCACTGG GAGATCCTTT GGGGGCTGGG AGGTCCTTCT 420
GTCCCAGGCT AAAGGAAAAG CTTCACAAGG GTAAGAGCCA CAGAACCCTC GGCAAGAAAG 480
GCCGGTCAGG GAGAATGAAT GGTACAGAGA GGAAAGGAAG GAAAGGGGGT GGAACAGAGG 540
TAGAAGGCAA GGAAGGGATC CCGCACTGGA GACCGATGGG GACACTCTAA TTGTGCAAGA 600
GGGAGGATCT TCCTTCTTGA ATGCTGAACA CAGCTAGTCT GAACCTTCCT TGGAAAGTCC 660
AGCTGTTTGC CCATGCATAG GGCCAACTCT CCCTGCAAAG CAGCAAATGT GGCTTCTATC 720
AGGAAGGAAA AGTATCCATC AGTGTGACAA GAGGTCACCT TCGAACTTGC ATGAACTCCT 780
TGCGCAGCCA CAAAGAGTCC TGGTAGAAGT GAGGATCGCC TAGTCTTACG GCTGTCCGTT 840
TATAGAAGTA GCAGTACAAC ACTGCTGCTA GTCTCTGGAA TACAAACAGC ATTTGAAGTC 900
CATCTGTCCA TATGAAGCTG TTGGAGTTTT TCCAGCGTAA GTTCATGACC CAGACATGAA 960
GGGAGATGCT GAGGGCAAAG TACACAGCTG TCAGGATGAT GGTCCCTTTG AACTTATGGA 1020
ATAGGAGGTT GACCAGGCCA GCCTGGAAGA CGAAGGTGTT GAAGAACATG AGGAAAATCA 1080
TGATGATGTT GAAGAGGACT GCAATATCCT GGATGCACTG AGGGAGAGGY TTCTAGTTCC 1140
TTTGAATGAG AGCTGTTTCC CTTGCTCTAA GGCAAGCACC TCCAA 1185
(2) INFORMATION FOR SEQ ID NO: 284:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1634 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 284:
AGGGAAAGGG GAGGGTAGCG GGAGGGTAGC AGGTGAGTTC CTAGGGCTGG AAGGTTTAGC 60
AGCAGCCTGG TGCAGTGCCC TCTCATCAAG ACAAACCCAC GGTCCTMCTG GGTGCCTACC 120
AAGCTTGGTT TGTACAAAAG CAAGGTGGGA GTCTATTTTT GTACATGAGA TACATCACAC 180
TTACCTCTGG GCCAGTATTG TGAAGTGAGT CTGAGTTGTT TACACTGATG CCTTCCCTGC 240
CCACCACAAA TTCTCTACAT AGTCTTCAGA TGATACCACC CCTTTCCCCA GCTCCCAACC 300
AAGAGCTGGT TCTAGGCCTG TGTTATATCT CATATTTAGC STTTTTATAT ATCACCTTTG 360
ATTTCTGTTG TTTGTATTTT AGCACAGTGT ATGCACCTTC ATTTAAATAC ATCTGTGTGC 420
ATACAGATAC GCATATATGT GTGTGCGTAT GCATATATCT CTCATCTGTA GTTTCCAAGA 480
GTTCAGCTGA AGCAGATGGA GTCCTGCAGC CCAGGAGACA CCCTGCATCC CTGCTAATAG 540
TGTTTGCCAC AAGTATTAGT GAGTCTTCCT TATTAATATT TTCATTTCAG AAGACTGAAG 600
CAAAGCTGAT AGTGTTTGCT GTTTCTTTGG CAGCTAAGTG AGGGTCTTGG GATGACTTGC 660
TGTGTTCCTC AAGCTGCACT TTGGGGCCAT CTCTGCAGTA TTAGCCCCCT TTTTGCTTGG 720
TGGTACTCTG TCTGTGCCTG TGTGTGTGTG TGATAGTCAC TCTTGCATGG CTTCCATGTC 780
TGGTTTGTGG CATTTGGGGA TAAGGTGCTG AAGCCAGAGC ATTTGCAGTT TGTTTGAGGC 840
CTCGTTGCCA ATGATAGATC ACTCCTGTTG ACCTGGTATG TCTGCTTGCT TCCTGCTTTT 900
CCTTGCTTTC TCTTGGAAGA GGAAAGGACT CTGGTCAGCC CCAGGCTGAG TGAGATGAGC 960
TCCAGCTGGC TCATGGCCTT CTTAGAGCAG AGAGAGGAGT ATGTCATTTT ACTAAGTTCC 1020
TAAACAAACA TTTATGCAGG CAACACTCCT TGCAGATCCA GAAACTGAGG CACAATAGGG 1080
TTATGACTTG CTCAAGAATA TGTAGCTGCT AGGGGGTAAA TCAAGGCATC ACAATTTCTG 1140
TTCAGCGGGC AGGAATAGGC TGTGAATTGC TAGCACTTTT TTTTTTTAAG CAATTACTTT 1200
TTGACTTGTT CCTCTGAAAG TGCAAGAGGC GTACACCTTT CCCAAATGTA GACTAGAATC 1260
TGCAGGATGC CACCCACTGT ATAGTTCTGC TTTCCCAGAG AGGAAGAACT TTTAGAAACC 1320
AAATGATCTT AATTGTTATT GCCCACCCCT GGCTTTTCCG GGTAGAAAAT TCACAGTAGG 1380
AATGATTGTT AAGAGAGAGT GCTTGGAACC ATGGGTTAAC AGGAAAGGCT ACCTAACTTC 1440
ACATATCTCC AACCAGAGCA GCCACCAAGC ATTACTTAGC AGCAGGAAAA TGATTGTATT 1500
TCAGTTCCTG TGTGTCCAAA ACTCAGGCAC CATGTTCTTT GAAAACATGC CACCTCAAGG 1560
CTGGGCGCGG TGGCTCACAC CTGTAATCCC AGCAYTTTGG GGAGGCCSAG GGCGGGGCGG 1620 KTTCACCGGG GGTC 1634
(2) INFORMATION FOR SEQ ID NO: 285:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1795 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 285: TTCCCCCCAG GTTGGCTTCC TTCGATTCCT TTTCTTGGTA TCAACGTTTG ATTGGAAGAA 60
CAACCCCCTC TTTGTCAACC TCAATAATGA GCTCACTGTG GAGGAGCAGC TCGGGCACAG 120
CTCMCCGTYA TGGTCATTGT TACCCCCCAA GACCGCAAAA ACTCTGTGTG GACACAGGAT 180
GGACCCTCAG CCCAGATCCT GCAGCAGCTT GTGGTCCTGG CAGCTGAAGC CCTGCCCATG 240
TTAGAGAAGC AGCTCATGGA TCCCCGGGGA CCTGGGGACA TCAGGACAGT GTTCCGGCCG 300 CCCTTGGACA TTTACGACGT GCTGATTCGC CTGTYTCCTC GCCATATCCC GCGGCACCGC 360
AGGCTTGTGG ACTCGCCAGY TGCCTCCTTC TGCCGGGGCC TGCTCAGCCA GCCGGGGCCC 420
TCATCCCTGA TGCCCGTGCT GGGTNATGAT CCTNCTCAGC TCTATCTGAC GCAGCTCAGG 480
GAGGCCTTTG GGGATCTGGC CCTTTTCTTC TATGACCAGC ATGGTGGAGA GGTGATTGGT 540
GTCCTCTGGA AGCCCACCAG CTTCCAGCCG CAGCCCTTCA AGGCCTCCAG CACAAAGGGG 600 CGCATGGTGA TGTCTCGAGG TGGGGAGCTA GTAATGGTGC CCAATGTTGA AGCAATCCTG 660
GAGGACTTTG CTGTGCTGGG TGAAGGCCTG GTGCAGACTG TGGAGGCCCG AAGTGAGAGG 720
TGGACTGTGT GATCCCAGCT CTGGAGCAAG CTGTAGACGG ACAGCAGGAC ATTGGACCTC 780
TAGAGCAAGA TCTCAGTAGG ATCACCTCCA CCCTCCTTGG ACATGAATCC TCCATGGAGG 840
GCCTGCTGGC TCAACATGCT GAATCATCTC CAACAAAACC CAGCCCCAAC TTTCTCTCTC 900 ATGCTCCAGC ATTGGGGCAG GGGCATGGTG GCCCATGTAG TCTCCTGGGC CTCACCATCC 960
CAGAAGAGGA GTGGGAGCCA GCTCAGAGAA GGAACTGAAC CCAGGAGATC CATCCACCTA 1020
TTAGCCCTGG GCCTGGACCT CCCTGCGATT TCCCACTCCT TTCTTAGTCT TCTTCCAGAA 1080
ACAGAGAAGG GGATGTGTGC CTGGGAGAGG CTCTGTCTCC TTCCTCCTGC CAGGACCTGT 1140
GCCTAGACTT AGCATGCCCT TCACTGCAGT GTCAGGCCTT TAGATGGGAC CCAGCGAAAA 1200 TGTGGCCCTT CTGAGTCACA TCACCGACAC TGAGCAGTGG AAAGGGGCTA TATGTGTATG 1260
AATAGACCAC ATTGAAGGAG CACAATCCCC TCCTGTGTTG ATCCCACTTC CCAGGGTGGA 1320 GACAGTGGAA AAGAACCGAG GACAGGAAAG GATTGGGTAG GTCAAGGGGT CAGGGGACTG 1380 GTAGTCACCC AATCTTGGAG AGGTGCAAAA AGCACTGGGG GCTACCCGTT AGCTGCATCT 1440 GCCCTGGCTG TTTGCCCGTT CATGTCACAA ACTGCCACTA CTATGTACCT GCAGTGGGGT 1500 TGCAGAGATG GGGGAGACTC AAGTCTTACT CCCCAGGAGC TCCCAGGGCC CAAGGAGGAG 1560 AATGCTGCCT CCTTTCAGTC TGGTCTACAC CCACTTTCTG GTAGCCTCTC TGCTTCCTGT 1620 AATTCTGGCT GTTTTTCCAG ACTCAGCTCA AATAGTGCCC CTCCTTAAGC CCATCCCTCG 1680 CCCCCAGCCT GAGGTGATCT TTCCCTCCTC TGAACTATTA GAGCAGTTAC TGTCTGTTCA 1740 GTTCGTTTGG CAGGCACACA CAGTGGCATA AATTCTATTG TTTTGAACTC TGATT 1795
(2) INFORMATION FOR SEQ ID NO: 286:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 858 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 286:
TCTGCTTTCG GTGCTGCGTG TACTGCTGGG CGGCTTCTTC GCGCTCGTGG GGTTGGCCAA 60
GCTCTCGGAG GAGATCTCGG CTCCAGTTTC GGAGCGGATG AATGCCCTGT TCGTGCAGTT 120
TGCTGAGGTG TTCCCGCTGA AGGTATTTCG CTACCAGCCA GATCCCCTGA ACTACCAAAT 180
AGCTGTGGGC TTTCTGGAAC TGCTGGCTGG GTTGCTGCTG GTCATGGGCC CACCGATGCT 240
GCAAGAGATC AGTAACTTGT TCTTGATTCT GCTCATGATG GGGGCTATCT TCACCTTGGC 300
AGCTCTGAAA GAGTCACTAA GCACCTGTAT CCCAGCCATT GTCTGCCTGG GGTTCCTGCT 360
GCTGCTGAAT GTCGGCCAGC TCTTAGCCCA GACTAAGAAG GTGGTCAGAC CCACTAGGAA 420
GAAGACTCTA AGTACATTCA AGGAATCCTG GAAGTAGAGC ATCTCTGTCT CTTTATGCCA 480
TGCAGCTGTC ACAGCAGGAA CATGGTAGAA CACAGAGTCT ATCATCTTGT TACCAGTATA 540
ATATCCAGGG TCAGCCAGTG TTGAAAGAGA CATTTTGTCT ACCTGGCACT GCTTTCTCTT 600
TTTAGCTTTA CTACTCTTTT GTGAGGAGTA CATGTTATGC ATATTAACAT TCCTCATGTC 660
ATATCAAAAT ACAAAATAAG CAGAAAAGAA ATTTAAATCA ACCAAAATTC TGATCCCCCA 720
AATAACCACT TTTAATCCCT TGGTGTAAGT ATACCTCTCA ACTTTTTTCT GTGCCTTTAA 780
ACAGATATAT ATTTTTTTTT AATGAAAATA AAACCATATA TCCTATTTTA TTTCCTCCTT 840
TTAAAACCTT ATAAACTA 858
(2) INFORMATION FOR SEQ ID NO: 287:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 915 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 287: GAATTCGCCA CGAGCGCGGC CATGGCGCTC CTGCTTTCGG TGCTGCGTGT ACTGCTGGGC 60 GGCTTCTTCG CGCTCGTGGG GTTGGCCAAG CTCTCGGAGG AGATCTCGGC TCCAGTTTCG 120 GAGCGGATGA ATGCCCTGTT CGTGCAGTTT GCTGAGGTGT TCCCGCTGAA GGTATTTGGC 180 TACCAGCCAG ATCCCCTGAA CTACCAAATA GCTGTGGGCT TTCTGGAACT GCTGGCTGGG 240 TTGCTGCTGG TCATGGGCCC ACCGATGCTG CAAGAGATCA GTAACTTGTT CTTGATTCTG 300 CTCATGATGG GGGCTATCTT CACCTTGGCA GCTCTGAAAG AGTCACTAAG CACCTGTATC 360 CCAGCCATTG TCTGCCTGGG GTTCCTGCTG CTGCTGAATG TCGGCCAGCT CTTAGCCCAG 420 ACTAAGAAGG TGGTCAGACC CACTAGGAAG AAGACTCTAA GTACATTCAA GGAATCCTGG 480 AAGTAGAGCA TCTCTGTCTC TTTATGCCAT GCAGCTGTCA CAGCAGGAAC ATGGTAGAAC 540 ACAGAGTCTA TCATCTTGTT ACCAGTATAA TATCCAGGGT CAGCCAGTGT TGAAAGAGAC 600 ATTTTGTCTA CCTGGCACTG CTTTCTCTTT TTAGCTTTAC TACTCTTTTG TGAGGAGTAC 660 ATGTTATGCA TATTAACATT CCTCATGTCA TATGAAAATA CAAAATAAGC AGAAAAGAAA 720 TTTAAATCAA CCAAAATTCT GATGCCCCAA ATAACCACTT TTAATGCCTT GGTGTAAGTA 780 TACCTCTGAA CTTTTTTCTG TGCCTTTAAA CAGATATATA TTTTTTTTWA ATGAAAATAA 840 AACCATATAT CCTATTTTAT TTCCTCCTTT TAAAACCTTA TAAACTATAA MAAAAAAAAA 900 AAAAAAAAAA CTCGA 915
(2) INFORMATION FOR SEQ ID NO: 288:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1517 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 2E
CCTTGTGGCA ACTAGTCGGT CCCCCGGGCT GCAGNAATTC GGGCAGTGGT TCTGNGTCTG 60 AAGATACTCT GAGTTCCTCT GAGAGATCCA AAGGCTCCGG GAGCAGACCC CCAACCCCCA 120 AAAGCAGCCC TCAGAAGACC AGGAAGAGCC CTCAGGTGAC CAGGGGTAGC CCTCAGAAGA 180 CCAGCTGTAG CCCTCAGAAG ACCAGGCAGA GCCCTCAGAC GCTGAAGCGG AGCCGAGTGA 240 CCACCTCACT TGAAGCTTTG CCCACAGGAC AGTGCTGACA GACAAGAGTG GGCGACAGTC 300 GAAGCTGAAG TCCTTCCAGA CCAGGGACAA CCAGGGCATT CTCTATGAAG CTCCACCCAC 360 CTCCACCCTC ACCTGTGACT CAGGACCACA GAAGCAAAAG TTCTCACTCA AACTGGATGC 420 CAAGGATGGG CGCTTGTTCA ATGAGCAGAA CTTCTTCCAG CGGGCCGCCA AGCCTCTGCA 480 AGTCAACAAG TGGAAGAAGC TGTACTCGAC CCCACTGCTC GCCATCCCTA CCTCCATGGG 540 TTTCGGTGTT CACCAGGACA AATAGAGCTT CTTGGTGTTA CCCAGCCTGG GGAGGAGCCT 600 TCAGTCGGCC CTGGATGTCA GCCCAAAGCA TGTGCTGTGC AGAGAGGTCT GTGCTGCAGG 660 TGGCCTGCCG GCTGCTGGAT GCCCTGGAGT TCCTCCATGA GAATGAGTAT GTTCATGGAA 720 ATGTGACAGC TGAAAATATC TTTGTGGATC CAGAGGACCA GAGTCAGGTG ACTTTGGCAG 780 GCTATGGCTT CGCNTTCCGC TATTGCCCAA GTGGCAAACA CCTGGCCTAC CTCGAAGGCA 840 GCAGGAGCCY TCACGAGGGG GACCTTGAGT TTCATTAGCA TGGACCTGCA CAAGGGATGC 900 GGGCCCTCCC GCCGCRGYGA CCTCCAGAGC CTGGGYTAMT GCATCCTGAA GTCGYTCTAM 960 GGGTTTCTGC CATGGACAAA TTGCCTTCCA AMAMTGAGGA CATCATGAAG CAAAAACAGA 1020 AGTTGCCTTG GGATTCATTT TAATGTAAGC TKGACTTTGT CATGCCAGAA ACAAGGCTCG 1080 GTCACCGTCA GCAGTTTGCA GTTTTCCACC TCCWCCCAGT TCCTCCCTCT GGTTGACCCA 1140 GATATCTCCG TTATGCAGCC GCCTCCGGGG GACCACCTCC CTCCCTTTGA GTCAGCCACA 1200 GACAGCCTAC TTCACGGCCC CGCTGGCCCC CACATTCCAC TGAACTCTGC GGATGCCACA 1260 GTGACCCCCT CTCAGGCACA GCATGACCTC CTGAAGTCGA GCCTGCTTGC TTTCAACCTA 1320 CCAGTTAAAA TCTCCTCAAA ATGTTTGGAT ACCGCCCATT GCCCCCTCAC AGCCACGAGC 1380 TCCCTGACCA GTGTGCGTGT GTGTGTGTGT GTGTGTCTGT GTGTGTGCTT GGGACGGGTG 1440 GGGAGGTCAC CTTTGGGTGT GCGGTGTGCC CCCAGGACCT GTAAGTAATA AAATCTTTAT 1500 TTCCAAAAAA AAAAAAA 1517
(2) INFORMATION FOR SEQ ID NO: 289:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 3865 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 289:
TGGAGGGGGG GAGCTTCCTT GAGCAGTGGG CCCAGGCCTG GCCCTCCACA CTTCATTCTC 60
TGACCTTTCT CTCTCCTCAT TTCGGTGCAT GTCCTTTCTG CAGCTGCCTT TCAGCACAGG 120
TGGTTCCACT GGGGGCAGCT AACGCTGAGT GACAAGGATG GGAAGCCACA GGTGCATTTT 180
ACTCAAGTCT TCTCTAGTCA ATGAGGGGCA CCCAGTGCTT CTAGGGCAGC CTGGGTGGTG 240
GTCCCCTAGG TATCAGCCTC TCTTACTGTA CTCTCCGGGA ATGTTAACCT TTCTATTTTC 300
AGCCTCTCCC ACCTGTCTAG GCAAGCTGGC TTCCCCATTG GCCCCTGTGG GTCCACAGCA 360
GCGTGGCTSC CCCCCAGGGC CACCGCTTCT TTCTTGATCC TCTTTCCTTA ACAGTGACTT 420
GGGCTTCAGT CTGGCAAGGA ACCTTGCTTT TAGCTTCACC ACCAAGGAGA GAGGTTGACA 480
TGACCTCCCC GCCCCCTCAC CAAGGCTGGG AACAGAGGGG ATGTGGTGAG AGCCAGGTTC 540
CTCTGGCCCT CTCCAGGGTG TTTTCCACTA GTCACTACTG TCTTCTCCTT GTAGCTAATC 600
AATCAATATT CTTCCCTTGC CTGTGGGCAG TNGGAGAGTG CTGCTGGGTG TACGCTGCAC 660
CTGCCCACTG AGTTGGGGAA AGAGGATAAT CAGTGAGCAC TCTTCTGCTC AGAGCTCCTG 720
ATCTACCCCA CCCCCTAGGA TCCAGGACTG GGTCAAAGCT GCATGAAACC AGGCCCTGGC 780
AGCAACCCTG GGAATGGCTG GAGGTGGGAG AGAACCTGAC TTCTCTTTCC CTCTCCCTCC 840
TCCAACATTA CTGGAACTCT ATCCTGTTAG GATCTTCTGA GCTTGTTTCC CTGCTGGGTG 900
GGACAGAGGA CAAAGGAGAA GGGAGGGTCT AGAAGAGGCA GCCCTTCTTT GTCCTCTGGG 960
GTAAATGAGC TTGACCTAGA GTAAATGGAG AGACCAAAAG CCTCTGATTT TTAATTTCCA 1020
TAAAATGTTA GAAGTATATA TATACATATA TATATTTCTT TAAATTTTTG AGTCTTTGAT 1080
ATGTCTAAAA ATCCATTCCC TCTGCCCTGA AGCCTGAGTG AGACACATGA AGAAAACTGT 1140
GTTTCATTTA AAGATGTTAA TTAAATGATT GAAACTTGGC TGTGGCTACT GCTTCTTAAT 1200
GTTGGGGGGA CAGGGCAGTG GTCTGGGCCC ACATTTAGAA GGGAAAATGT TTTGCCTGCT 1260
GCACACATTG GACCCAAGTA TGGGCCTCTT CTGCCTAGTA CTGCCAAAGG GACTGTTAAG 1320
GTGTCTTGTC CATCTTCTAC CCCCCACCCC CCATTACGGG TAAAGGRAAC CCCAGACTAG 1380
GTGAGGGGCC AGCAGCTGCC TCACATTGTG TTCTCTCCTG AGATGGTCCA GCTCACATCC 1440
AGACACCTTG TTCAGACATT TTATTTGAAT TTATCACAGT GATGGGGATT TGACTCAGAT 1500
GCCTTATGGA GAAGTACCCC ACCCTCTATC AAGACAGAAT CACTCTCTCC CATTCATTCT 1560
GCCTGATGCT AACAACACGC AGCTGATTTA GGGAGTGTCC CAGCCTAGCT GGATCAAGGG 1620
AAATTCCAGG AGCCCTGGGG CAGGCCCTGG NCCCCAGTGC CAAGCCTCAG AGTAAGCAGA 1680
CATTGGGAAA GTTGCCAACC ACTTGGTAGA CCACTAGGTT CTCTGTTTTC CCTTCCCTTT 1740
CCTTTTCAAA TCCCACAGTT TCCTGTTGGG GAGAAGCTGT AATTAGCCTA GTCCAGGTAC 1800
CAGATCCCAG CTAGGGGCGC AGCTGNCTTG GATAACTCCA AGAAAACCTC GGCACCAGTA 1860
TTTTTCCAAT TATAAGGACT GTGGCATAAA TTTTTAAATG AGTTATATTG AAACCAGATT 1920
TCTCCAGCTG CCAAGGGAAG AAGGTAGGGC TGGACTCCCT GCTGTGGCCC AGCCCTTGTT 1980
AGGGGTTGGT CTCTCACTGC AGCCAGACAG GATGATCCTG GGTTCTGGGG AGGGTAAGCT 2040
GCCCCTTGCC GAGTTCTGCA CCGAATAAAG AGTCCAAACC CGCTGCTTCC GTGTCCTCAG 2100
AGATGGGTAA ATGGGTGATG GATGGAGCAG ACTGAAGAGA CAGCAGATGA CTCAGTGGTG 2160
GAAGAAGGGG GGAAGATGCT GGGCTGGCTA GCTAATGTTC CCCCCTTTCA GCGATTTACA 2220
GGAAATGGAG CCCAGCTTGG TCATGAAGTT GGTTTGCTTC CACTGTGCGA TGCACTCCTC 2280
AGAAATTTTG AAGTCAGCCT GCAACTTCTC GAAGACTTTC TTCTTGGGCT TGAGCTCCTC 2340
ATCTGGTTGG CCCTTTTCAT AGCCCTTCAC AAACACGTGC TCACCAGGAG CAGAGCCTGC 2400
CGGAGGGTCC AGAGGTTCAA CTGGCGGTTT ATCCCTTCTA TAGAAGCACA CAGAAGCATG 2460
CCTTGGGACT CGACTCCTCT CATCTTCTGG GGTTTCAGGT TGCACAGCAC CACTACCAGC 2520
CTGTCCTGCA GTTCCTCCTT GGGCACGAAC TGTACCAGGC CGCTCACCAC AGTCCGTGGT 2580
TCAGCTTCCC CCACGTCAAT CTTCTCTACA TACAGGCTGT CTGCATCTGG GTGCTTCTCC 2640
ACAGTGATGA TTTTCCCCAC ACGGATATCC AGCCGGGATC GGATGACCTC CTCTGGTTCT 2700
GAATTCTTGG CAGGCCTTTG GCCATTGGCT TCTGCTTTGA GGGATCTGGG TAGGCAGCGC 2760
TGGCCAGTTT TTTCAGGGCA GGGGTATTAA ACTTTTCCCG GATTGGATCC AGCAACTTGT 2820
TCAGTGCGAC TTCAACAGAA TTCTTCAGGT CTCCAGGATG TACAACCTCA GCAGCAAAGT 2880
CCTTTTCCAG GTCCACGTAA GCTGTGTAGG TTTTGTTTCC ACCCCATTTC TCATCTCGTA 2940
GGATCACAAA CTCGGACTTA AGGGGAAAAA GGACATGCTT GATGAAGCAC AGAACCCCAT 3000
TGTTCTCCAC ATTTCCTGGC TCACAGAAGG CCTTCTTCAG TTTTTTCTTC ACATCCTCCT 3060
TCCGATCAAG GAGATCAATC TTGGACTCCT CTTCTGAAGA GCTCATTTTG CTCCCTCTTA 3120
ATCCTGGAAC CATAGGATTC ATCAGATGGA CCCGTTTTGA ATAGCCAAGT GCAGGGAGGT 3180
ACTTCTCTCC AAAGGTGAAA ATCTTTCTCT GATCAATGCC TCCAAATTGG GCATCTACTT 3240
TTAAATACTC TTCATCCAAA GCCTGCAGTC CGGGGTATAA GAGGCCACTC AGCAAAGGGT 3300
GCTCCACCTG CTTTACCACC TCAGCTCCAG CCTTCTTCGA ATCGTGCTGT GTGACCACGG 3360
AGGAGAGTCT GTACACATCT AGTCTCTACT CTTTCCTGAG CTGGTAATCA GTGCCTTTGA 3420
TGAACTTGAG CTTCTCCAAG GGCACACCAA TGCTCTCCAG CATTGCTTTG ATCACATTCT 3480
CATAGTAACT GACTCGGAGT TCTAGAAGTT CCCATGGGGC TTTCATGTTA TCCAGGTATG 3540 CGTGGAGGTC CGCAAACAGA ATTCTTACCT CACACCCTCC CTTTAAGAAG TCTGCAATCT 3600 TTGACATGGG CACAAAGTAA GCCACATCTG GTTTCCCCGT GGTTGCCGTT CCCCAGTAAA 3660 TTTTAAGTTC CCGCTCCTTC AGTATCTCCT TCAGCTTCTC TTCCCCCAGA ACCTCCTGCA 3720 GGTTCCGGGT GATAAGGTGC AGTTTCTCTT CAGGGCTGGG AGCGTCCCCC ATCCTCCGCT 3780 ACCCCTGCTT CCCCCGCTCA GCCCGGCACC AGAGCCCCTT CCTGGGTCAC CGTCGCCGCC 3840 GCGTGCCGGG AACTGTCACG CGAGT 3865
(2) INFORMATION FOR SEQ ID NO: 290:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1910 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 290:
AGGGAGAGGA GGAGAGGGGG TCTGCGCGCG GCCGCTACCC AGAAGCCAGC GGACGGCAGC 60
ACGGAGTGGG CTGTCCCCGA GCCCAGCCCC GAGCGAGCCC CCCCCCCGCC CCCGMAGGAC 120
GCGCCTYCCA GCCACCCCGA CTCCTAGGAG GAGGGGAGGC GGGAAAGCAG CTCAAGCCTC 180
ACCCACCGCC CTGCCCCCAG CCCCGCCACT CCCAGGCTCC TCGGGACTCG GCGGGTCCTC 240
CTGGGAGTCT CGGAGGGGAC CGNCTGTGCA GACGCCATGG AGTTGGTGCT GGTCTTCCTC 300
TGCAGCCTGC TGGCCCCCAT GGTCCTGGCC AGTGCAGCTG AAAAGGAGAA GGAAATGGAC 360
CCTTTTCATT ATGATTACCA GACCCTGAGG ATTGGGGGAC TGGTGTTCGC TGTGGTCCTC 420
TTCTCGGTTG GGATCCTCCT TATCCTAAGT CGCAGGTGCA AGTGCAGTTT CAATCAGAAG 480
CCCCGGGCCC CAGGAGATCA GGAAGCCCAG CTCGAGAACC TCATCACCGC CAATGCAACA 540
GAGCCCCAGA AAGCAGAGAA CTGAAGTGCA GCCATCAGGT GGAAGCCTCT GGAACCTCAG 600
GCGGCTCCTT GAACCTTTGG ATGCAAATGT CGATCCTTAA GAAAACCGGC CACTTCAGCA 660
ACAGCCCTTT CCCCAGGAGA AGCCAAGAAC TTGTGTGTCC CCCACCCTAT CCCCTCTAAC 720
ACCATTCCTC CACCTGATGA TGCAACTAAC ACTTGCCTCC CCACTGCAGC CTGCGGTCCT 780
GCCCACCTCC CGTGATGTGT GTGTGTGTGT GTGTGTGTGT GACTGTGTGT GTTTGCTAAC 840
TCTCGTCTTT GTGGCTACTT GTTTGTGGAT GGTATTGTGT TTGTTAGTGA ACTGTGGACT 900
CGCTTTCCCA GGCAGGGGCT GAGCCACATC GCCATCTGCT CCTCCCTGCC CCCGTGGCCC 960
TCCATCACCT TCTGCTCCTA GGAGGCTGCT TGTTGCCCGA GACCAGCCCC CTCCCCTGAT 1020
TTAGGGATGC GTAGGGTAAG AGCACGGGCA GTGGTCTTCA GTCGTCTTGG GACCTGGGAA 1080
GGTTTGCAGC ACTTTGTCAT CATTCTTCAT GGACTCCTTT CACTCCTTTA ACAAAAACCT 1140
TGCTTCCTTA TCCCACCTGA TCCCAGTCTC AAGGTCTCTT AGCAACTGGA GATACAAAGC 1200
AAGGAGCTGG TCAGCCCAGC GTTGACGTCA GGCAGGCTAT GCCCTTCCGT GGTTAATTTC 1260
TTCCCAGGGG CTTCCACGAG GAGTCCCCAT CTGCCCCGCC CCTTCACAGA GCGCCCGGGG 1320
ATTCCAGGCC CAGGGCTTCT ACTCTGCCCC TGGGGAATGT GTCCCCTGCA TATCTTCTCA 1380
GCAATAACTC CATGGGCTCT GCGACCCTAC CCCTTCCAAC CTTCCCTGCT TCTGAGACTT 1440
CAATCTACAG CCCAGCTCAT CCAGATGCAG ACTACAGTCC CTCCAATTGG GTCTCTGGCA 1500
GGCAATAGTT GAAGGACTCC TGTTCCGTTG GGGCCAGCAC ACCGGGATGG ATGGAGGGAG 1560
AGCAGAGGCC TTTCCTTCTC TGCCTACGTC CCCTTAGATG GGCAGCAGAG GCAACTCCCG 1620
CATCCTTTGC TCTGCCTGTC GGTGGTCAGA GCGGTGAGCG AGGTGGGTTG GAGACTCAGC 1680
AGGCTCCGTG CAGCCCTTGC GAACAGTGAG AGGTTGAAGG TCATAACGAG AGTGGGAACT 1740
CAACCCAGAT CCCGCCCCTC CTGTCCTCTG TGTTCCCGCG GAAACCAACC AAACCGTGCG 1800
CTGTGACCCA TTGCTGTTCT CTCTATCGTG ATCTATCCTC AACAACAACA GAAAAAAGGA 1860
ATAAAATATC CTTTGTTTCM TAAAAAAAAA AAAAAAAAAA AGGGGGGGGG 1910
(2) INFORMATION FOR SEQ ID NO: 291:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 3276 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 291: GCGACCGTCG TTTGAGTCGT CGCTGCCGCT GCCGCTGCCA CTGCCACTGC CACCTCGCGG 60 ATCAGCAGCC AGCGTTGTTC GCCCGACGCC TCGCTCCCGG TGGGAGGAAG CGAGAGGGAA 120 GCCGCTTGCG GGTTTGTCGC CGCTGCTCGC CCACCGCCTG GAAGAGCCGA GCCCCGGCCC 180 AGTCGGTCGC TTGCCACCGC TCGTAGCCGT TACCCGCGGG CCGCCACAGC CGCCGGCCGG 240 GAGAGGCGCG CGCCATGGCT TCTGGAGCCG ATTCAAAAGG TGATGACCTA TCAACAGCCA 300 TTCTCAAACA GAAGAACCGT CCCAATCGGT TAATTGTTGA TGAAGCGATC AATGAGGACA 360 ACAGTGTGGT GTCCTTGTCC CAGCCCAAGA TGGATGAATT GCAGTTGTTC CGAGGTGACA 420 CAGTCTTGCT GAAAGGAAAG AAGAGACGAG AAGCTGTTTG CATCGTCCTT TCTCATCATA 480
CTTGTTCTGA TGAGAAGATT CGGATGAATA GAGTTGTTCG GAATAACCTT CGTGTACGCC 540
TAGGGGATGT CATCAGCATC CAGCCATGCC CTGATGTGAA GTACGGCAAA CGTATCCATG 600
TGCTGCCCAT TCATGACACA GTGGAAGGCA TTACTCGTAA TCTCTTCGAG GTATACCTTA 660
AGCCGTACTT CCTGGAAGCG TATCGACCCA TCCGGAAAGG AGACATTTTT CTTGTCCGTG 720
GTGGGATGCG TGCTGTGGAG TTCAAAGTGG TGGAAACAGA TCCTAGCCCT TATTGCATTG 780
TTGCTCCAGA CACAGTGATC CACTGCGAAG GGGAGCCTAT CAAACGAGAG GATGAGGAAG 840
AGTCCTTGAA TGAAGTAGGG TATGATGACA TTGGTGGCTG CAGGAAGCAG CTAGCTCAGA 900
TAAAGGAGAT GGTGGAACTG CCCCTGAGAC ATCCTGCCCT CTTTAAGGCA ATTGGTGTGA 960
AGCCTCCTAG AGGAATCCTG CTTTACGGAC CTCCTGGAAC AGGAAAGACC CTGATTGCTC 1020
GAGCTGTAGC AAATGAGACT GGAGCCTTCT TCTTCTTGAT CAATGGTCCT GAGATCATGA 1080
GCAAATTGGC TGGTGAGTCT GAGAGCAACC TTCGTAAAGC CTTTGAGGAG GCTGAGAACA 1140
ATGCTCCTCC CATCATCTTC ATTGATGAGC TAGATGCCAT CGCTCCCAAA AGAGAGAAAA 1200
CTCATGGCGA GGTGGAGCGG CGCATTGTAT CACAGTTGTT GACCCTCATG GATGGCCTAA 1260
AGCAGAGGGC ACATGTGATT GTTATGGCAG CAACCAACAG ACCCAACAGC ATTGACCCAG 1320
CTCTACGGCG ATTTGGTCGC TTTGACAGGG AGGTAGATAT TGGAATTCCT GATGCTACAG 1380
GACGCTTAGA GATTCTTCAG ATCCATACCA AGAACATGAA GCTGGCAGAT GATGTGGACC 1440
TGGAACAGTA GCCAATGAGA CTCACGGGCA TGTGGGTGCT GACTTAGCAG CCCTGTGCTC 1500
AGAGGCTGCT CTGCAAGCCA TCCGCAAGAA GATGGATCTC ATTGACCTAG AGGATGAGAC 1560
CATTCATGCC GAGGTCATGA ACTCTCTAGC AGTTACTATG GATCACTTCC GGTGGGCCTT 1620
GAGCCAGAGT AACCCATCAG CACTGCGGGA AACCGTGGTA GAGGTGCCAC AGGTAACCTG 1680
GGAAGACATC GGGGGCCTAG AGGATGTCAA ACGTGAGCTA CAGGAGCTGG TCCAGTATCC 1740
TGTGGAGCAC CCAGACAAAT TCCTGAAGTT TGGCATGACA CCTTCCAAGG GAGTTCTGTT 1800
CTATGGACCT CCTGGCTCTG GGAAAACTTT GTTGGCCAAA GCCATTGCTA ATGAATGCCA 1860
GGCCAACTTC ATCTCCATCA AGGGTCCTGA GCTGCTCACC ATGTGGTTTG GGGAGTCTGA 1920
GGCCAATGTC AGAGAAATCT TTGACAAGGC CCGCCAAGCT GCCCCCTGTG TCCTATTCTT 1980
TGATCAGCTG GATTCGATTG CCAAGGCTCG TGGAGGTAAC ATTGGAGATC GTGGTGGGGC 2040
TGCTGACCGA GTCATCAACC AGATCCTGAC AGAAATGGAT GGCATGTCCA CAAAAAAAAA 2100
TGTGTTCATC ATTGGCGCTA CCAACCGGCC TGACATCATT GATCCTGCCA TCCTCAGACC 2160
TGGCCGTCTT GATCAGCTCA TCTACATCCC ACTTCCTGAT GAGAAGTCCC GTGTTGCCAT 2220
CCTCAAGGCT AACCTGCGCA AGTCCCCAGT TGCCAAGGAT GTGGACTTGC AGTTCCTGGC 2280
TAAAATCACT AATGGCTTCT CTCGAGCTCA CCTGACAGAG ATTTGCCAGC GTGCTTGCAA 2340
GCTGGCCATC CGTCAATCCA TCGAGAGTGA GATTAGGCGA GAACGAGAGA GCCAGACAAA 2400
CCCATCAGCC ATGGAGGTAG AAGAGGATGA TCCAGTGCCT GAGATCCGTC GAGATCACTT 2460
TGAAGAAGCC ATGCGCTTTG CGCGCCGTTC TGTCAGTGAC AATCACATTC GGAAGTATGA 2520
GATGTTTGCC CAGACCCTTC AGCAGAGTCG GGGCTTTGGC AGCTTCAGAT TCCCTTCAGG 2580
GAACCAGGGT GGAGCTGGCC CCAGTCAGGG CAGTGGAGGC GGCACAGGTG GCAGTGTATA 2640
CACAGAAGAC AATGATGATG ACCTGTATGG CTAAGTGGTG GTGGCCAGCG TGCAGTGAGC 2700
TGGCCTGCCT GGACCTTGTT CCCTGGGGGT GGGGGCGCTT GCCCAGGAGA GGGACCAGGG 2760
GTGCGCCCAC AGCCTCCTCC ATTCTCCAGT CTGAACAGTT CAGCTACAGT CTGACTCTGG 2820
ACAGGGGGTT TCTGTTGCAA AAATACAAAA CAAAAGCGAT AAAATAAAAG CGATTTTCAT 2880
TTGGTAGGCG GAGAGTGAAT TACCAACAGG GAATTGGGCC TTGGGCTATG CCATTTCTCT 2940
TGTAGTTTGG GGCAGTGCAG GGGACCTGTG TGGGGTGTCA ACCAAGCCAC TACTGCCACC 3000
TGCCACAGTA AAGCATCTGC ACTTGACTCA ATGCTGCCCG AGCCCTCCCT TCCCCCTATC 3060
CAACCTGGGT AGGTGGGTAG GGGCCACAGT TGCTGGATGT TTATATAGAG AGTAGGTTGA 3120
TTTATTTTAC ATGCTTTTGA GTTAATGTTC GAAAACTAAT CACAAGCAGT TTCTAAACCA 3180
AAAAATCACA TGTTGTAAAA GGACAATAAA CGTTGGCTCN AAATGGGWRA AAAAAAAAAA 3240
AAAAAAGGGG GGCCCCTCTA AAGNNCCANN CTTCGT 3276
(2) INFORMATION FOR SEQ ID NO: 292:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1695 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO : 292: TTGCAATGCT TCAATTCCCC TCCTCACGCC AGCCTAGGAG AAGAAGTTCG TAGTCCCAGA 60 GGTCAGGCAG GAGGCGGCAG TTTCTGGCGG GTCAGGGCGG AGCTCAAGTG ACAGCGGAGG 120 CGGAAGCAAC GGTCGGTGGG GCGGAGAAGG GGGCTGGCCC CAGGAGGAGG AGGAAACCCT 180 TCCGAGAAAA CAGCAACAAG CTCAGCTGCT GTGACAGAGG GCAACAAGAT GGCGGCGCCG 240 AAGGGAGCCT CTGGGTGAGG ACCCAACTGG GGCTCCCGCC GCTCCTGCTG CTCACCATGG 300 CCTTGGCCGG AGGTTCGGGG ACCGCTTCGG CTGAAGCATT TGACTCGGTC TTGGGTGATA 360
CGGCGTCTTG CCACCGGGCC TGTCAGTTCA CCTACCCCTT GCACACCTAC CCTAAGGAAG 420
AGGAGTTCTA CGCATGTCAG AGAGGTTCCA GGCTGTTTTC AATTTGTCAG TTTGTGGATG 480
ATGGAATTGA CTTAAATCGA ACTAAATTCG AATGTGAATC TGCATCTACA GAAGCATATT 540
CCCAATCTGA TCAGCAATAT GCTTCCCATC TTGGTTGCCA GAATCAGCTG CCATTCGCTC 600
AACTGAGACA AGAACAACTT ATGTCCCTGA TGCCAAAAAT GCACCTACTC TTTCCTCTAA 660
CTCTGGTGAG GTCATTCTGG AGTGACATGA TGGACTCCGC ACAGAGCTTC ATAACCTCTT 720
CATCGACTTT TTATCTTCAA GCCGATCACG GAAAAATAGT TATATTCCAG TCTAAGCCAG 780
AAATCCAGTA CGCACCACAT TTGGAGCAGG AGCCTACAAA TTTGAGAGAA TCATCTCTAA 840
GCAAAATGTC CTATCTGCAA ATGAGAAATT CACAAGCGCA CAGGAATTTT CTTGAAGATG 900
GAGAAACTCA TGGCTTTTTA AGATGCCTCT CTCTTAACTC TGGGTGGATT TTAACTACAA 960
CTCTTGTCCT CTCGGTGATG GTATTGCTTT GGATTTGTTG TGCAACTGTT GCTACAGCTG 1020
TGGAGCAGTA TGTTCCCTCT GAGAAGCTGA GTATCTATGG TCACTTGGAG TTTATGAATG 1080
AACAAAAGCT AAACAGATAT CCAGCTTCTT CTCTTGTGGT TGTTAGATCT AAAACTGAAG 1140
ATCATGAAGA AGCAGGGCCT CTACCTACAA AAGTGAATCT TGCTCATTCT GAAATTTAAG 1200
CATTTTTCTT TTAAAAGACA AGTGTAATAG ACATCTAAAA TTCCACTCCT CATAGAGCTT 1260
TTAAAATGGT TTCATTGGAT ATAGGCCTTA AGAAATCACT ATAAAATGCA AATAAAGTTA 1320
CTCAAATCTG TGAAGACTGT ATTTGCTATA ACTTTATTGG TATTGTTTTT GTAGTAATTT 1380
AAGAGGTGGA TGTTTGGGAT TGTATTATTA TTTTACTAAT ATCTGTAGCT ATTTTGTTTT 1440
TTGCTTTGGT TATTGTTTTT TTCCCTTTTC TTAGCTATGA GCTGATCATT GCTCCTTCTC 1500
ACCTCCTGCC ATGATACTGT CAGTTACCTT AGTTAACAAG CTGAATATTT AGTAGAAATG 1560
ATGCTTCTGC TCAGGAATGG CCCACAAATC TCTAATTTCA AATTTAGCAG GAAATGACCT 1620
TTAATGACAC TACATTTTCA GGAACTGAAA TCATTAAAAT TTTATTTGAA TAATTAAAAA 1680
AAAAAAAAAA AANCT 1695
(2) INFORMATION FOR SEQ ID NO: 293:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1501 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 293: CACTTTCAGC AGTCCTTTGC TCTCTTTCCT TCTACCTCAA ATAGCCCCAG GAGTGGGCTT 60
TAGTCTCCAA TATGGAGCAT CTGAAGCTTC TCCTGGGGGA TGGGGATTGG GATGGGCAGA 120
ATCTGTTTTG GWTCTCCGGG TTATTTCCAG TGGGTGTAAA AGCAGAGCTG GGCCTTTCCC 180
TCTCTTATCC CTGAGGGTGG GTAAGAAGGA CTGTATCTAC ACCTGTTCTT CCCTACCTTC 240
TCTTTTGTTA GGGAGGCCTC ATTCTAAGTT CCTCAAGAGA GTCCTTGGCT TAAAGCTGTA 300
GCAAGGGTGT GCTAGGTGGG GGATTTGGAG CAAAACCGTC GAGTAGGCAT GATACTGGTA 360
TGGAGTGGGC CTGCAAAATC AGACAGAAAT GGCTTGAGAA GCCGCAGGGG AGCATGCCTG 420
TCTCTCAGTG ATAGAGTATG GGAGGGACCT CCCTAGCTTG GAAAATGAGA ATTGAAGGGG 480
TTATGAACAA ATAGGATGCC TAGTTGAGGA TGTTCCCAAA GTTTTGTCCA ATCTTATCAT 540
TAGTAGATTT TATAAGCCAC AGAGACAAAC CAGAAACGGA ATAATGTTAC TTTGGATGCT 600
TTATTTTTTT GTTCTAGGTG TGGCTTTGTA CATGCAGAAG AATGCTATAT GCTGCACATT 660
TTGCCTTTAA AGTCTTACGA CTTTCCCCAT TTTAGTCTAA TGGGAAGATA CAGATGTGCA 720
AGTCTGCTTT TTTGTTTTTT GTTATTATTT TTTTTTTTTT GCTCTGTGTT ATGGACATTT 780
TCAGACATGC ACAGAAGTGG AGAGGATGGT CCTTGGACCC MATGTGTCCA TCACCTAGCT 840
GCATCACTTA TCAGCTATGG TCAACCTGGT TTCATCTGTA TCTCTCTCTT TTCACCTGTA 900
TTCTTTATTG AAAATCCAAG ACACTATGCC AATGCAACCG TGACTACTTT GGGAGATTGG 960
TAGTCTCTTT TGATGGTGAT AGTGATCGGG TGCACTATCA TAATCACATC AGGTCTGCTT 1020
TTTGCTTTTA ATGTTAACTA ATGAAGTTCC AGAGATGGGC CTTAGAAATG TGTTTTAAGA 1080
ATTAACAAGG AGTCTCAAAA AGAAATGAGA GGGATGCTTC CTTTNCCCTT GCATCTACAA 1140
AACMAGAGAG AGACTGTTCT GTTGTAAAAC TCTTTCAAAA ATTCTGATAT GGTAAGGTAC 1200
TTGAGACCCT TCACCAGAAT GTCAATCTTT TTTTCTGTGT AACATGGAAA CTTGTGTGAC 1260
CATTAGCATT GTTATCAGCT TGTACTGGTC TCATAACTCT GCTTTTGGAA GAATAATTTG 1320
GAAATTGTTG CTGTGTTCTG TGAAAATAAC CTCCCCAAAA TAATTAGTAA CTCGTTGTTC 1380
TACTTGGTAA TTTGACACCC TGTTAATAAC GCAATTATTT CTGTGTTCTT AAACAGTATA 1440
AATAGTTGTA AGTTTGCATG CATGATGGAA AAATAAAAAC CTGTATCTCT GTTAAAAAAA 1500
A 1501
(2) INFORMATION FOR SEQ ID NO: 294:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2683 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 294:
TGANTGTGGT CCCGGGTCCN GATTGGCAGN GCCTCCGCCG CGGCTCGTGG TTCTCCCGCC 60
ATCGCACTGT CGCGGGGGCT GCCCCGGGAG CTCGCTGAGG CGGTGGCCGG GGGCCGGGTR 120
CTGGTGGTGG GGGCGGGCGG CATCGGCTGC GAGCTCCTCA AGAATCTCGT GCTCACCGGT 180
TTCTCCCACA TCGACCTGAT TGATCTGGAT ACTATTGATG TAAGCAACCT CAACAGACAG 240
TTTTTCTTTC AAAAGAAACA TGTTGGAAGA TCAAAGGCAC AGGTTGCCAA GGAAAGTGTA 300
CTGCAGTTTT ACCCGAAAGC TAATATCGTT GCCTACCATG ACAGCATCAT GAACCCTGAC 360
TATAATGTGG AATTTTTCCG ACAGTTTATA CTGGTTATGA ATGCTTTAGA TAACAGAGCT 420
GCCCGAAACC ATGTTAATAG AATGTGCCTG GCAGCTGATG TTCCTCTTAT TGAAAGTGGA 480
ACAGCTGGGT ATCTTGGACA AGTAACTACT ATCAAAAAGG GTGTGACCGA GTGTTATGAG 540
TGTCATCCTA AGCCGACCCA GAGAACCTTT CCTGGCTGTA CAATTCGTAA CACACCTTCA 600
GAACCTATAC ATTGCATCGT TTGGGCAAAG TACTTGTTCA ACCAGTTGTT TGGGGAAGAA 660
GATGCTGATC AAGAAGTATC TCCTGACAGA GCTGACCCTG AAGCTGCCTG GGAACCAACG 720
GAAGCCGAAG CCAGAGCTAG AGCATCTAAT GAAGATGGTC ACATTAAACG TATTTCTACT 780
AAGGAATGGG CTAAATCAAC TGGATATGAT CCAGTTNAAA CTTTTTACCA AGCTTTTTAA 840
AGATGACATC AGGTATCTGT TGACAATGGA CAAACTATGG CGGAAAAGGA AACCTCCAKT 900
TCCGTTGGAC TGGGCTGAAG TACAAAGTCA AGGAGAAGAA ACGAATCCAT CAGATCAACA 960
GAATGAACCC CAGTTAGGCC TGAAAGACCA GCAGGTTCTA GATGTAAAGA GCTATGCACG 1020
TCTTTTTTCA AAGAGCATCG AGACTTTGAG AGTTCATTTA GCAGAAAAGG GGGATGGAGC 1080
TGAGCTCATA TGGGATAAGG ATGACCCATC TGCAATGGAT TTTGTCACCT CTCCTGCAAA 1140
CCTCAGGATG CATATTTTCA GTATGAATAT GAAGAGTAGA TTTGATATCA AATCAATGGC 1200
AGCGAACATT ATTCCTGCTA TTGCTACTAC TAATGCAGTA ATTGCTGGGT TGATAGTATT 1260
GGAAGGATTG AAGATTTTAT CAGGAAAAAT AGACCAGTCC AGAACAATTT TTTTGAATAA 1320
ACAACCAAAC CCAAGAAAGA AGCTTCTTGT GCCTTGTGCA CTGGATCCTC CCAACCCCAA 1380
TTGTTATGTA TGTGCCAGCA AGCCAGAGGT GACTGTGCGG CTGAATGTCC ATAAAGTGAC 1440
TGTTCTCACC TTACAAGACA AGATAGTGAA AGAAAAATTT GCTATGGTAG CACCAGATGT 1500
CCAAATTGAA GATGGGAAAG GAACAATCCT AATATCTTCC GAAGAGGGAG AGACGGAAGC 1560
TAATAATCAC AAGAAGTTGT CAGAATTTGG AATTAGAAAT GGCAGCCGGC TTCAAGCAGA 1620
TGACTTCCTC CAGGACTATA CTTTATTGAT CAACATCCTT CATAGTGAAG ACCTAGGAAA 1680
GGACGTTGAA TTTGAAGTTG TTGGTGATGC CCCGGAAAAA GTGGGGSCCA AACAAGCTGA 1740
AGATGCTGCC AAAAGCATAA CCAATGGGCA GTGATCATGG AGCTCAGCCC TCCACCTCCA 1800
CAGCTCAAGA GCAAGATGAC GTTCTCATAG TTGATTCGGA TGAAGAAGAT TCTTCAAATA 1860
ATGCCGACGT CATGAAGAAG AGAGAAGCCG CAAGAGGAAA TTAGATGAGA AAGAGAATCT 1920
CAGTGCAAAG AGGTCACGTA TAGAACAGAA GGAAGAGCTT GATGATGTCA TAGCATTAGA 1980
TTGAACAGAA ATGCCTCTAA ACAGAACCCT CTTACTATTT AGTTTATCTG GGCAGAACCA 2040
GATTGTTATG TCCTTTGTTC CAAAGGGAAA AAATTGACAG CAGTGACTTG AAAATGATTC 2100
TGCTCCCTTT GAAAGCATTC ATTTTGCTAG AACTGTTAGA CACATTGCAG TATGCTGTAT 2160
TGAAAGTAGG AATATAGTTT TAAAAACCCT TTGAACAAAG TGTGTGCATA ACCAGTCATG 2220
AGATAAAACA ACACAATGCA TGTTGCCTTT TTAATGTAAA TACCCTTAGG TATCATTAAT 2280
AGTTTCAAAA TATTGTGGTT TAGTAAAGTT GATACCTGGT TATAAATATT ATGCCTTTAT 2340
TTTTGGCTAG AAGAAGAATT ATTTTTAGCC TAGATCTAAC CATTTTCATA CTCTTAACTG 2400
ATTGAAACAG ATTCAAAGAA GTATCGAGTG CTATGCATTG AAACTTGTTT TTAAATGTTA 2460
GATGGCACTA TGTATATTAA TGTAAAACAA TGTTAATTTA CTCAAGTTTT CAGTTTGTAC 2520
CGCCTGGTAT GTCTGTGTAA GAAGCCAATT TTTGTGTATT GTTACAGTTT CAGGTTATTT 2580
ATATTCGATG TTTTGTAAAA CTCAAATAAC GACTATACTT ATGGACCAAA TAAATGGCAY 2640
TGCATTCTKG TKAAAAAAAN NACAGAAAAA AAAAAAAACA AGA 2683
(2) INFORMATION FOR SEQ ID NO: 295:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1454 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO : 295: GGACTCGGGG TGGCTCTAAG GGGCAGGGAT AGGGCTGGGG AGCGCCGGCC TGTGGCCCTG 60 ACCAGCCCCT TCTCGTCCAG GTTCCACCCC GATGCAGGTC GTCACGTCGT TGACGCGGGA 120 CAGCTACCTG ACGCACTGCT TCCTCCAGCA CCTCATGGTC GTGCTGTCCT CTCTGGAACG 180 CACGCCCTCG CCGGAGCCTC TTGACAAGGA CTTCTACTCC GAGTTTGGGA ACAAGACCAC 240 AGGGAAGATG GAGAACTACG AGCTGATCCA CTCTAGTCGC GTCAAGTTTA CCTACCCCAG 300 TGAGGAGGAG ATTCGGGACC TGACGTTCAC TGTGGCCCAA AAGATGGCTG AGCCAGAGAA 360 GGCCCCAGCC CTCAGCATCC TGCTGTACGT GCAGGCCTTC CAGGTGGGCA TGCCACCCCC 420
TGGGTGCTGC AGGGGCCCCC TGCGCCCCAA GAGACTCCTC CTCACCAGCT CCGAGATCTT 480
CCTCCTGGAT GAGGACTGTG TCCACTACCC ACTCCCCGAG TTTGCCAAAG AGCCGCCGCA 540
GAGAGACAGG TACCGGCTGG ACGATGGCCG CCGCGTCCGG GACCTGGACC GAGTGCTCAT 600
GGGCTACCAG ACCTACCCGC AGCCCTCACC CTCGTYTTCG ATGACGTGCA AGGTCATGAC 660
CTCATGGGCA GTGTCACCCT GGACCACTTT GGGGAGGTGC CAGGTGGCCC GGCTAGAGCC 720
AGCCAGGGCC GTGAAGTCCA GTGGCAGGTG TTTGTCCCCA GTGCTGAGAG CAGAGAGAAG 780
CTCATCTCGC TGTTGGCTCG CCAGTGGGAG GCCCTGTGTG GCCTGAGCTG CCTGTCGAGC 840
TCACCGGCTA GCCCAGGCCA CAGCCAGCCT GTCGTGTCCA GCCTGACGCC TACTGGGGCA 900
GGGCAGCAGG CTTTTGTGTT CTCTAAAAAT GTTTTATCCT CCCTTTGGTA CCTTAATTTG 960
ACTGTCCTCG CAGAAATGTG AACATGTGTG TGTGTTGTGT TAATTCTTTC TCATGTTGGG 1020
AGTGAGAATG CCGGGCCCCT CAGGGCTGTT CGGTGTGCTG TCAGCCTCCC ACAGGTGGTA 1080
CAGCCGTGCA CACCAGTGTC GTGTCTGCTG TTGTGGGACC GTTGTTAACA CGTGACACTG 1140
TGGGTCTGAC TTTYTCTTCT ACACGTCCTT TCCTGAAGTG TCGAGTCCAG TCCTTTGTTG 1200
CTGTTGCTGT TGCTGTTGCT GTTGCTGTTC GCATCTTGCT GCTAATCCTG AGGCTGGTAG 1260
CAGAATGCAC ATTGGAAGCT CCCACCCCAT ATTGTTCTTC AAAGTGGAGG TCTCCCCTGA 1320
TCCAGACAAG TGGGAGAGCC CGTGGGGGCA GGGGACCTGG AGCTGCCAGC ACCAAGCGTG 1380
ATTCCTGCTG CCTGTATTCT CTATTCCAAT AAAGCAGAGT TTGACACCGW MAAAAAAAAA 1440
AAAAAAAAAA AACN 1454
(2) INFORMATION FOR SEQ ID NO: 296:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 828 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 296: ACCCTGGCAT GCCCCACAAA CAGATCACCA GCCAGCTTAC ACAGGCATTA ACTCTCCTCA 60 ATGAGGAAGA ATCATTCACA ACTGAGCAAG ACATTCATAT GATCATTTAA GGAAGTGTTT 120 CCCTTATGTG TTAGCAAGTA TAATCGGCTA ACTCCTAAAT CCCAATGAAT AGTCCTAGGC 180 TGGACAGCAA TGGGCTGCAA TTAGGCAGAT AAAGACATCA GTCCCAGTAA ATGAATCCAT 240 AGACTGATCT AGCACCAACT ACCATTAGCA CTATGTTAGG AGCTGCAAGG CCCCAAAGTA 300
GAAGATGTGC ATAATGTCTG CTCTTGTGTA GCTCAGGAGA CAATTCCAGC ACAGACACTA 360
CAGTTAACGC TGAACTGCAG CTGCAAGTAA TAGCAWGAAC AGTCAGAAAA ATACCTTATG 420 AGGGGGCAGG GCTGAAGCTG GGCCTTGAAG GATGGATGAA ATTTGGATAG AGAATGAGGA 480
AGACAGAGGG NCTCCAAGTG AGAGAAGCAT GAAAAATGAG CARGGGCCTG GATCAGTGGG 540
GTGTATTCAG AGCACCTYTC CAGATGCACC ATGCATGCTG ACAGTCCCTT GCCTATGTGT 600
GGCAGAGTGT CCCAGCCAGA TGTGTGCCCC CACCCCATGT CCATTTACAT GTCCTTCAAT 660
GCCCACCTCA AAAGGYACYT CTTCTGTAAA GCTTTCCCTK GGTATCAGGA ATCAAAATTA 720 ATCAGGGATC TTTTCACACT GCTCTTTTTT CCTCTTTGGT CCTTCTATCA CTAAAACTCA 780
TCTCATTCAG CCTTACAGCA TAACTAATTA TTTGTTTTCC TCACTACA 828
(2) INFORMATION FOR SEQ ID NO: 297:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 2416 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 297:
TCAATTTCCA TTAACTCAGA TCAGCCATTG TGATTCACCA TTTGTCAGGC TCTCAGGTTT 60 AACAAAACCT ACTATCACCA TCATCCTTCA ACAGCCACAG TCTGAATTCA GCCAACATTT 120
TTTTTTCTTT GAGAAAGAAG TGGACTGGGG CACAACTTTT AGTCTGAGGG GAGCTAGTGG 180
AAATCTAGAC AATAGAAGTC ATCGATAGCA GCTTTTCCTC AAATGTGTGA CTCCTCAGGG 240 GCTAAACTGC TCTTAGCTTA GAATTATGCT TTACTAGAGA TCTAGCAGAT AAGTGGGTTA 300
ATCACTACCA TCCTGTAACT AGTTATATAG CTTCCAGACA TGAGGGAGAC ATCAAACAGG 360
GATGGAAGCA ACCCCAAGGA TATGCAAGAA GGGCATGATG AACCCCCTTC CCTCTGGCAG 420
GAGAACAAGG CCAACCAAGG GACAGACTGG AAAGCACTTA GATGTTTAAG GAGGAGAAAG 480
GGGAAGCTTT GACCAGTCCT TGCCTTTTCC CAAGTTCAGC CAGTTCTCCG CTGCTTGCAA 540 CCTCTAGCGC AGTAACATTT GCAGAATTGC AGATTTTCCC CCAGATACTA GGAGGAAAGG 600
GACTTTGGGG GGTGGGGAAG GGGTCGTGGT GTTTTAAAAG CATAAGTTAC CTGTTTGCAC 660
TGTTTTAAGA TAGGAAAAAA AAATAGTGGG CAAGGTGAAC ATCAGACGTA AATTTGTGTG 720
TTTTTATTTT GTCATGCTCT TGAAAATCTT TGACCATTTG TAGTATACAC AGTGAAACTT 780
GATTCTCTGT TGCATAAAAC ACTATATTTT TTTGGAAATG TTACTGTCCA AAAGCCTCTT 840 CCCTCCCTTT CCTTTTCCTA TGTACTTCCT TCATACTTGC TTTACTGATC AGCCAGGCAA 900
TAGCCATCCA AGAGCTAGAG CATGAAACAG GGCCCTTTCC AAGTAGGCTC TGGGTGTCCT 960
AAGCCAGCGT GTGCCCTCTG GTTTAGTGAG TCTAATAGAG TCCCTGGCAC CTTTCTTTCC 1020
AAATCAGGCT AACAGACCAG ACTCCAGCAA GTTATCAGAT TCCTCAATCA GATGCACTAG 1080
GAGTGAGGAG CCCAGGGATG GAGGGGGTTC CTGAAGTATT GCAGTTGGCT GTAGTAGCTG 1140
AGTTCTTTTC CATGTTACCG AAACTGTAGC CAGTTACAGT TTACTCAGGA AAACGGTAGA 1200
TCAATTCAGC CATGGTAGTG CTGGTTGGCA GGGATTGGTA ACGGAGAGAA CTGCTCATCA 1260
GCCAAAACTC AAGCCTTGCC TTTTAGGAGG CCACCAGCAG AGGGACTTGG TCCTCCTTGT 1320
CTGGTACTTG TGTACATGCC GGTGACCTCA GGACTCCACT CACACTGGCG AGCAAAAAGG 1380
GAGCAGTGAT TCTCTTTTCT CTCCCCACCC CCTGCCCTTT GTTACCAACA CCAGTTTCCC 1440
AGGGGGTACA TCAGTTTCTG AATTTTTAAA AAATCTTTTT GGTTTGGTTT TTCTGGGGAC 1500
TGATAAGTGC TTTAAGCAAT GTCCATACCC CGTCAAGACT CCCAGCTTAG TCATTTTCTT 1560
GTATTTTTCT GTTCACAGTA TTTGTGTGTG TCCTTGTTTT GGCAGCTCAT TTTGGCTGTA 1620
TTATATATTC AGTGATGAAT TGATCCTCTT TTTTCCCTAA GGGATATGAA TTGTTTTTCT 1680
TGTGTTATAT TCTGCTTGTG AATAGCTGGA GCAAACCTGG GGCTGACACG CGTAAGSTAG 1740
GGCTGCAAAR CGAGAAGAGA GCCGGTCGAG TGTACTTGTC CCTGACAGGC TGACCTACCT 1800
GAGTCTCTGA GCTTTTCAGT CCAAATCTTT GCAAGGCTCA AAATCCCACA GAACCTCTCC 1860
TCTTCTCCCC ACTCCCCATG GCAGGGACCG GACCATCCCT ACATGCAACA TGCTGTTCCT 1920
CCAGCCCCTC CCATTGCCAT GGCAAAACAG GTACCTTTGG GGCATGGGGG CATTACATGG 1980
GATGCTTGTG TAATCGACCA CCTAGCCTTC TCTCTCCCCT CCCGTCCTCC CCCAGAATCA 2040
CTTCCTAGGA CACCCGAGCT GCTTGCCCAG GGTCCTGTTT CCCTGCTAAC TCCAGAGAAG 2100
CATCCCAGGG CTTTGTGACA GTCTCTAATT CCCTTCCCTT CTCGTTAAGA ATCATATTGT 2160
ATAGTAGCTT TCAGACCATA CAGTATTCAT TGGGTTACTC CTATTATTAT CAAGTAGCTG 2220
GAATTGTCAA GGTCGGAGTA GTTAGATCTT TAGCTTTTAT TCCTTATTTT TTTGTATTAC 2280
TCTCCATGTG TATAAATTAT TGATCATGTT GCTGGCTTTT ATAAACTCTA AGCGAAGGAG 2340
GAGCACTGCC TCAGCCTTTG CACATGGTAA TGAAGCACTG TTTTTAAATA AAAGRGRGAA 2400
MCMCCAAAAA AAAAAA 2416
(2) INFORMATION FOR SEQ ID NO: 298:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 545 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 298:
GAATTCGGCA CGAGCCATGC YTGGCCTCTC CTTGATTCTT ACAGTCACTT TCTTGGCTGT 60 TTCTGACTCA GCAGCTACCT GCATTGTGGC CAAAGGATGA CCTATTCCTT CTCAGGAGGG 120
CAAAAATGTG GAATAGTCTC TGTCCATGCC TCTCCTCATG GGCTACCACC TCTCCCACCG 180
TGGTTAATCA GTAACAACCA GGAGAGAAGC TGCTGGAACT GACCTCTGGG AACTCCCTCG 240 ATCGTTTGGT GCAGGAATGT AGTAGGCATA CACGTGGTTG CGTGGATCTG GGCCCTCCTG 300
ATGTGAGTAG AGAGGTAAAA GGSCACCATC TCCTTGACCT YTGGGGAACT CATCCACAAA 360
GAAGATGTTT CCAAGATCCT TCTGAAGATT GSCTAAAAAT AGCCGGTTTC CACCCCCGTG 420
AATGCATCCA TTCTAGAATG CTCCTTCACC AGGACCAGAG AACTGATTTA CAGAAGTGAC 480
ATGAAAACAT TCCATCCCAG AATTTCCANT ACCTCAAATT NAATTTCTAC CTATTAAAAA 540 NAAAA 545
(2) INFORMATION FOR SEQ ID NO: 299:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1530 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 299: GGCTCTGCTG GGCATCATAC TTCTCACTGG GTAAACACTT TGCCCACTTA CCGCAGATCA 60
AGCTGCTTGC CAGGGCTCTC CGGCTCTGTG AGTTTGGGAG GCAGGCATCT TCCAGGAGGC 120
TGGTGGCTGG CCAGGGATGT GTGGGGCCCC GGCGAGGGTG CTGCGCTCCC GTCCAGGTGG 180
TTGGGCCCAG GGCTGATCTC CCACCCTGTG GAGCCTGCAT TACTGGAAGG ATCATGCGGC 240
CAGATGATGC CAACGTGGCC GGCAATGTCC ACGGGGGGAC CATCCTGAAG ATGATCGAGG 300 AGGCAGGCGC CATCATCAGC ACCCGGCATT GCAACAGCCA GAACGGGGAG CGCTCTCTGG 360
CCGCCCTGGC TCGTGTCGAG CGCACCGACT TCCTGTCTCC CATGTGCATC GGTGAGGTGG 420
CGCATGTCAG CGCGGAGATC ACCTACACCT CCAAGCACTC TGTGGAGGTG CAGGTCAACG 480
TGATGTCCGA AAACATCCTC ACAGGTGCCA AAAAGCTGAC CAATAAGGCC ACCCTGTGGT 540
ATGTGCCCCT GTCGCTGAAG AATGTGGACA AGGTCCTCGA GGTGCCTCCT GTTCTGTATT 600 CCCGGCANGA GCAGGAGGAG GAGGGCCGGA AGCGCTATGA AGCCCAGAAG CTCGAGCGCA 660
TGGAGACCAA GTGGAGGAAC GGGGACATCG TCCAGCCAGT CCTCAACCGA GAGCCGAACA 720
CTGTCAGCTA CAGCCAGTCC AGCTTGATCC ACCTGGTGGG GCCTTCAGAC TGCACCCTGC 780
ACGGCTTTGT GCACGGAGGT GTCACCATGA AGCTCATGGA TGAGGTCGCC GGGATCGTGG 840
CTGCACGCCA CTGCAAGACC AACATCCTCA CAGCTTCCGT GGACGCCATT AATTTTCATG 900
ACAAGATCAG AAAAGGCTCC GTCATCACCA TCTCGGGACG CATGACCTTC ACGAGCAATA 960
AGTCCATGGA GATCGAGGTG TTGGTGGACG CCGACCCTGT TGTGGACAGC TCTCAGAAGC 1020
GCTACCGGGC CGCCAGTGCC TTCTTCACCT ACGTGTCGCT GAGCCAGGAA GGCAGGTCGC 1080
TGCCTGTGCC CCAGCTGGTG CCCGAGACCG AGGACGAGAA GAAGCGCTTT GAGGAAGGCA 1140
AAGGGCGGTA CCTGCAGATG AAGGCGAAGC GACAGGGCCA CGCGGAGCCT CAGCCCTAGA 1200
CTCCCTGCTC CTGCCACTGG TGCCTCGAGT AGCCATGGCA ACGGGCCCAG TGTCCAGTCA 1260
CTTAGAAGTT CCCCCCTTGG CCAAAAACCC AATTCACATT GAGAGCTGGT GTTCTCTGAA 1320
GTTTTCGTAT CACAGTGTTA ACCTGTACTC TCTCCTGCAA ACCTACACAC CAAAGCTTTA 1380
TTTATATCAT TCCAGTATCA ATGCTACACA GTGTTGTCCC GAGCGCCGGG AGGCGTTGGG 1440
CAGAAACCCT CGGGAATGCT TCCGAGCACG CTGTAGGGTA TGGGAAGAAC CCAGCACCAC 1500
TMATAAAGCT GNTGCTTGGC TGGGGAAGNA 1530
(2) INFORMATION FOR SEQ ID NO: 300:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 997 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 300: AGGTAGTGAG AGACACATTA CACCTAACCA ACAAGAAGAA GGATCCTCCC CCTTATAATT 60 TAACTATGTT TACAGGGAAT GCGTACATTG TGGCTTCCCG AGNATTTCGT CCAACATGTT 120 TTCAAGAACC CTAAATCCCA ACAACTCATT GAATCGGTAA AAGACACTTA TAGCCCAGAT 180 GAACACCTCT GGGCCACCCT TCAGCGTGCA CGCTCGATGC CTCGCTCTGT TCCCAACCAC 240 CCCAAGTACG ACATCTTCAG ACATCACTTC TATTGCCAGG CTGGTCAAGT GGCAGGGTCA 300 TGAGGGAGAC ATCGATAAGC GTGCTCCTTA TCCTCCCTGC TCTGGAATCC ACCAGCGGGC 360 TATCTCCGTT TATGGGGCTG GGGACTTGAA TTGGATCCTT CAAAACCATC ACCTGTTGGC 420 CAACAAGTTT GACCCAAAGG TAGATGATAA TGCTCTTCAG TGCTTAGAAG AATACCTACG 480
TTATAAGGCC ATCTATGGGA CTGAACTTTC AGACACACTA TCAGAGCGTT GCTACCTGTG 540
GGGCAAGAGC ATGTACAAAC ATGCTCAGAA CTTGCTGGGA CAGTGTGGGT GGGAGACCAG 600 GGCTTTGCAA TTCGTCGCAT CCTTTAGGAT AAGAGGGCTG MTATTAGATT GTGGGTAAGT 660
AGATCTTTTG CCTTGCAAAT TGCTGCCTGG GTGRATCCTG CTTGTTCTCT CACCCCTAAC 720
CCTAGTAGTT CCTCCACTAA CTTTCTCACT AAGTGAGAAT GAGAACTGCT GTGATAGGGA 780
GAGTGAAGGA GGGATATGTG GTAGAGCACT TGATTTCAGT TGAATGCCTG CTGGTAGCTT 840
TTCCATTCTG TGGAGCTGCC GTTCCTAATA ATTCCAGGTT TGGTAGCGTG GAGGAGAACT 900 TTGATGGAAA GAGAACCTTC CCTTCTGTAC TGTTAACTTA AAAATAAATA GCTCCTGATT 960
CAAAGTAAGG AAAAARAAAA AAAGAAAAAA AACTCGA 997
(2) INFORMATION FOR SEQ ID NO: 301:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 2345 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 301:
TTGAGGCCGA CGCTAGGGGC CCGGAAGRAA ACTGCGAGGC GAAGGTGACC GGGGACCGAG 60 CATTTCAGAT CTGCTCGGTA GACCTGGTGC ACCACCACCA TGTTGGCTGC AAGGCTGGTG 120
TGTCTCCGGA CACTACCTTC TAGGGTTTTC CACCCAGCTT TCACCAAGGC CTCCCCTGTT 180
GTGAAGAATT CCATCACGAA GAATCAATGG CTGTTAACAC CTAGCAGGGA ATATGCCACC 240 AAAACAAGAA TTGGGATCCG GCGTGGGAGA ACTGGCCAAG AACTCAAAGA GGCAGCATTG 300
GAACCATCGA TGGAAAAAAT ATTTAAAATT GATCAGATGG GAAGATGGTT TGTTGCTGGA 360
GGGGCTGCTG TTGCTCTTCG AGCATTCTGC TACTATGGCT TGGGACTGTC TAATGAGATT 420
GGAGCTATTG AAAAGGCTGT AATTTGGCCT CAGTATGTCA AGGATAGAAT TCATTCCACC 480
TATATGTACT TAGCAGGGAG TATTGGTTTA ACAGCTTTGT CTGCCATAGC AATCAGCAGA 540 ACGCCTGTTC TCATGAACTT CATGATGAGA GGCTCTTGGG TGACAATTGG TGTGACCTTT 600
GCAGCCATGG TTGGAGCTGG AATGCTGGTA CGATCAATAC CATATGACCA GAGCCCAGGC 660
CCAAAGCATC TTGCTTGGTT GCTACATTCT GGTGTGATGG GTGCAGTGGT GGCTCCTCTG 720
ACAATATTAG GGGGTCCTCT TCTCATCAGA GCTGCATGGT ACACAGCTGG CATTGTGGGA 780
GGCCTCTCCA CTGTGGCCAT GTGTGCGCCC AGTGAAAAGT TTCTGAACAT GGGTGCACCC 840 CTGGGAGTGG GCCTGGGTCT CGTCTTTCTG TCCTCATTGG GATCTATGTT TCTTCCACCT 900
ACCACCGTGG CTGGTGCCAC TCTTTACTCA GTCGCAATGT ACGCTCGATT AGTTCTTTTC 960
AGCATCTTCC TTCTGTATGA TACCCAGAAA GTAATCAAGC GTCCAGAAGT ATCACCAATG 1020
TATGGAGTTG AAAAATATGA TCCCATTAAC TCGATCCTGA GTATCTACAT GGATACATTA 1080
AATATATTTA TGCGAGTTCC AACTATGCTC GCAACTGGAG GCAACAGAAA GAAATGAAGT 1140
GACTCAGCTT CTGGCTTCTC TGCTACATCA AATATCTTGT TTAATGGGGC AGATATGCAT 1200
TAAATAGTTT GTACAAGCAG CTTTCGTTGA AGTTTAGAAG ATAAGAAACA TCTCATCATA 1260
TTTAAATGTT CCGGTAATGT GATGCCTCAG GTCTGCCTTT TTTTCTGGAG AATAAATGCA 1320
GTAATCCTCT CCCAAATAAG CACACACATT TTGAATTCTC ATCTTTGAGT GATTTTAAAA 1380
TGTTTTGGTG AATGTGAAAA CTAAAGTTTC TGTCATGAGA ATGTAAGTCT TTTTTCTACT 1440
TTAAAATTTA GTAGGTTCAC TGAGTAACTA AAATTTAGCA AACCTGTGTT TGCATATTTT 1500
TTTGGAGTCC AGAATATTGT AATTAATGTC ATAAGTGATT TGGAGCTTTG GTAAAGGGAC 1560
CAGAGAGAAG GAGTCACCTG CAGTCTTTTC TTTTTTTAAA TACTTAGAAC TTAGCACTTG 1620
TGTTATTGAT TAGTGAGGAG CCAGTAAGAA ACATCTGGGT ATTTGGAAAC AAGTGGTCAT 1680
TGTTACATTC ATCTGCTGAA CTTAACAAAA CTGTTCATCC TGAAACAGGC ACAGGTGATG 1740
CATTCTCCTG CTGTTGCTTC TCAGTGCTCT CTTTCCAATA TAGATGTGGT CATGTTTGAC 1800
TTGTACAGAA TGTTAATCAT ACAGAGAATC CTTGATGGAA TTATATATGT GTGTTTTACT 1860
TTTGAATGTT ACAAAAGGAA ATAACTTTAA AACTATTCTC AAGAGAAAAT ATTCAAAGCA 1920
TGAAATATGT TGCTTTTTCC AGAATACAAA CAGTATACTC ATGATTGCTA AGTGTTTTTT 1980
TATTTTTGCA TATTTATTGA ACTCTCTAAT TGAATACAGC TTGCTCTTGT CACCTCTTCA 2040
AGCTTTCAAG CCTTTATAGA AAAGCTTCTT TGTGGCTTAC ACTGGAAATT ATCAAAGCAG 2100
TTTTTCTCCT AAGACTTTTG GTTTCTCGCA TTGCCTCTCA GACTAAGCAC TAAAAAGCAA 2160
AGCAAAACAG AACTAGTNCT GTCTTAATGA AATATATCAA CCCAAAAGTG TAATGAGGAA 2220
AATGCTTCAT TAGTTTCCCC TAGCAGACTT TTACTTCTCT TACACTGCTA CACCATTACT 2280
TTCTTGAGAC ATTTGTAAGT CCTTTGATAC AGAAGAGTTA TATTTAGGAG GNCTTTAATG 2340
AAGGG 2345
(2) INFORMATION FOR SEQ ID NO: 302:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2369 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 302:
TTTTTTTTTT TTTTTTTTTT TTTTTNCAAG ATCATTGTTT ATTTATTACT TCAGATAAAA 60
AGATAGTATA CATATTAGGG AATCCCTTAA AATTCAACTC TAGAGTTATA CACCATCTAG 120
TACTTTTGCA ATGAATGTTA ACAACAACAA AAAAAATCTC TAAACACCTG AAAGCCCCAC 180
TATTAACATG GACTATGGTA ATAAAAAATT TTGACATTTA ATTTGTTCAA CATATAGTAT 2 0
TTACATTATG AAACCAATGG TGATGATACA ATAAAGTGAT AAAGAAATAG TAAAAATAAA 300
CTTTAAAAAG CAAAGGTTTA TAGTCTGACA ATGCTAATTA TCCTAATTGT ATATAAAAAA 360
TTAAAACATA GAGCTTTCTG TTACAAAATT CTTAATCCTC TGGGTTGTAA TCATTACTTG 420
CTACCAATTT ACATGCAACA TCTGCTAGGA CTGACATTTG ATTTTTTTCC CCAAGAATGT 480
GTGAGTAGAT AAATGACATT TCAGAGCAGA TATTAATTTA CTTGTGGACA GAAAAAGAAA 540
CTCAAGATTC GTACTGGTCA CAAGCCTCTT CCCAATAGAA ATTATAAAAA CAGTAAGATA 600
AAATTTAAAA AAAATCTAAA AAGGGGATGC ATAGGCAAAG AGTACCATAA ATGGCACAGC 660
TCAAAAAATC CCAGGACCAA TCAGACACAC ATCTTTTCTC TCTCCTTCAG CGACAAGAGG 720
TCGATTTTGC CATCAAATAA CCATGATTGA AGCAAGCGAG GGGCACCAGG TGTACAACTG 780
ATTAGATCTT GCAAAATACT AAGATGGGAG CAGGGGTGGC CAGAAGAAGG GCTAATTTAT 840
ATATAATTCA AACTATATAC AGCATAAATG GAATGCAGCC CATCCCAAAC TGGCTCTGTG 900
AAACAATTGG ACCTTTATAG TTAAAATTAT AACAAGTGTA ATAATACAAT AGATTTACAT 960
GGGAAGCAAA ATCCAAGGGA CATTTTATAT TAAGTATTTA CTCTGCTCTT TCAATTTAAA 1020
AATAATTTTG CTAAGTATAC ATCTCAACTG AAGTCTATGT AAAAAATCTC CTAATAGATA 1080
CAGATATTTA CCTTTGGTCA GTTGAAGGCC TTTTTGTGAC TTCTCTCTGA ACTGTAGGCA 1140
GAATCCTAGA TGTACATGCA CATATGGAGA AACTCAAGCT GAGGTCATCC AAAAGCTGTG 1200
CGTATGAGGA GGCTGGAGGT ACTTTGAAAG TCAAAGTAGA CCAGAAACCC AAAACAGGTA 1260
ACAGTGAGGA TGGCAACAGG GAATGGAATG CCAATATGGC AGTAAAACTT TTTTTAAAAA 1320
CAGAAAGAGG AAGGCCTCTC GTACCAGCAG AATCCTGTAC ACGTACAAAA AAGAAAAAGC 1380
CACCCACCAT TTTGTAAAAC AGAAGCCAAT TATAGTGTCG GAAAGTACAA ATTACAGAAA 1440
ACCAGAAGTC AACAGAAGAA AAACTACTGG TTTACTTGAG AGAAAGGAGA ATGGTTCACC 1500
CCGAGCAGAG TTACTTGGTC AACGCCGCCA CCACCGCCCA CAGAACCTCA TTGGTGTTGG 1560
CCTTCAGACA TTCCACTTCA GGGTCTAAGT CGAGAARNTG CCGCACTCTC TTGGTAGCCA 1620
AATCATACTG CTCGTCCAGA AGAGGAGCAA AAGCATTCTC CAGGACGTCC GAGGCATGAG 1680
CCAGGTAAAT GAGGGCCAGC AAGCGCCTGT CCATGCGGTG AGGGTCATTC ACCCATTTGT 1740
CAAGAACGGC TTCCTGTACT TTCTTGATGA GGCGCTGCTT AATGTTCTTA TTGGTGAGGG 1800
GATGTGTTGT CATGTCAAAA AGTAGGAAGT TCTGTTTCTC TGTTGTCAAT ACACCCTTTT 1860
CCACCAGGTT TTTAGCTAAT CGTTCCCGTA CATTTCTTAA CTGATAATGC AATTTTAATG 1920
GATTCCATGT CTCACCACTA AGTAATTCAA TCCAGTTCTG GACCGTTTCT GGAGGCTGAG 1980
TTTCCTTAAC ATGCTTCAGA GCTTCATCAA GAAGAACATC CCCTGTTGGA GCATCTGACT 2040
TACAGATTAC CTTTCTTGTT AATAGACTTT TACGTCTCAT TCCACAAGCC TCTAGTTGTA 2100
ACCTTCCTCT CAATGCTAAT TCAATTAACA TACAGCCACG TAATCCAGAT GATATACAGT 2160
CATTCCAAAA TCATCTCTAA ACCTTCGCGG TCCTTGAGGC CCAGCAGGAG CACTTCCTCC 2220
ATCAGGGTCA GCCGCGTTTC CTTGGAGTCG CCCTTGTCGT CGTCGTCCTG CTCGTCGCGG 2280
CGGCTCTGCG CGTCGTCCTC GCTGCTAGCC GCGCCGCCGC CCGCCGCCCG CTCCTTGTCG 2340
GCGGCGTTGC GGGAGGCCTC GGTGCGCCG 2369
(2) INFORMATION FOR SEQ ID NO: 303:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1181 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS : double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 303:
GGGACGTGTG GTTTCAGCTC GTGCGCCTCC CCGTGGGTTT GCGACGTTTA GCGACTATTG 60
CGCCTGCGCC ACGCCGGCTG CGAGACTGGG GCCGTGGYTG CTGGTCCCGG GTGATGCTAG 120
GCGGCTCCCT GGGCTCCAGG CTGTTGCGGG GTGTAGGTGG GAGTCACGGA CGGTTCGGGG 180
CCCGAGGTGT CCGCGAAGGT GGCGCACATG GGCGGCAGGG GAGAGCATGG CTCAGCGGAT 240
GGTCTGGGTG GACCTGGAGA TGACAGGATT GGACATTGAG AAGGACCAGA TTATTGAGAT 300
GGCCTGTCTG ATAACTGACT CTGATCTCAA CATTTTGCCT GAAGGTCCTA ACCTGATTAT 360
AAAACAACCA GATGAGTTCC TGGACAGCAT GTCAGATTGG TGTAAGGAGC ATCACGGGAA 420
GTCTGGCCTT ACCAAGGCAG TGAAGGAGAG TACAATTACA TTCCAGCAGG CAGAGTATGA 480
ATTTCTGTCC TTTGTACGAC AGCAGACTCC TCCAGGGCTC TGTCCACTTG CAGGAAATTC 540
AGTTCATGAA GATAAGAAGT TTCTTGACAA ATACATGCCC CAGTTCATGA AACATCTTCA 600
TTATAGAATA ATTGATGTGA GCACTGTTAA AGAACTGTGC AGACGCTGGT ATCCAGAAGA 660
ATATGAATTT GCACCAAAGA AGGCTGCTTC TCATAGGGCA CTTGATGACA TTAGTGAAAG 720
CATCAAAGAG CTTCAGTTTT ACCGAAATAA CATCTTCAAG AAAAAAATAG ATGAAAAGAA 780 GAGGAAAATT ATAGAAAATG GGGAAAATGA GAAGACCGTG AGTTGATGCC AGTTATCATG 840 CTGCCACTAC ATCGTTATCT GGAGGCAACT TCTGGTGGTT TTTTTTTCTC ACGCTGATGG 900 CTTGGCAGAG CMCTTCGGTT AACTTGCATC TCCAGATTGA TTACTCAAGC AGACAGCACA 960 CGAAATACTA TTTTTCTCCT AATATGCTGT TTCCATTATG ACACAGCAGC TCCTTTCTAA 1020 GTACCAGGTC ATGTCCATCC CTTGGTACAT ATATGCATTT GCTTTTAAAC C^TTTCTTTT 1080 GTTTAAATAA ATAAATAAGT AAATAAAGCT AGTTCTATTG AAATGCAAAA AAAAAAAAAA 1140 AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA N 1181
(2) INFORMATION FOR SEQ ID NO: 304:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1537 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 304:
CTTTTTCTGT TCCGGCCGAT CCCACCTCTC CTCGACCCTG GACGTCTACC TTCCGGAGGC 60
CCACATCTTG CCCACTCCGC GCGCGGGGCT AGCGCGGGTT TCAGCGACGG GAGCCCTCAA 120
GGGACATGGC AACTACAGCG GCGCCGGCGG GCGGCGCCCG AANATGGAGC TCGCCCGGAA 180
TCGGGAGGGT TCGAAGAAAA CATCCAGGGC GGAGGCTGAG CTGTGATTGA CATGGAGAAC 240
ATGGATGATA CCTCAGGCTC TAGCTTCGAG GATATGGGTG AGCTGCATCA GCGCCTGCGC 300
GAGGAAGAAG TAGACGCTGA TGCAGCTGAT GCAGCTGCTG CTGAAGAGGA GGATGGAGAG 360
TTCCTGGGCA TGAAGGGCTT TAAGGGACAG CTGAGCCGGC AGGTGGCAGA TCAGATGTGG 420
CAGGCTGGGA AAAGACAAGC CTCCAGGGCC TTCAGCTTGT ACGCCAACAT CGACATCCTC 480
AGACCCTACT TTGATGTGGA GCCTGCTCAG GTGCGAACAG GGCTCCTGGA GTCCATGATC 540
CCTATCAAGA TGGTCAACTT CCCCCAGAAA ATTGCAGGTG AACTCTATGG ACCTCTCATG 600
CTGGTCTTCA CTCTGGTTGC TATCCTACTC CATCGGATCA AGACGTCTGA CACTATTATC 660
CGGGAGGGCA CCCTGATGGG CACAGCCATT GGCACCTGCT TCGGCTACTG GCTGGGAGTC 720
TCATCCTTCA TTTACTTCCT TGCCTACCTG TGCAACGCCC AGATCACCAT GCTGCAGATG 780
TTGGCACTGC TGGGCTATGG CCTCTTTGGG CATTGCATTG TCCTGTTCAT CACCTATAAT 840
ATCCACCTCC ACGCCCTCTT CTACCTCTTC TGGCTGTTGG TGGGTGGACT GTCCACACTG 900
CGCATGGTAG CAGTGTTGGT GTCTCGGACC GTGGGCCCCA CACAGCGGCT GCTCCTCTGT 960
GGCACCCTGG CTGCCCTACA CATGCTCTTC CTGCTCTATC TGCATTTTGC CTACCACAAA 1020
GTGNTAGAGG GGATCCTGGA CACACTGGAG GGCCCCAACA TCCCGCCCAT CCAGAGGGTC 1080
CCCAGAGACA TCCCTGCGAT GCTCCCTGCT GCTCGGCTTC CCACCACCGT CCTCAACGCC 1140
ACAGCCAAAG CTGTTGCGGT GACCCTGCAG TCACACTGAC CCCACCTGAA ATTCTTGGCC 1200
AGTCCTCTTT CCCGCAGCTG CAGAGAGGAG GAAGACTATT AAAGGACAGT CCTGATGACA 1260
TGTTTCGTAG ATGGGGTTTG CAGCTGCCAC TGAGCTGTAG CTGCGTAAGT ACCTCCTTGN 1320
AGCTGTCGGC ACTTCTGAAA GCACAAGGCC AAGAACTCCT GGCCAGGACT GCAAGGCTCT 1380
GCAGCCAATG CAGAAAATGG GTCAGCTCCT TTGAGAACCC CTCCCCACCT ACCCCTTCCT 1440
TCCTCTTTAT CTCTCCCACA TTGTCTTGCT AAATATAGAC TTGGTAATTA AAAAAAAAAA 1500
AAAAAAAAAA AAAAAAAAAA AAAAAAGGGG GGNCCCC 1537
(2) INFORMATION FOR SEQ ID NO: 305:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1493 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 305:
TCCATGCCAA AACCAATGCC TGCCAAACAA AATCTTAGAC ATCCCAATAT AATATGTTAG 60
TTATATTTCT ATTCACATCA TTATTGAAAA TACCCAGCTC AGTGCCTGGC TTAATAAATG 120
TTTAATTCCC TTACCTACTC TTGCTCTATT TTTTTATTTG AAATGGAGAT GAGCAAAATA 180
ACACATTCAT GGCTGAAGCA ATTTTTTGGA CATTTCTTCT TACCAAAAGA TCTATAATCA 240
GGATGATCCT GAGCTGTTCA AACAAGCTGT ATATAAACAG ACAATGAAAC TCTTTGCAGA 300
GCTGGAAATT AAAAGGAAAG AGAGAGAAGC CAAAGAGATG CATGAAAGGA AACGACAAAG 360
GGAAGAAGAG ATTGAAGCTC AAGAAAAAGC CAAACGGGAA AGAGAGTGGC AGAAAAACTT 420
TGAGGAAAGT CGAGATGGTC GTGTGGACAG CTGGCGAAAC TTCCAAGCCA ATACGAAGGG 480
GAAGAAAGAG AAGAAAAATC GGACCTTCCT GAGACCACCG AAAGTAAAAA TGGAGCAACG 540
TGAGTGACCG CCCAAGGTCA CAGGCACAGA ACCTTTCCCC TGCTATCTCC CTTCCTGCTT 600
CGAAGGACTC ATTCTTTCCT CCCACTTCCA CCCCAACATA GAGTAGTATT TGCTTTTTAG 660
TCCATTTTGT TTTCAATACG ATTTAATATC GATCAGAGTA ATTCTTTTGT ACATTGAAAT 720
GAGGGGCTTG GTTTAAAAAA AGACCTTTCC CTCTCCCTGC CCCTAGAACA ACCAGTATTA 780
GAAGGTGCCA CCATTGGTGC TGCCTTCTCT TCCCACAGCC TGTAACTCAG TGTTTTGTAC 840
TTCACTGAAT TCTGATGGTT AGAAACTTCG TGGATAGTTT GTCGAAATCA TCCAATTAAA 900
CATACTGCTT AAAACAGTGT TGCTGTGACT TCAGAGACAA GCCTGGAAGG GGCACCTTAG 960
GAAGCCCCTT CGCTTCAGTT GCTCGCTTCT GGGTGTGCTC CCTTCGAAGG CCCAGATAAG 1020
ACAGGGAACA CTTGTGAGCA CACAGAGCAG CATCTGATGC CCTGTGGTGT TTGGCATGTG 1080
CCCCCTGTCT ACTGACCAAT CAGTGTGGCA TGAGGCCCAC GCCACCCAAA CCTTTCACTT 1140
TCCAAAGAGC TAGCCGTCCT CCACCCAGTA CCATGTCCTA GCCTGTCTGC ATTTGTTAGT 1200
GGTAATATTC TTTATGTATA ATAAATTTTT ATACCCAAGC CATTGATGTA CTTTTCCTTG 1260
TACTCTCCCT TGTGGGTCCC TTGTCTGGCT TGGCTGAACC CCAAAATGCT TTGGGGTTGG 1320
ACAGACCTGG CTGAACCTTA GTTTCTTCAT CTATGAAATC GGAATATGAA TTACTGCAGC 1380
AGCTTTTAGG GCAGATTTGC CATGGCATAT ACAAGGTAAC TACCATAGTG CTCCTTGGGT 1440
ATTGCCAATA TCCTATTATT TCTGTGTAAA ATGAAGATAC TCATTGTTTT GAG 1493
(2) INFORMATION FOR SEQ ID NO: 306:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 577 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO 306: AATTCGGCAG AGGNATTATA TACACTATAC TGGCATTTAC TGTTTCACCC AGCCCGGAAA 60 GTCAGAGATG TATATTGGAA AATTTACAAC TGCATCTACA TTGGTTCCCA GGACGCTCTC 120 ATAGCACATT ACCCAAGAAT CTACAACGAT GATAAGAACA CCTATATTCG TTATGAACTT 180 GACTATATCT TATAATTTTA TTGTTTATTT TGTGTTTAAT GCACAGCTAC TTCACACCTT 240 AAACTTGCTT TGATTTGGTG ATGTAAACTT TTAAACATTG CAGATCAGTG TAGAACTGGT 300 CATAGAGGAA GAGCTAGAAA TCCAGTAGCA TCATTTTTAA ATAACCTGTC TTTCTTTTTG 360 ATGTTAAACA GTAAATGCCA GTAGTGACCA AGAACACAGT GATTATATAC ACTATACTGG 420 AGGGATTTCA TTTTTAATTC ATCTTTATGA AGATTTAGAA CTCATTCCTT GTGTTTAAAG 480 GGAATGTTTA ATTGAGAAAT AAACATTTGT GWACAAAATC YTAAAAAAAA AAAAAAAAAA 540 AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AACTCGA 577
(2) INFORMATION FOR SEQ ID NO: 307:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2860 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 307:
GTCTNGACCG CTCTCNCAAT ATGCCTCCCC CGGGCTGGCA GRWRKTCRGT CWCKRGTGGC 60
TAGCCTGTCC TCACAGGGGA GAGTTAAGCT CCCGTTCTCC ACCGTCCCGC CTCGCCAGGT 120
GGGCTGAGGG TGACCGAGAG ACCAGAACCT GCTTGCTGGA GCTTAGTGCT CAGAGCTGGG 180
GAGGGAGGTT CCGCCGCTCC TCTGCTGTCA GCGCCGGCAG CCCCTCCCGG CTTCACTTCC 240
TCCCGCAGCC CCTGCTACTG AGAAGCTCCG GGATCCCAGC AGCCGCCACG CCCTGGCCTC 300
AGCCTGCGGG GCTTCCAGTC AGGCCAACAC CGAGGCGCAC TGGGGAGGAA GACAGGACCC 360
TTGACATCTC CATCTGCACA GAGGTCCTGG CTGGAACCGA GCAGCCTCCT CCTCCTAGGA 420
TGACCTCACC CTCCAGCTCT CCAGTTTTCA GGTTGGAGAC ATTAGATGGA GGCCAAGAAG 480
ATGGCTCTGA GGCGGACAGA GGAAAGCTGG ATTTTGGGAG CGCGCTGCCT CCCATGGAGT 540
CACAGTTCCA GGGCGAGGAC CGGAAATTCG CCCCTTCAGA TAAGAGTCAA CCTCCAACTA 600
CCGAAAGGGA ACAGGTGCCA GTCAGCCGGA TCCAAACCGA TTTCACCGAG ATCGGCTCTT 660
CAATGCGGTC TCCCGGGGTG TCCCCGAGGA TCTCGCTGGA CTTCCAGAGT ACCTGAGCAA 720
GACCAGCAAG TACCTCACCG ACTTCGGAAA TACACAGAGG GCTCCACAGG TAAGACGGCC 780
TGATGAAGGC TGTGCTGAAA CCTTAAGGAC GGGGTCAATG CCTGCATTCT GCCACTCCTG 840
CAGATCGACC GGGACTCTGG CAATCCTCAG CCCCTGGTAA ATGCCCAGTG CACAGATGAC 900
TATTACCGAG GCCACAGCGC TCTGCACATC GCCATTGAGA AAGAGGAGTC TGCAGTCTCT 960
GAAGCTCCTG GTGGAGAATG GGGCCAATGT GCATGCCCGG GTCTGCGGCG ACTTCTTCCA 1020
GAAGGGCCAA GGGACTTGCT TTTATTTCGG TGAGCTACCC CTCTCTTTGG CCGCTTGCAC 1080
CAAGCAGTGG GATGTGGTAA GCTACCTCCT GGAGAACCCA CACCAGCCCG CCAGCCTGCA 1140
GGCCACTCAC TCCCAGGGCA ACACAGTCCT GCATGCCCTA GTGGATGATC TCGGACAACT 1200
CAGCTGAGAA CATTGCACTC GTGACCAGCA TGTATGATGG GCTCCTCCAA GCTKGCGSCC 1260
SCCYTCTGCC CTACCGTCCA GCTTGAGCAC ATCCGCAACC TGCAGGATCT CACGCCTCTC 1320
AAGCTGGCCG CCAAGGAGGG CAAGATCGAG ATTTTCAGCC ACATCCTGCA GCGGGAGTTT 1380
TCAGGACTCA GCCACCTTTC CCGAAAGTTC ACCGAGTGGT GCTATGGGCC TGTCCGGGTG 1440
TCGCTGTATG ACCTGGCTTC TGTGGACAGC TGTGAGGAGA ACTCAGTGCT GGAGATCATT 1500
GCCTTTCATT GCAAGAGCCC GCACCGACAC CGAATCGTCG TTTTGGAGCC CCTGAACAAA 1560
CTCCTGCAGG CGAAATGGGA TCTGCTCATC CCCAAGTTCT TCTTAAACTT CCTGTGTAAT 1620
CTGATCTACA TCTTCATCTT CACCGCTGTT GCCTACCATC AGCCTACCCT GAAGAAGCAG 1680
GCCGCCCCTC ACCTGAAAGC GGAGGTTCGA AACTCCATGC TGCTGACGGG CCACATCCTT 1740
ATCCTCCTAG GGGGGATCTA CCTCCTCGTG GGGCCAGCTG TGGTACTTCT GGCGGCGCCA 1800
CGTGTTCATC TGGATCTCGT TCATAGACAG CTACTTTGGA AATCCTCTTC CTGTTCCAGG 1860
CCCTGCTTCA CAGTGGTGTC CCAGGTGCTG TGTTTCCTGG GCCATCGAGT GGTACCTGCC 1920
CCTCCTTGTG TCTGCGCTCG TGGCTGGGCT GGCTGAACCT GCTTTACTAA TACACGTGGC 1980
GTTCCAGCAC ACAGGCAGTC TACAGTTTCA TGWTCCCTGA AGCCCTGGTG AGCCTGAGCC 2040
AGGAGGCTTG GCGCCCCGAA GCTCCTACAG GCCCCAATGC CACAGAGTCA GTCCAGCCCA 2100
TCGAGGGACA GGAGGACGAG GGCAACGGGG CCCAGTACAG GGGTATCCTG GAAGCCTCCT 2160
TGGAGCTCTT CAAATTCACC ATCGGCATGG GCGAGCTGGC CTTCCAGGAG CAGCTGCACT 2220
TCCGCGGCAT GGTGCTGCTG CTGCTGCTGG CCTACGTGCT GCTCACCTAC ATCCTCCTGC 2280
TCAACATGCT CATCGCCCTC ATGAAGCGAA CGTCACAGTC TCGCCACTGA CAGCTGGAGC 2340
ATCTGGAAGC TGCAGAAAGC CATCTCTGTC CTGGAGATGG AGAATGGCTA TTGGTGGTGC 2400
AGGAAAAAGC AGCGGGCAGG TGTGATGCTG ACCGTTGGCA CTAAGCCCAG ATGGCAGCCC 2460
CGATGAGCGC TCGTGCTTCA GGGTGGAGGA GGTGAACTGG GCTTCATGGG GAGCAGACGC 2520
TGCCTACGCT GTCTGAGGAC CCGTCAGGGG CAGGTGTCCC TCGAACTCTC GAGAACCCTG 2580
TCCTGGCTTC CCCTCCCAAG GAGGATGAGG ATGGTGCCTC TGAGGAAAAC TATGTGCCCG 2640
TCCAGCTGCT CCAGTCCAAC TGATGGCCCA GATGCAGCAG GAGGCCAGAG GACAGAGCAG 2700
AGGATCTTTC GAACCACATC TGCTGGCTCT GGGGTCCCAG TGAATTCTGG TGGCAAATAT 2760
ATATTTTCAC TAACTCAAAA AAAAAAAAAA AAAAAAAAAA AAAAVGAGGG GGGGCCCGKT 2820
ASCCAAWTTC GCCCTATAAG TGAGTGCCWA TTACGATAAA 2860
(2) INFORMATION FOR SEQ ID NO: 308:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 876 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 308:
CTGCTTGTGT CTGCGCTGGT GCTGGGCTGG CTGAACCTGC TTTACTATAC ACGTGGCTTC 60
CAGCACACAG GCATCTACAG TGTCATGATC CAGAAGCCCT GGTGAGCCTG AGCCAGGANN 120 TTGGCGCCCC GAAGCTCCTA CAGGCCCCAA TGCCACAGAG TCAGTGCAGC CCATGGAGGG 180
ACAGGAGGAC GAGGGCAACG GGGCCCAGTA CAGGGGTATC CTGGAAGCCT CCTTGGAGCT 240
CTTCAAATTC ACCATCGCCA TGGGCGAGCT GGCCTTCCAG GAGCAGCTGC ACTTCCGCGG 300
CATGGTGCTG CTGCTGCTGC TGGCCTACGT GCTGCTCACC TACATCCTGC TGCTCAACAT 360
GCTCATCGCC CTCATGNAGC GAGACCGWCA ACAGTGTCGC CACTGACAGC TGGAGCATCT 420 GGAAGCTGCA GAAAGCCATC TCTGTCCTGG AGATCGAGAA TGGCTATTGG TGGTGCAGGA 480
AGAAGCAGCG GGCAGGTCTG ATGCTGACCG TTGGCACTAA GCCAGATGGC AGCCCCGATG 540
AGCGCTGGTG CTTCAGGGTG GAGGAGGTGA ACTGGGCTTC ATGGGAGCAG ACGCTGCCTA 600
CGCTGTCTGA GGACCCGTCA GGGGCAGGTG TCCCTCGAAC TCTCGAGAAC CCTGTCCTGG 660
CTTCCCCTCC CAAGGAGGAT GAGGATGGTG CCTCTGAGGA AAACTATGTG CCCGTCCAGC 720 TCCTCCAGTC CAACTGATGG CCCAGATGCA GCAGGAGGCC AGAGGACAGA GCAGAGGATC 780
TTTCCAACCA CATCTGCTGG CTCTGGGGTC CCAGTGAATT CTGGTGGCAA ATATATATTT 840
TCACTAAMWM AAAAAAAAAA AAAAAAAAAA ACTCGA 876
(2) INFORMATION FOR SEQ ID NO: 309:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2025 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 309:
CATGACCCGC CTGATGCGAT CCCGCACAGC CTCTGGTTCC AGCGTCACTT CTCTGGATGG 60
CACCCGCAGC CGCTCCCACA CCAGCGAGGG CACCCGAAGC CGCTCCCACA CCAGCGAGGG 120
CACCCGCAGC CGCTCGCACA CCAGCGAGGG GGCCCACCTG GACATCACCC CCAACTCGGG 180 TGCTGCTGGG AACASGCCGG GCCCAAGTCC ATGGAGGTCT CCTGCTAGGC GGCCTGCCCA 240
GCTGCCGCCC CCGGACTCTG ATCTCTGTAG TGGCCCCCTC CTCCCCGGCC CCTTTTCGCC 300
CCCTGCCTGC CATACTGCGC CTAACTCGGT ATTAATCCAA AGCTTATTTT GTAAGAGTGA 360
GCTCTGGTGG AGACAAATGA GGTCTATTAC GTGGGTGCCC TCTCCAAAGG CGGGGTGGCG 420
GTGGACCAAA GGAAGGAAGC AAGCATCTCC GCATCGCATC CTCTTCCATT AACCAGTGGC 480 CGGTTGCCAC TCTCCTCCCC TCCCTCAGAG ACACCAAACT GCCAAAAACA AGACGCGTAC 540
AGCACACACT TCACAAAGCC AAGCCTAGGC CGCCCTGAGC ATCCTGCTTC AAACGGGTGC 600
CTGGTCAGAA GGCCAGCCGC CCACTTCCCG TTTCCTCTTT AACTGAGGAG AAGCTGATCC 660
AGTTTCCGGA AACAAAATCC TTTTCTCATT TGGGGAGGGG GGTAATAGTG ACATGCAGGC 720
ACCTCTTTTA AACAGGCAAA ACAGGAAGGG GGAAAAGGTG GGATTCATGT CGAGGCTAGA 780
GGCATTTGGA ACAACAAATC TACGTAGTTA ACTTGAAGAA ACCGATTTTT AAAGTTGGTG 840
CATCTAGAAA GCTTTGAATG CAGAAGCAAA CAAGCTTGAT TTTTCTAGCA TCCTCTTAAT 900
GTGCAGCAAA AGCAGGCRAC AAAATCTCCT GCCTTTACAG ACAAAAATAT TTCAGCAAAC 960
GTTGGGCATC ATGGTTTTTG AAGGCTTTAG TTCTGCTTTC TGCCTCTCCT CCACAGCCCC 1020
AACCTCCCAC CCCTGATACA TCAGCCACTG ATTATTCTTG TTCAGGGAGA AGATCATTTA 1080
GATTTGTTTT GCATTCCTTA GAATGGAGGG CAACATTCCA CAGCTGCCCT GGCTGTGATG 1140
AGTGTCCTTG CAGGGGCCGG AGTAGGAGCA CTGGGGTGGG GGCGGAATTG GGGTTACTCC 1200
ATGTAAGGGA TTCCTTGTTG TTGTGTTGAG ATCCAGTGCA GTTGTGATTT CTGTGGATCC 1260
CAGCTTGGTT CCAGGAATTT TGTGTGATTC GCTTAAATCC AGTTTTCAAT CTTCGACAGC 1320
TGGGCTGGAA CGTGAACTCA GTAGCTGAAC CTGTCTGACC CGGTCACGTT CTTGGATCCT 1380
CAGAACTCTT TGCTCTTGTC GGGGTGGGGG TGGGAACTCA CGTGGGGAGC GGTGGCTGAG 1440
AAAATGTAAG GATTCTGGAA TACATATTCC ATGGGACTTT CCTTCCCTCT CCTGCTTCCT 1500
CTTTTCCTGC TCCCTAACCT TTCGCCGAAT GGGGCAGCAC CACTGACGTT TCTGGGCGGC 1560
CAGTGCGGCT GCCAGGTTCC TGTACTACTG CCTTGTACTT TTCATTTTGG CTCACCGTGG 1620
ATTTTCTCAT AGGAAGTTTC GTCAGAGTGA ATTGAATATT GTAAGTCAGC CACTGGGACC 1680
CGAGGATTTC TGGGACCCCG CAGTTGGGAG GAGGAAGTAG TCCAGCCTTC CAGGTGGCGT 1740
GAGAGGCAAT GACTCGTTAC CTGCCGCCCA TCACCTTGGA GGCCTTCCCT GGCCTTGAGT 1800
AGAAAAGTCG GGGATCGGGG CAAGAGAGGC TGAGTACGGA TGGGAAACTA TTGTGCACAA 1860
GTCTTTCCAG AGGAGTTTCT TAATGAGATA TTTGTATTTA TTTCCAGACC AATAAATTTG 1920
TAACTTTGCA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA AAAAAAACTC 1980
GAGGGGGGCC CGTACCCAAT TCGCCGTATA TGATCGTAAA CAATC 2025
(2) INFORMATION FOR SEQ ID NO: 310:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 3026 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 310:
TAGGCAGCAC TGAAATATCC TAACCCCCTA AGCTCCAGGT GCCCTGTGGN ACGAGCAACT 60
GGACTATAGC AGGGCTGGGC TCTGTCTTCC TGGTCATAGG CTCACTCTTT CCCCGAAATC 120
TTCCTCTGGA GCTTTGCAGC CAAGGTGCTA AAAGGAATAG GTAGGAGACC TCTTCTATCT 180
AATCCTTAAA AGCATAATGT TGAACATTCA TTCAACAGCT GATGCCCTAT AACCCCTGCC 240
TCGATTTCTT CCTATTAGGC TATAAGAAGT AGCAAGATCT TTACATAATT CAGAGTGGTT 300
TCATTGCCTT CCTACCCTCT CTAATGGCCC CTCCATTTAT TTGACTAAAG CATCACACAG 360
TGGCACTAGC ATTATACCAA GAGTATGAGA AATACAGTGC TTTATGGCTC TAACATTACT 420
GCCTTCAGTA TCAAGGCTGC CTGGAGAAAG GATGGCAGCC TCAGGGCTTC CTTATGTCCT 480
CCACCACAAG AGCTCCTTGA TGAAGGTCAT CTTTTTCCCC TATCCTGTTC TTCCCCTCCC 540
CGCTCCTAAT GCTACGTGGG TACCCAGGCT GGTTCTTGGG CTAGGTAGTG GGGACCAAGT 600
TCATTACCTC CCTATCAGTT CTAGCATAGT AAACTACGGT ACCAGTGTTA CTGGGAAGAG 660
CTGGGTTTTC CTAGTATACC CACTGCATCC TACTCCTACC TGGTCAACCC GCTGCTTCCA 720
GGTATGGGAC CTGCTAAGTG TGGAATTACC TGATAAGGGA GAGGGAAATA CAAGGAGGGC 780
CTCTGGTGTT CCTGGCCTCA GCCAGCTGCC CACAAGCCAT AAACCAATAA AACAAGAATA 840
CTGAGTCAGT TTTTTATCTG GGTTCTCTTC ATTCCCACTG CACTTGGTGC TGCTTTGGCT 900
GACTGGGAAC ACCCCATAAC TACAGAGTCT GACAGGAAGA CTGGAGACTG TCCACTTCTA 960
GCTCGGAACT TACTGTGTAA ATAAACTTTC AGAACTGCTA CCATGAAGTG AAAATGCCAC 1020
ATTTTGCTTT ATAATTTCTA CCCATGTTGG GAAAAACTGG CTTTTTCCCA GCCCTTTCCA 1080
GGGCATAAAA CTCAACCCCT TCGATAGCAA GTCCCATCAG CCTATTATTT TTTTAAAGAA 1140
AACTTGCACT TGTTTTTCTT TTTACAGTTA CTTCCTTCCT GCCCCAAAAT TATAAACTCT 1200
AAGTGTAAAA AAAAGTCTTA ACAACAGCTT CTTGCTTGTA AAAATATGTA TTATACATCT 1260
GTATTTTTAA ATTCTGCTCC TGAAAAATGA CTCTCCCATT CTCCACTCAC TCCATTTGGG 1320
GCCTTTCCCA TTGGTCTGCA TGTCTTTTAT CATTGCAGGC CAGTGGACAG AGGGAGAAGG 1380
GAGAACAGGG GTCGCCAACA CTTGTCTTCC TTTCTCACTG ATCCTGAACA AGAAAGAGTA 1440
ACACTGAGGC GCTCGCTCCC ATGCACAACT CTCCAAAACA CTTATCCTCC TGCAAGAGTG 1500
GGCTTTCCAG GGTCTTTACT GGGAAGCAGT TAAGCCCCCT CCTCACCCCT TCCTTTTTTC 1560
TTTCTTTACT CCTTTGGCTT CAAAGGATTT TGGAAAAGAA ACAATATGCT TTACACTCAT 1620
TTTCAATTTC TAAATTTGCA GGGGATACTG AAAAATACGG CAGGTGGCCT AAGGCTGCTG 1680
TAAAGTTGAG GGGAGAGGAA ATCTTAAGAT TACAAGATAA AAAACGAATC CCCTAAACAA 1740
AAAGAACAAT AGAACTGGTC TTCCATTTTG CCACCTTTCC TCTTCATGAC AGCTACTAAC 1800
CTGGAGACAG TAACATTTCA TTAACCAAAG AAAGTGGGTC ACCTGACCTC TGAAGAGCTG 1860
AGTACTCAGG CCACTCCAAT CACCCTACAA GATGCCAAGG AGGTCCCAGG AAGTCCAGCT 1920
CCTTAAACTG ACGCTAGNMA ATAAACCTGG GCAAGTGAGG CAAGAGAAAT GAGGAAGAAT 1980
CCATCTGTGA GGTGAYAGGC AAGGATGAAA GACAAAGAAG GAAAAGAGTA TCAAAGGCAG 2040
AAAGGAGATC ATTTAGTTGG GTCTGAAAGG AAAAGTCTTT GCTATCCGAC ATGTACTGCT 2100
AGTACCTGTA AGCATTTTAG CTCCCAGAAT GCAAAAAAAA ATCAGCTATT GGTAATATAA 2160
TAATGTCCTT TCCCTGGAGT CAGTTTTTTT AAAAAGTTAA CTCTTAGTTT TTACTTGTTT 2220
AATTCTAAAA GAGAAGGGAG CTGAGGCCAT TCCCTGTAGG AGTAAAGATA AAAGGATAGG 2280
AAAAGATTCA AAGCTCTAAT AGAGTCACAG CTTTCCCAGG TATAAAACCT j&AAATTAAGA 2340
AGTACAATAA GCAGAGGTGG AAAATGATCT AGTTCCTGAT AGCTACCCAC AGAGCAAGTG 2400
ATTTATAAAT TTGAAATCCA AACTACTTTC TTAATATCAC TTTGGTCTCC ATTTTTCCCA 2460
GGACAGGAAA TATGTCCCCC CCTAACTTTC TTGCTTCAAA AATTAAAATC CAGCATCCCA 2520
AGATCATTCT ACAAGTAATT TTGCACAGAC ATCTCCTCAC CCCAGTGCCT GTCTGGAGCT 2580
CACCCAAGGT CANCCAAACA ACTTGGTTGT GAACCCAACT GCCTTAACCT TCTGGGGGAG 2640
GGGGATTAGC TAGACTAGGA GACCCAGAAG TGAATGGGAA AGGGTGAGGA CTTCACAATG 2700
TTGGCCTGTC AGAGCTTGAT TAGAAGCCAA GACAGTGGCA GCAAAGGAAG ACTTGGCCCA 2760
GGAAAAACCT GTGGGTTGTC CTAATTTCTG TCCAGAAAAT AGGGTGGACA GAAGCTTGTG 2820
GGGTGCATGG AGGAATTGGG ACCTGGTTAT GTTGTTATTC TCGGACTGTG AATTTTGGTG 2880
ATGTAAAACA GAATATTCTG TAAACCTAAT GTCTGTATAA ATAATGAGCG TTAACACAGT 2940
AAAATATTCA ATAAGAAGTC AAAAAAAAAA AAAAAAAACT CGAGGGGGGG CCCGGTACCC 3000
AATTTNCCAA ATAGAGATNG TATTAC 3026
(2) INFORMATION FOR SEQ ID NO: 311:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 712 base pairs
(B) TYPE: nucleic acid (C) STRANDEDNESS: double
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 311: GCAGGCTTTG TGCTCACCTA CAAGCTGGGT GAGCAGGGTC CCAGCAGCCT GTTTCCTCTT 60
CTCCTGCTGG ACCACGGCGT TTCTGCTCCC GAGTTGGGAC TGTGGAATGG TGTGGGTGCT 120
GTGGTCTGCT CCATCGCTGG CTCCTCCCTG GGTGGGACCT TGCTGGCCAA GCACTGGAAA 180
CTGCTGCCTC TGTTGARGTC GGTGCTGCGC TTCCGCCTCG GGGGCCTAGC CTGTCAGACT 240
GCCTTGGTCT TCCACCTGGA CACCCTGGGG GCCAGCATGG ACGCTCGCAC AATCTTGAGA 300 GGGTCAGCCT TGCTGAGCCT ATGTCTGCAG CACTTCTTCG GAGGCCTGGT CACCACAGTC 360
ACCTTCACTG GGATGATGCG CTGCAGCCAG CTGGCCCCCA GGCCCTGCAG GCCACACACT 420
ACAGCCTTCT GGCCACGCTG GAGCTGCTGG GGAAGCTGCT GCTGGGCACT CTGCGGAGGC 480
CTGGCTGATG GGTTGGGGCC ACATCCCTGC TTCTTGCTCC TGCTCATCCT CTCTCCCTTT 540
CCCGTTCTGT ACCTGGACCT AGCACCCAGC ACCTTTCTCT GAGCTGAGTG GCTGGAGTGG 600 TCAATAAAGC CACATGTGCC TGTGGCCCAA AAAAAAAAAA AAAAAAAAAA AAAAAAAAAA 660
AACTGGAGGG GGGGCCCGGT ACCCAAATCG CCGGATATGA TCGTAAACAA TC 712
(2) INFORMATION FOR SEQ ID NO: 312:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 1289 base pairs
(B) TYPE: nucleic acid
(C) STRANDEDNESS: double
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 312:
CAAAATTTCA GAACTTTCAG GAGGGCAAGA GAATATCAAA CAAAGATTTC TGGAAGTATT 60 TTGCCAACCT TCTCGTTGAG CTGCAAGAAA ATATTTATGG TGAGAACTTT TCTGTTTCCC 120
GTTATTGGGT TTTTGGTTGG TTTTTCTTTG TTTTTTACTA TGCTTTGGTC TGTAAAAATA 180
TGCAACTGAA CTACATTCAG AAGGAAATAT TGTCTACATA GAATATTATA TCAAGTTGGT 240 ACATAATTCT GATGAGGAAA AAAAATCTTT GCAATTCTTT AAGCCATATT GTTGTTTTTC 300
TCTCTTGTTT TCCCTGGATG AAAATATCAG TATTAAGTAG ACAGCATATT ATTCAAGTGT 360
TTAGACTTAT TAATATGTTC TTGTCCTGTA TTTATACATA TGTGTATTTT GGAAAGTATT 420
GCCTTTTTTA AGGGAAGCTA TAATTCGATA CATAGTGAAA AAGGGAATGG TGACCCCTTT 480
GTGCCTCTTC CACTGAGGAT AACAAACAGC ATTGTAATCC ATTCTCTTGC ACCTTCTTCT 540 TCTTATCTTC TTATTACGGT TTTATTAATT TTGTAGAGGC ACAGGGAGTC GGCAAGGGGA 600
AGAAGCAGCT TATTTCACTA ACCAGCCCCT CTCTGGTCCA CCAGCGTCTT GGCTTGGTGG 660
GAGGGCTCTC AATCAGCAGG GCCCCAGGAG GGAAGAAGAA GTGGGGCAAA GCCTGGCCTC 720
GCCGCTCGGG AGCTTTGCCA TCTGAGCCAC GCCTCCTCCA GGCCATCCTC CTTGAACTTG 780
GAAATGTCAA CCGGAGCCCT TACACCAGCC CTCCAGCATC TAATAGACTT GAATCTACTC 840
TAAACGAATA TTTAATCCAA CCTCACTACA TTGTAGCTCA GTCCAACGAC TAACCCTGAA 900
ATGGGGGTGT TCCAGCCTTC AGCGAGATGG CCAAGCGGTC CCCTGGGGGC TGTGGCAGCG 960
GGCTTATCCT TCTCTGTTGC CAACCTTGCC GTCCGACCTC CTCCGCCCCC ATGCGGTGAC 1020
CCCCTCCCTC TCTGTGTCTG TCCATACGTG TGAGTCCAGC TAAAAAGACA AAACAGAACC 1080
CGTGGGCCCA GCTCGGAAGG TGCGTGGAGA AGGCTCCGAC GTCTCCGAAG TGCAGCCCTT 1140
GGGATGGCAT TGCGTTGTGT GCCTTATTCC TGGAGAATCT GTATACGGCT CGCCTATAGA 1200
AATATAGCCT CTTCATGCTG TATTAAAAGG ACTTTTAAAA GCAAAAAAAA AAAAAAAAAA 1260
CTTGAGGGGG GGNCCGGTAC CCAATTNTC 1289
(2) INFORMATION FOR SEQ ID NO: 313:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 22 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 313:
Met Phe Leu lie Phe Val Tyr Phe Leu Lys lie Leu Phe Ser Ser Ser 1 5 10 15
Leu Pro Phe Leu Trp Leu 20
(2) INFORMATION FOR SEQ ID NO: 314:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 128 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 314:
Met Met Phe Leu Thr Gin Gly Gly Pro Leu Pro Ser Thr Arg Ala Arg 1 5 10 15
Pro Thr Cys Gin Ala Gly Ala Leu Pro Lys Pro Ser Gly Leu Leu Gly 20 25 30
Val Thr Cys Trp Asn Gly Leu Lys Gly Pro Leu Cys Gly Asn Arg Cys 35 40 45
Ser Pro Asn Thr Leu Leu Leu Ala Ala Arg Gin Ala Leu Trp Lys Gly 50 55 60
Arg Gly Arg Thr His Gin Asp Leu Pro Gly Pro Leu Gin Gly Arg Gin
65 70 75 80
Leu Gly Pro Glu Pro Lys His Leu Ala Leu Leu Pro Pro Arg Gly Gin 85 90 95
Glu Ala Ser Trp Ala Ser Ser Leu Pro Gly Gin Gly Pro Leu Pro Leu 100 105 110
Pro His lie Asn Cys Thr Val Phe Ser Leu Lys Ala Ser Phe lie Lys 115 120 125
(2) INFORMATION FOR SEQ ID NO: 315:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 28 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 315: Met Gin Phe Leu Leu Thr Ala Phe Leu Leu Val Pro Leu Leu Ala Leu 1 5 10 15
Cys Asp Val Pro lie Ser Leu Gly Phe Ser Pro Ser 20 25
(2) INFORMATION FOR SEQ ID NO: 316: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 64 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 316:
Met Asp Gly Phe Ser Ser Arg Leu Phe Ser Ser Leu Pro Phe Val Ala 1 5 10 15
Leu Gin Trp Phe lie Val lie Ser His Leu Leu Ser Leu Ser Leu Ser 20 25 30
Ala Cys Cys Tyr Gin Thr His Cys Ser Leu Xaa Gin Leu Ser Ser Ala 35 40 45 Phe Ser Xaa Met Gly Glu Ser Cys Val Gly Glu Arg Glu Tyr Xaa Phe 50 55 60
(2) INFORMATION FOR SEQ ID NO: 317: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 317:
Met Pro Leu lie Asn Leu Leu Leu Leu Tyr Tyr Val Pro Asn Gly Gly 1 5 10 15
Lys Gin Asp Lys Lys 20
(2) INFORMATION FOR SEQ ID NO: 318:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 318:
Met Gly Arg His Leu Val Leu Val Met Phe lie Thr Thr Ser Leu His
1 5 10 15 Ser Gly Thr Pro Val Pro Glu Asn Val lie Cys Gly Val Thr Lys Gly 20 25 30
Pro Gin Gly Lys Lys Lys Lys 35
(2) INFORMATION FOR SEQ ID NO: 319: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 33 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 319:
Met Leu Trp Trp Ser Arg Asp Tyr Thr Met Val Phe Leu Leu Phe Thr 1 5 10 15
Met Val Phe Thr Gly Asp Leu Val lie Arg Gly Arg Thr Glu Leu Ser 20 25 30
Leu
(2) INFORMATION FOR SEQ ID NO: 320:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 88 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 320: Met Val Cys Ser Ser Leu Cys Asp lie Gly Gly lie lie Thr Pro Phe
1 5 10 15
He Val Phe Arg Leu Arg Glu Val Trp Gin Ala Leu Pro Leu He Leu 20 25 30
Phe Ala Val Leu Gly Leu Leu Ala Ala Gly Val Thr Leu Leu Leu Pro 35 40 45
Glu Thr Lys Gly Val Ala Leu Pro Glu Thr Met Lys Asp Ala Glu Asn 50 55 60
Leu Gly Arg Lys Ala Lys Pro Lys Glu Asn Thr He Tyr Leu Lys Val
65 70 75 80 Gin Thr Ser Glu Pro Ser Gly Thr
85
(2) INFORMATION FOR SEQ ID NO: 321:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 23 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 321:
Met Gin Pro Gly Ala Gly Val Leu Val Leu Gly Leu Leu Leu Pro Pro 1 5 10 15
Pro Gin Ser Pro Ser Leu Ser 20
(2) INFORMATION FOR SEQ ID NO: 322:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 322:
Met Thr Phe Thr Leu Gly Asp Ser Gin Val Leu Leu He Asn Leu Phe 1 5 10 15
Pro Ser Met Pro Ser Gly Ser Cys Ala Arg Pro 20 25
(2) INFORMATION FOR SEQ ID NO: 323:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 64 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 323: Met Cys Leu Glu Cys Trp Ala Glu Asn Leu Gly Pro His His Thr Ser
1 5 10 15
Ser Leu Leu Asn Pro Arg His Leu Pro Ser He Pro Ala Met Phe Pro 20 25 30
Val Ser Ser Gly Cys Phe Gin Glu Gin Gin Glu Met Asn Lys Ser Leu 35 40 45
Val Ser Cys Leu Phe Val Leu His Phe Val Leu His Cys He Phe Xaa 50 55 60
(2) INFORMATION FOR SEQ ID NO: 324:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 196 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 324: Met Leu Ser Thr Ser Glu Tyr Ser Gin Ser Pro Lys Met Glu Ser Leu 1 5 10 15
Ser Ser His Arg He Asp Glu Asp Gly Glu Asn Thr Gin He Glu Asp 20 25 30
Thr Glu Pro Met Ser Pro Val Leu Asn Ser Lys Phe Val Pro Ala Glu 35 40 45
Asn Asp Ser He Leu Met Asn Pro Ala Gin Asp Gly Glu Val Gin Leu 50 55 60
Ser Gin Asn Asp Asp Lys Thr Lys Gly Asp Asp Thr Asp Thr Arg Asp 65 70 75 80 Asp He Ser He Leu Ala Thr Gly Cys Lys Gly Arg Glu Glu Thr Val
85 90 95
Ala Glu Glu Val Cys He Asp Leu Thr Cys Asp Ser Gly Ser Gin Ala 100 105 110
Val Pro Ser Pro Ala Thr Arg Ser Glu Ala Leu Ser Ser Val Leu Asp 115 120 125
Gin Glu Glu Ala Met Glu He Lys Glu His His Pro Glu Glu Gly Ser 130 135 140
Ser Gly Ser Glu Val Glu Glu He Pro Glu Thr Pro Cys Glu Ser Gin 145 150 155 160 Gly Glu Glu Leu Lys Glu Glu Asn Met Glu Ser Val Pro Leu His Leu
165 170 175
Ser Leu Thr Glu Thr Gin Ser Gin Gly Leu Cys Leu Arg Arg His Pro 180 185 190
Lys Lys Lys Lys 195
(2) INFORMATION FOR SEQ ID NO: 325:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 252 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 325:
Met Gly Gly Asp Leu Val Leu Gly Leu Gly Ala Leu Arg Arg Arg Lys 1 5 10 15
Arg Leu Leu Glu Gin Glu Lys Ser Leu Ala Gly Trp Ala Leu Val Leu 20 25 30 Ala Xaa Xaa Gly He Gly Leu Met Val Leu His Ala Glu Met Leu Trp 35 40 45
Phe Gly Gly Cys Ser Ala Val Asn Ala Thr Gly His Leu Ser Asp Thr 50 55 60
Leu Trp Leu He Pro He Thr Phe Leu Thr He Gly Tyr Gly Asp Val 65 70 75 80
Val Pro Gly Thr Met Trp Gly Lys He Val Cys Leu Cys Thr Gly Val 85 90 95
Met Gly Val Cys Cys Thr Ala Leu Leu Val Ala Val Val Ala Arg Lys 100 105 110 Leu Glu Phe Asn Lys Ala Glu Lys His Val His Asn Phe Met Met Asp 115 120 125
He Gin Tyr Thr Lys Glu Met Lys Glu Ser Ala Ala Arg Val Leu Gin 130 135 140
Glu Ala Trp Met Phe Tyr Lys His Thr Arg Arg Lys Glu Ser His Ala 145 150 155 160
Ala Arg Xaa His Gin Arg Xaa Leu Leu Ala Ala He Asn Ala Phe Arg 165 170 175
Gin Val Arg Leu Lys His Arg Lys Leu Arg Glu Gin Val Asn Ser Met 180 185 190 Val Asp He Ser Lys Met His Met He Leu Tyr Asp Leu Gin Gin Asn 195 200 205
Leu Ser Ser Ser His Arg Ala Leu Glu Lys Gin He Asp Thr Leu Ala 210 215 220
Gly Lys Leu Asp Ala Leu Thr Glu Leu Leu Ser Thr Ala Leu Gly Pro 225 230 235 240
Arg Gin Leu Pro Glu Pro Ser Gin Gin Ser Lys Xaa 245 250
(2) INFORMATION FOR SEQ ID NO: 326:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 68 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 326:
Met Trp Arg Cys Arg Gly Lys Leu Ser Phe Pro Leu Phe Ala Val Val 1 5 10 15 He Val Ser Cys Arg Lys Asp Gly Pro Asp Ala Ala Ala Ala Pro Ala 20 25 30
Val He Lys Asn Asn Ser His Tyr Gin Thr Ser Lys Ala Leu Glu Leu 35 40 45
Glu Lys Thr Thr Glu Asn Lys Glu Ser Asn Pro Phe He Leu Gin Val 50 55 60
Asn Lys Leu Xaa 65
(2) INFORMATION FOR SEQ ID NO: 327:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 84 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 327:
Met Gly Glu Gly Lys Asn Gly Phe Gly Gly Phe Val His Thr Ala Asp 1 5 10 15 Ala Cys Trp Glu Gly Val His Ser Glu Pro Val Cys Arg Thr Val His 20 25 30
Thr Val His Thr Cys His His Gin Ala Phe Leu Val Leu He Gly Trp 35 40 45
Ser Lys Ser Gly Lys Glu Arg Lys Glu Ala Phe Leu Thr Ala He He 50 55 60
Leu Asn Ser Arg Ser He His He Ser Cys Ser Trp Pro Pro Ser Pro 65 70 75 80
Val Pro Gin Xaa
(2) INFORMATION FOR SEQ ID NO: 328:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 36 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 328: Met Leu Leu He Asn Leu Leu Trp Leu Val Thr Met He Lys Ser Val 1 5 10 15
He Asn Asn Asn He He Leu Phe Leu Lys Lys Lys Ser Leu Phe Phe 20 25 30
He Asp Ser Val 35
(2) INFORMATION FOR SEQ ID NO: 329:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 63 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 329:
Met Thr Phe Pro Phe Glu Lys Lys He Val Ala Phe Ser Ala Phe Tyr 1 5 10 15
Leu He Pro Gly Glu Ser Arg Leu Ala Pro Thr Phe Asn Pro Ser Ala 20 25 30 Asp Met Thr Val He Leu Arg Gly Arg Ala Gin His Lys Thr Ala Met 35 40 45
Leu Glu Ser Tyr Asn Trp Lys Val Ser Cys Gin Leu Arg Glu Xaa 50 55 60
(2) INFORMATION FOR SEQ ID NO: 330: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 35 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 330:
Met His Ser Lys Gly Ser Ser Leu Leu Leu Phe Leu Pro Gin Leu He 1 5 10 15
Leu He Leu Pro Val Cys Ala His Leu His Glu Glu Leu Asn Cys Cys 20 25 30
Phe His Arg 35
(2) INFORMATION FOR SEQ ID NO: 331:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 23 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 331:
Met Gly Ala Leu Val Leu Leu Leu Cys Leu Leu Val Gly Val Gin Gin 1 5 10 15
Ser Gly Ser Val Trp Asp Ser 20
(2) INFORMATION FOR SEQ ID NO: 332: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 40 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 332:
Met Gin Ser Ala Glu He Leu Ser Trp Thr Asp Val Leu His Asp Phe 1 5 10 15
Leu Phe Ser Leu Phe Leu Trp Pro Ala Phe Glu Asp Arg Ala Leu Leu 20 25 30
He Phe Thr Leu Asn Gin He Val 35 40
(2) INFORMATION FOR SEQ ID NO: 333:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 111 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 333: Met Gin Ser Leu Val Gin Trp Gly Leu Asp Ser Tyr Asp Tyr Leu Gin 1 5 10 15
Asn Ala Pro Pro Gly Phe Phe Pro Arg Leu Gly Val He Gly Phe Ala 20 25 30
Gly Leu He Gly Leu Leu Leu Ala Arg Gly Ser Lys He Lys Lys Leu 35 40 45
Val Tyr Pro Pro Gly Phe Met Gly Leu Ala Ala Ser Leu Tyr Tyr Pro 50 55 60
Gin Gin Ala He Val Phe Ala Gin Val Ser Gly Glu Arg Leu Tyr Asp 65 70 75 80 Trp Gly Leu Arg Gly Tyr He Val He Glu Asp Leu Trp Lys Glu Asn
85 90 95
Phe Gin Lys Pro Gly Asn Val Lys Asn Ser Pro Gly Thr Lys Xaa 100 105 110
(2) INFORMATION FOR SEQ ID NO: 334:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 106 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 334:
Met Ala Pro Ser Leu Leu Leu Leu Ala Pro Leu Cys Ser Leu Glu Ala 1 5 10 15
Val Leu Ser Ser Pro Leu Glu Lys Gin Cys Gin Leu Pro Gly He Phe 20 25 30
Cys Gin Leu Gin Leu Pro Cys Pro Leu Leu Leu Ser Ala Gin Leu Leu 35 40 45 Lys Gly He Val Xaa Pro Arg Cys Pro Ala Ser Leu Pro Gin Pro Pro 50 55 60
His Pro Ala Pro Ser Trp His Leu Pro Leu His Cys Thr Glu Arg Xaa 65 70 75 80
Pro His His Leu Pro Leu Gin Gly Gly Ser Ser Asn Met Glu Glu Xaa 85 90 95
Asn Tyr Arg Gly Tyr Xaa Asp Ala Gin Leu 100 105
(2) INFORMATION FOR SEQ ID NO: 335:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 50 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 335:
Met Thr Thr Cys Leu Phe Gly Leu Leu Ser Cys Glu Met Ser Ala Gin 1 5 10 15 Val Ser Gin Lys Ser Cys Val Tyr Asp Glu Ser Glu Cys Phe Ser Ser 20 25 30
Val Gly Gin Leu Leu Ala Leu Leu He Leu Val Tyr Val Leu Pro Ser 35 40 45
He Xaa 50
(2) INFORMATION FOR SEQ ID NO: 336:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 48 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 336:
Met Leu Trp Lys Cys Ser Gin Asn He Ala Arg Cys Leu Leu Leu Leu 1 5 10 15
Leu Ala Leu Val Glu He Lys Leu Glu Asp Leu Gin Ser Gin Leu His 20 25 30 Pro Thr Trp Lys Ser He Pro Gly Pro Ser Pro Arg Asn Gin His Arg 35 40 45
(2) INFORMATION FOR SEQ ID NO: 337: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 41 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 337:
Met Leu He Pro Leu Gin Cys Leu Phe Ser Ser Asp Arg Met Leu Thr 1 5 10 15
Phe Leu Thr Pro Trp Gin Lys Gly Glu Lys Cys Val Leu Gly Trp Val 20 25 30
Thr Lys Phe Leu Ser Glu He Ser Xaa 35 40
(2) INFORMATION FOR SEQ ID NO: 338:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 76 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 338: Met Thr Phe Ser Ser Leu Lys Leu Phe Val Leu Thr Cys He He Lys 1 5 10 15
Gly Leu Glu Arg Phe He He Leu Arg Glu Val Cys Asn Gin Glu He 20 25 30
Gin Arg Ser Leu Ser Ser Asn Leu Val His Val Leu Leu Gin Pro Ala 35 40 45
Thr Phe Lys Asp Val Leu Val Thr Glu He He Cys Leu Cys Met Cys 50 55 60
Leu Tyr Ser He Lys Tyr Met Pro Pro Gin Lys Lys 65 70 75
(2) INFORMATION FOR SEQ ID NO: 339:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 31 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 339: Lys Val Tyr He Phe Leu He Phe Met Val Leu He Leu Pro Ser Leu 1 5 10 15
Gly Leu Thr Arg Tyr Met Pro Pro Xaa Ser Xaa Leu Asn Ser Glu 20 25 30
(2) INFORMATION FOR SEQ ID NO: 340: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 42 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 340:
Met Ala Lys He Ser Pro Phe Glu Val Val Lys Arg Thr Ser Val Pro 1 5 10 15
Val Leu Val Gly Leu Val He Val He Val Ala Thr Glu Leu Met Val 20 25 30
Pro Gly Thr Ala Ala Ala Val Thr Gly Lys 35 40
(2) INFORMATION FOR SEQ ID NO: 341:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 26 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 341: Met Arg Leu Phe Phe He Gly Phe Leu Leu Leu Phe Ser Phe Gly Leu 1 5 10 15
Leu Arg Gin Pro Ser Leu Ser Ala Glu His 20 25
(2) INFORMATION FOR SEQ ID NO: 342: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 26 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 342:
Met Val Phe Ser Val Ser Ser Ala Leu Ala Leu Leu Leu Met Leu Leu 1 5 10 15
Arg Ser Ser Asp Leu Ala Lys Lys Thr Glu 20 25
(2) INFORMATION FOR SEQ ID NO: 343:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 157 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 343:
Met Ser Leu Glu Phe Tyr Gin Lys Lys Lys Ser Arg Trp Pro Phe Ser 1 5 10 15 Asp Glu Cys He Pro Trp Glu Val Trp Thr Val Lys Val His Val Val 20 25 30
Ala Leu Ala Thr Glu Gin Glu Arg Gin He Cys Arg Glu Lys Val Gly 35 40 45
Glu Lys Leu Cys Glu Lys He He Asn He Val Glu Val Met Asn Arg 50 55 60
His Glu Tyr Leu Pro Lys Met Pro Thr Gin Ser Glu Val Asp Asn Val 65 70 75 80
Phe Asp Thr Gly Leu Arg Asp Val Gin Pro Tyr Leu Tyr Lys He Ser 85 90 95 Phe Gin He Thr Asp Ala Leu Gly Thr Ser Val Thr Thr Thr Met Arg 100 105 110
Arg Leu He Lys Asp Thr Leu Pro Ser Glu Arg Arg Trp He Ser Gly 115 120 125
Ser Ser Leu Met Ala Pro Arg Pro Trp Leu Leu Gly He Ala Leu Leu 130 135 140
Gly Leu Trp Ala Leu Glu Pro Ala Leu Gly His Trp Xaa 145 150 155
(2) INFORMATION FOR SEQ ID NO: 344:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 520 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 344:
Met Phe Leu Leu Pro Leu Pro Ala Ala Gly Arg Val Val Val Arg Arg 1 5 10 15 Leu Ala Val Arg Arg Phe Gly Ser Arg Ser Leu Ser Thr Ala Asp Met
20 25 30
Thr Lys Gly Leu Val Leu Gly He Tyr Ser Lys Glu Lys Glu Asp Asp 35 40 45
Val Pro Gin Phe Thr Ser Ala Gly Glu Asn Phe Asp Lys Leu Leu Ala 50 55 60
Gly Lys Leu Arg Glu Thr Leu Asn He Ser Gly Pro Pro Leu Lys Ala 65 70 75 80
Gly Lys Thr Arg Thr Phe Tyr Gly Leu His Gin Asp Phe Pro Ser Val 85 90 95 Val Leu Val Gly Leu Gly Lys Lys Ala Ala Gly He Asp Glu Gin Glu 100 105 110
Asn Trp His Glu Gly Lys Glu Asn He Arg Ala Ala Val Ala Ala Gly 115 120 125
Cys Arg Gin He Gin Asp Leu Glu Leu Ser Ser Val Glu Val Asp Pro 130 135 140
Cys Gly Asp Ala Gin Ala Ala Ala Glu Gly Ala Val Leu Gly Leu Tyr 145 150 155 160
Glu Tyr Asp Asp Leu Lys Gin Lys Lys Lys Met Ala Val Ser Ala Lys 165 170 175 Leu Tyr Gly Ser Gly Asp Gin Glu Ala Trp Gin Lys Gly Val Leu Phe 180 185 190
Ala Ser Gly Gin Asn Leu Ala Arg Gin Leu Met Glu Thr Pro Ala Asn 195 200 205
Glu Met Thr Pro Thr .Arg Phe Ala Glu He He Glu Lys Asn Leu Lys 210 215 220
Ser Ala Ser Ser Lys Thr Glu Val His He Arg Pro Lys Ser Trp He 225 230 235 240
Glu Glu Gin Ala Met Gly Ser Phe Leu Ser Val Ala Lys Gly Ser Asp 245 250 255 Glu Pro Pro Val Phe Leu Glu He His Tyr Lys Gly Ser Pro Asn Ala 260 265 270
Asn Glu Pro Pro Leu Val Phe Val Gly Lys Gly He Thr Phe Asp Ser 275 280 285
Gly Gly He Ser He Lys Ala Ser Ala Asn Met Asp Leu Met Arg Ala 290 295 300
Asp Met Gly Gly Ala Ala Thr He Cys Ser Ala He Val Ser Ala Ala 305 310 315 320
Lys Leu Asn Leu Pro He Asn He He Gly Leu Ala Pro Leu Cys Glu 325 330 335 Asn Met Pro Ser Gly Lys Ala Asn Lys Pro Gly Asp Val Val Arg Ala
340 345 350
Lys Asn Gly Lys Thr He Gin Val Asp Asn Thr Asp Ala Glu Gly Arg 355 360 365
Leu He Leu Ala Asp Ala Leu Cys Tyr Ala His Thr Phe Asn Pro Lys 370 375 380
Xaa He Leu Asn Ala Ala Thr Leu Thr Gly Ala Met Asp Val Ala Leu 385 390 395 400
Gly Ser Gly Ala Thr Gly Val Phe Thr Asn Ser Ser Trp Leu Trp Asn 405 410 415 Lys Leu Phe Glu Ala Ser He Glu Thr Gly Asp Arg Val Trp Arg Met 420 425 430
Pro Leu Phe Glu His Tyr Thr Arg Gin Val Val Asp Cys Gin Leu Ala 435 440 445
Asp Val Asn Asn He Gly Lys Tyr Arg Ser Ala Gly Ala Cys Thr Ala 450 455 460
Ala Ala Phe Leu Lys Glu Phe Val Thr His Pro Lys Trp Ala His Leu 465 470 475 480
Asp He Ala Gly Val Met Thr Asn Lys Asp Glu Val Pro Tyr Leu Arg 485 490 495 Lys Gly Met Thr Gly Arg Pro Thr Arg Thr Leu He Glu Phe Leu Leu 500 505 510
Arg Phe Ser Gin Asp Asn Ala Xaa 515 520
(2) INFORMATION FOR SEQ ID NO: 345: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 345:
Thr He Leu Phe Leu Phe Leu Gin Leu Ser Ala Leu Arg Leu He Val 1 5 10 15
Gly Lys Asp Ser He Asp He Asp He Ser Ser Arg Arg Arg Glu Asp 20 25 30
Gin Ser Leu Arg Leu Asn Ala 35
(2) INFORMATION FOR SEQ ID NO: 346:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 234 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 346:
Met Thr Ser Glu Leu Asp He Phe Val Gly Asn Thr Thr Leu He Asp 1 5 10 15
Glu Asp Val Tyr Arg Leu Trp Leu Asp Gly Tyr Ser Val Thr Asp Ala 20 25 30
Val Ala Leu Arg Val Arg Ser Gly He Leu Glu Gin Thr Gly Ala Thr 35 40 45
Ala Ala Val Leu Gin Ser Asp Thr Met Asp His Tyr Arg Thr Phe His 50 55 60
Met Leu Glu Arg Leu Leu His Ala Pro Pro Lys Leu Leu His Gin Leu 65 70 75 80 He Phe Gin He Pro Pro Ser Arg Gin Ala Leu Leu He Glu Arg Tyr
85 90 95
Tyr Ala Phe Asp Glu Ala Phe Val Arg Glu Val Leu Gly Lys Lys Leu 100 105 110
Ser Lys Gly Thr Lys Lys Asp Leu Asp Asp He Ser Thr Lys Thr Gly 115 120 125
He Thr Leu Lys Ser Cys Arg Arg Gin Phe Asp Asn Phe Lys Arg Val 130 135 140
Phe Lys Val Val Glu Glu Met Arg Gly Ser Leu Val Asp Asn He Gin 145 150 155 160 Gin His Phe Leu Leu Ser Asp Arg Leu Ala Arg Asp Tyr Ala Ala He
165 170 175
Val Phe Phe Ala Asn Asn Arg Phe Glu Thr Gly Lys Lys Lys Leu Gin 180 185 190
Tyr Leu Ser Phe Gly Asp Phe Ala Phe Cys Ala Glu Leu Met He Gin 195 200 205
Asn Trp Thr Leu Gly Pro Val Asp Ser Gin Met Asp Asp Met Asp Met 210 215 220
Asp Leu Asp Arg Asn Phe Ser Arg Thr Xaa 225 230
(2) INFORMATION FOR SEQ ID NO: 347:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 169 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 347: Met Ala Ala Ala Val Ala Gly Met Leu Arg Gly Gly Leu Leu Pro Gin
1 5 10 15
Ala Gly Arg Leu Pro Thr Leu Gin Thr Val Arg Tyr Gly Ser Lys Ala 20 25 30
Val Thr Arg His Arg Arg Val Met His Phe Gin Arg Gin Lys Leu Met 35 40 45
Ala Val Thr Glu Tyr He Pro Pro Lys Pro Ala He His Pro Ser Cys 50 55 60
Leu Pro Ser Pro Pro Ser Pro Pro Gin Glu Glu He Gly Leu He Arg 65 70 75 80 Leu Leu Arg Arg Glu He Ala Ala Val Phe Gin Asp Asn Arg Met He
85 90 95
Ala Val Cys Gin Asn Val Ala Leu Ser Ala Glu Asp Lys Leu Leu He 100 105 110
Ala Thr Pro Ala Ala Glu Thr Gin Asp Pro Asp Glu Gly Leu Pro Gin 115 120 125
Pro Gly Pro Glu Ser Pro Ser Trp Arg He Pro Ser Thr Lys He Cys 130 135 140
Cys Pro Phe Leu Trp Gly Thr Thr Cys Cys Trp Ser Val Lys Ser Pro 145 150 155 160 Arg Ser Arg Arg Trp Tyr Gly Ser Xaa
165
(2) INFORMATION FOR SEQ ID NO: 348:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 43 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 348:
Met Lys Arg Ser Phe Leu Leu Pro Leu Leu Leu Val Gly Phe Leu Asp 1 5 10 15
Thr Ala His Leu He Leu Leu Glu Thr Leu Ser Val Cys Leu Trp Leu 20 25 30
Pro Ser Leu He Asp Ser Arg Cys Val Met Ser 35 40
(2) INFORMATION FOR SEQ ID NO: 349:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 78 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 349:
Met Lys Glu Gly Pro Pro Cys Lys Arg His His Tyr Tyr Gin Asn Cys 1 5 10 15 Gly Ala Lys Leu Leu Val Ser Leu Phe Gly Glu Thr Asn Gin He His 20 25 30
Leu Leu Glu Thr Gin Val Gly Thr Glu Lys Gly Gly Glu Arg He Trp 35 40 45
Glu Glu Lys Trp Arg He Ser Ser Thr Val Leu Phe He Ser Val Asn 50 55 60
Ser Tyr Val Glu Gly Ser Val Leu Glu He Lys Leu Phe Tyr 65 70 75
(2) INFORMATION FOR SEQ ID NO: 350:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 24 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 350:
Met Ser Glu He Leu Ser Leu Leu Phe Cys Leu Leu Gly Pro Ala Leu 1 5 10 15 Asp Glu Arg Arg Glu Glu Lys Asp 20
(2) INFORMATION FOR SEQ ID NO: 351:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 274 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 351:
Met Ser Ser Ala Gly Thr Ala Thr Pro Leu Glu Met Asp His Lys Leu 1 5 10 15
Thr Ser Gin Pro Gly Arg Pro Ser Phe Tyr Cys Asn Ser Arg His Ser 20 25 30
He Val Gly Ser Ser His Gin Leu Gly Phe Trp Phe Ser His Leu Glu 35 40 45
Ser Ser Gly Leu Lys Val Phe Gin Val Ser Leu Pro Cys Glu Cys Val 50 55 60 Asn Leu Pro Thr Arg He Ala Ser Val Val Leu Ser Leu Met Ser Leu 65 70 75 80
Leu Val Val Gly Gin Ala Pro Ala Trp Glu Gly Ser Leu Leu Arg Gly 85 90 95
Arg Pro Ala Gly Gly Ala His Leu Cys Ala Met Xaa Val He Glu Gly 100 105 110
Leu Val Val Asp Val Gly Glu Arg He Leu His Gly Gin Arg Glu Val 115 120 125
Gly Gin Val Ser Gin Val Leu Pro Ala Leu Ser Leu Gly Leu Val Phe 130 135 140 Leu Cys Gin Gly Thr Val Glu Lys Val Ser Gly Ala Ala His Cys Ser 145 150 155 160
Ser Leu Leu Cys Cys Leu Pro Trp Gin Cys Ser Gly Gly Gly Phe Pro 165 170 175
Thr Xaa Arg Cys Ser Arg Pro Tyr Phe Ser Ser His Lys Gly Val Ala 180 185 190
Ala Thr Leu Ala Leu Thr Cys His Cys Asp Lys Val His Val Ala Gly 195 200 205
Leu Gly Lys Asp Trp Ala He Glu Gin Arg Arg Arg Thr Cys Glu Ser 210 215 220 Asp Xaa Glu Xaa Xaa Pro Phe Thr Leu Ala Gly Leu Val Leu Val Leu 225 230 235 240
Arg Phe Cys Gin Val Val Leu Val Trp He Pro Gin Leu Gly Asp Lys 245 250 255
His Trp Arg Gly Met Thr Arg Leu Gly Arg Val Ser Leu Thr Ser Ser 260 265 270
He Xaa
(2) INFORMATION FOR SEQ ID NO: 352:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 47 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 352:
Met He Phe Thr Ser Val Thr Lys Gly He Leu Leu He Ala Leu Trp 1 5 10 15 Val Pro Leu Phe His Phe Met Leu He Asp Ser He Leu Gly Pro Ser 20 25 30
Arg Leu Leu Thr Asp Gly Val Pro Phe Asn Pro Trp His Val Xaa 35 40 45
(2) INFORMATION FOR SEQ ID NO: 353: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 3 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 353:
Met Lys Thr
1
(2) INFORMATION FOR SEQ ID NO: 354:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 52 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 354:
Met Ser He Ser Gly Thr Asp Gly Leu He Leu Leu Leu Val Gly Leu 1 5 10 15
Glu Ala Xaa Val Arg Ser Ser Lys Lys Trp He Pro Lys Ala Leu Xaa 20 25 30 Val Thr Gin Ala Lys Trp Asn Ser Trp Pro Ser Arg Arg Asn Ala Gly 35 40 45
Phe Ala Leu His 50
(2) INFORMATION FOR SEQ ID NO: 355: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 132 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 355:
Met Glu His Cys Leu Tyr His Ser Val His Gly He Asn Pro Tyr He 1 5 10 15
His Lys Asn Thr His Pro Ser He Asn He Tyr Met Val Trp Asp Glu 20 25 30
Gin Val Asn Ser Phe Glu Arg Glu Phe Val Pro Phe Phe Phe Leu He 35 40 45 He Leu Leu Asn Cys Cys Gin Leu Ser Asn Lys Gin Thr Glu Lys Leu 50 55 60
Phe Gly Lys Thr Leu His Thr Pro Phe Leu Ser Ser Ala Leu Lys Tyr 65 70 75 80
Arg Leu Asn Thr His He Leu Pro Val Phe Ser Tyr Ser Asp Ser He 85 90 95
Leu Thr Cys His Leu He Leu Ala Ser Tyr Phe Ser His Val Tyr Leu 100 105 110
Pro Val Thr Cys He Cys Tyr Leu Asn Arg Lys Lys Asn He Gin Lys 115 120 125
Lys Lys Asn Xaa 130
(2) INFORMATION FOR SEQ ID NO: 356:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 204 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 356:
Met Gly Ser Arg Asp His Leu Phe Lys Val Leu Val Val Gly Asp Ala 1 5 10 15
Ala Val Gly Lys Thr Ser Leu Val Gin Asp Tyr Ser Gin Asp Ser Phe 20 25 30
Ser Lys His Tyr Lys Ser Thr Val Gly Val Asp Phe Ala Leu Lys Val 35 40 45
Leu Gin Trp Ser Asp Tyr Glu He Val Arg Leu Gin Leu Trp Asp He 50 55 60 Ala Gly Gin Glu Arg Phe Thr Ser Met Thr Arg Leu Tyr Tyr Arg Asp 65 70 75 80
Ala Ser Ala Cys Val He Met Phe Asp Val Thr Asn Ala Thr Thr Phe 85 90 95
Ser Asn Ser Gin Arg Trp Lys Gin Asp Leu Asp Ser Lys Leu Thr Leu 100 105 110
Pro Asn Gly Glu Pro Val Pro Cys Leu Leu Leu Ala Asn Lys Cys Asp 115 120 125
Leu Ser Pro Trp Ala Val Ser Arg Asp Gin He Asp Arg Phe Ser Lys 130 135 140 Glu Asn Gly Phe Thr Gly Trp Thr Glu Thr Ser Val Lys Glu Asn Lys 145 150 155 160
Asn He Asn Glu Ala Met Arg Val Leu He Glu Lys Met Met Arg Asn 165 170 175
Ser Thr Glu Asp He Met Ser Leu Ser Thr Gin Gly Asp Tyr He Asn 180 185 190
Leu Gin Thr Lys Ser Ser Ser Trp Ser Cys Cys Xaa 195 200
(2) INFORMATION FOR SEQ ID NO: 357:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 47 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 357:
Met He Ser Leu He Phe Gin Leu Glu Glu Glu Lys Leu Val Glu Lys 1 5 10 15 Phe Phe Phe Phe Leu Phe Phe Phe Leu Lys Lys Gly Ser Gin Gly Ser 20 25 30
Asn Leu Lys He Val Pro Arg His Met Arg Val Val Leu Arg Gly 35 40 45
(2) INFORMATION FOR SEQ ID NO: 358: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 73 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 358:
Met Thr Tyr Val Thr Cys Leu His Val Cys Leu Leu Val Glu Phe Leu 1 5 10 15
Asn Ser Gin Leu Thr Asn His Arg Lys Tyr Tyr Phe Leu Ser Tyr Gly 20 25 30
Phe Trp Phe Thr Gly Leu Arg Gly Phe Ser Glu Tyr Leu Trp Pro Gin 35 40 45 Gin His Thr Ser Phe His Pro Asn Arg Asn Glu He Asn Phe Val Ser 50 55 60
Thr Asp Asn Arg He Trp Val Thr Xaa 65 70
(2) INFORMATION FOR SEQ ID NO: 359: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 102 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 359:
Met Ser Asp Gin Glu Ala Lys Pro Ser Thr Glu Asp Leu Gly Asp Lys 1 10 15
Lys Glu Gly Glu Tyr He Lys Leu Lys Val He Gly Gin Asp Ser Ser 20 25 30
Glu He His Phe Lys Val Lys Met Thr Thr His Leu Lys Lys Leu Lys 35 40 45 Glu Ser Tyr Cys Gin Arg Gin Gly Val Pro Met Asn Ser Leu Arg Phe
50 55 60
Leu Phe Glu Gly Gin Arg lie Ala Asp Asn His Thr Pro Lys Glu Leu 65 70 75 80
Gly Met Glu Glu Glu Asp Val He Glu Val Tyr Gin Glu Gin Thr Gly 85 90 95
Gly His Ser Thr Val Xaa 100
(2) INFORMATION FOR SEQ ID NO: 360:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 48 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 360:
Met Gly Phe Pro Gin Trp His Leu Gly Asn His Ala Val Glu Pro Val 1 5 10 15 Thr Ser He Leu Leu Leu Phe Leu Leu Met Met Leu Gly Val Arg Gly 20 25 30
Leu Leu Leu Val Gly Leu Val Tyr Leu Val Ser His Leu Ser Gin Arg 35 40 45
(2) INFORMATION FOR SEQ ID NO: 361:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 179 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 361:
Met Ser Ala Glu Val Lys Val Thr Gly Gin Asn Gin Glu Gin Phe Leu 1 5 10 15
Leu Leu Ala Lys Ser Ala Lys Gly Ala Ala Leu Ala Thr Leu He His 20 25 30 Gin Val Leu Glu Ala Pro Gly Val Tyr Val Phe Gly Glu Leu Leu Asp 35 40 45
Met Pro Asn Val Arg Glu Leu Ala Glu Ser Asp Phe Ala Ser Thr Phe 50 55 60
Arg Leu Leu Thr Val Phe Ala Tyr Gly Thr Tyr Ala Asp Tyr Leu Ala 65 70 75 80
Glu Ala Arg Asn Leu Pro Pro Leu Thr Glu Ala Gin Lys Asn Lys Leu 85 90 95
Arg His Leu Ser Val Val Thr Leu Ala Ala Lys Val Lys Cys He Pro 100 105 110 Tyr Ala Val Leu Leu Glu Ala Leu Ala Leu Arg Asn Val Arg Gin Leu 115 120 125
Glu Asp Leu Val He Glu Ala Val Tyr Ala Asp Val Leu Arg Gly Ser 130 135 140
Leu Asp Gin Arg Asn Gin Arg Leu Glu Val Asp Tyr Ser He Gly Arg 145 150 155 160
Asp He Gin Arg Gin Asp Leu Ser Ala He Ala Arg Thr Leu Xaa Lys 165 170 175
Asn His Xaa
(2) INFORMATION FOR SEQ ID NO: 362:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 25 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 362: Met Lys Ser Ser Ser Leu Phe Phe Phe Phe Leu Ala His Phe He His 1 5 10 15
Ser His Asp Leu Pro Gly Leu Cys Arg 20 25
(2) INFORMATION FOR SEQ ID NO: 363: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 224 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 363:
Met Lys Phe Ala Ala Ser Gly Xaa Phe Leu His His Met Ala Gly Leu 1 5 10 15
Ser Ser Ser Lys Leu Ser Met Ser Lys Ala Leu Pro Leu Thr Lys Val 20 25 30
Val Gin Asn Asp Ala Tyr Thr Ala Pro Ala Leu Pro Ser Ser He Arg 35 40 45 Thr Lys Ala Leu Thr Asn Met Ser Arg Thr Leu Val Asn Lys Glu Glu 50 55 60
Pro Pro Lys Glu Leu Pro Ala Ala Glu Pro Val Leu Ser Pro Leu Glu 65 70 75 80
Gly Thr Lys Met Thr Val Asn Asn Leu His Pro Arg Val Thr Glu Glu 85 90 95
Asp He Val Glu Leu Phe Cys Val Cys Gly Ala Leu Lys Arg Ala Arg 100 105 110
Leu Val His Pro Gly Val Ala Glu Val Val Phe Val Lys Lys Asp Asp 115 120 125 Ala He Thr Ala Tyr Lys Lys Tyr Asn Asn Arg Cys Leu Asp Gly Gin 130 135 140
Pro Met Lys Cys Asn Leu His Met Asn Gly Asn Val He Thr Ser Asp 145 150 155 160
Gin Pro He Leu Leu Arg Leu Ser Asp Ser Pro Ser Met Lys Lys Glu 165 170 175
Ser Glu Leu Pro Arg Arg Val Asn Ser Ala Ser Ser Ser Asn Pro Pro 180 185 190
Ala Glu Val Asp Pro Asp Thr He Leu Lys Ala Leu Phe Lys Ser Ser 195 200 205 Gly Ala Ser Xaa Thr Thr Gin Pro Thr Glu Phe Lys He Lys Leu Xaa 210 215 220
(2) INFORMATION FOR SEQ ID NO: 364: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 349 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 364:
Met Ser Lys Asn Cys He Lys Leu Leu Cys Glu Asp Pro Val Phe Ala 1 5 10 15
Glu Tyr He Lys Cys He Leu Met Asp Glu Arg Thr Phe Leu Asn Asn 20 25 30
Asn He Val Tyr Thr Phe Met Thr His Phe Leu Leu Lys Val Gin Ser 35 40 45 Gin Val Phe Ser Glu Ala Asn Cys Ala Asn Leu He Ser Thr Leu He 50 55 60
Thr Asn Leu He Ser Gin Tyr Gin Asn Leu Gin Ser Asp Phe Ser Asn 65 70 75 80
Arg Val Glu He Ser Lys Ala Ser Ala Ser Leu Asn Gly Asp Leu Arg 85 90 95
Ala Leu Ala Leu Leu Leu Ser Val His Thr Pro Lys Gin Leu Asn Pro 100 105 110
Ala Leu He Pro Thr Leu Gin Glu Leu Leu Ser Lys Cys Arg Thr Cys 115 120 125
Leu Gin Gin Arg Asn Ser Leu Gin Glu Gin Glu Ala Lys Glu Arg Lys 130 135 140
Thr Lys Asp Asp Glu Gly Ala Thr Pro He Lys Arg Arg Arg Val Ser 145 150 155 160
Ser Asp Glu Glu His Thr Val Asp Ser Cys He Ser Asp Met Lys Thr 165 170 175
Glu Thr Arg Glu Val Leu Thr Pro Thr Ser Thr Ser Asp Asn Glu Thr 180 185 190
Arg Asp Ser Ser He He Asp Pro Gly Thr Glu Gin Asp Leu Pro Ser 195 200 205 Pro Glu Asn Ser Ser Val Lys Glu Tyr Arg Met Glu Val Pro Ser Ser 210 215 220
Phe Ser Glu Asp Met Ser Asn He Arg Ser Gin His Ala Glu Glu Gin 225 230 235 240
Ser Asn Asn Gly Arg Tyr Asp Asp Cys Lys Glu Phe Lys Asp Leu His
245 250 255
Cys Ser Lys Asp Ser Thr Leu Ala Glu Glu Glu Ser Glu Phe Pro Ser 260 265 270
Thr Ser He Ser Ala Val Leu Ser Asp Leu Ala Asp Leu Arg Ser Cys 275 280 285 Asp Gly Gin Ala Leu Pro Ser Gin Asp Pro Glu Val Ala Leu Ser Leu 290 295 300
Ser Cys Gly His Ser Arg Gly Leu Phe Ser His Met Gin Gin His Asp 305 310 315 320
He Leu Asp Thr Leu Cys Arg Thr He Glu Ser Thr He His Val Val 325 330 335
Thr Arg He Ser Gly Lys Gly Asn Gin Ala Ala Ser Xaa 340 345
(2) INFORMATION FOR SEQ ID NO: 365:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 467 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 365:
Met Leu His Gin Asp His He Thr Phe Ala Met Leu Leu Ala Arg He 1 5 10 15 Lys Leu Lys Gly Thr Val Gly Glu Pro Thr Tyr Asp Ala Glu Phe Gin
20 25 30
His Phe Leu Arg Gly Asn Glu He Val Leu Ser Ala Gly Ser Thr Pro 35 40 45
Arg He Gin Gly Leu Thr Val Glu Gin Ala Glu Ala Val Val Arg Leu 50 55 60
Ser Cys Leu Pro Ala Phe Lys Asp Leu He Ala Lys Val Gin Ala Asp 65 70 75 80
Glu Gin Phe Gly He Trp Leu Asp Ser Ser Ser Pro Glu Gin Thr Val 85 90 95 Pro Tyr Leu Trp Ser Glu Glu Thr Pro Ala Thr Pro He Gly Gin Ala 100 105 110
He His Arg Leu Leu Leu He Gin Ala Phe Arg Pro Asp Arg Leu Leu 115 120 125
Ala Met Ala His Met Phe Val Ser Thr Asn Leu Gly Glu Ser Phe Met 130 135 140
Ser He Met Glu Gin Pro Leu Asp Leu Thr His He Val Xaa Thr Glu 145 150 155 160
Val Lys Pro Asn Thr Pro Val Leu Met Cys Ser Val Pro Gly Tyr Asp
165 170 175 Ala Ser Gly His Val Glu Asp Leu Ala Ala Glu Gin Asn Thr Gin He 180 185 190
Thr Ser He Ala He Gly Ser Ala Glu Gly Phe Asn Gin Ala Asp Lys 195 200 205
Ala He Asn Thr Ala Val Lys Ser Gly Arg Trp Val Met Leu Lys Asn 210 215 220
Val His Leu Ala Pro Gly Trp Leu Met Gin Leu Glu Lys Lys Leu His 225 230 235 240
Ser Leu Gin Pro His Ala Cys Phe Arg Leu Phe Leu Thr Met Glu He 245 250 255 Asn Pro Lys Val Pro Val Asn Leu Leu Arg Ala Gly Arg He Phe Val 260 265 270
Phe Glu Pro Pro Pro Gly Xaa Lys Ala Asn Met Leu Arg Thr Phe Ser 275 280 285
Ser He Pro Val Ser Arg He Cys Lys Ser Pro Asn Glu Arg Ala Arg 290 295 300
Leu Tyr Phe Leu Leu Ala Trp Phe His Ala He He Gin Glu Arg Leu 305 310 315 320
Arg Tyr Ala Pro Leu Gly Trp Ser Lys Lys Tyr Glu Phe Gly Glu Ser 325 330 335 Asp Leu Arg Ser Xaa Cys Asp Thr Val Asp Thr Trp Leu Asp Asp Thr
340 345 350
Ala Lys Gly Arg Gin Asn He Ser Pro Asp Lys He Pro Trp Ser Ala 355 360 365
Leu Lys Thr Leu Met Ala Gin Ser He Tyr Gly Gly Arg Val Asp Asn 370 375 380
Glu Phe Asp Gin Arg Leu Leu Asn Thr Phe Leu Glu Arg Leu Phe Thr 385 390 395 400
Thr Arg Ser Phe Asp Ser Glu Phe Lys Leu Ala Cys Lys Val Asp Gly 405 410 415 His Lys Asp He Gin Met Pro Asp Gly Met Gin Ala Arg Gly Val Cys 420 425 430
Ala Val Gly Gly Val Ala Pro Arg His Pro Asp Ala Leu Leu Ala Gly 435 440 445
Pro Ala Gin Gin Arg Arg Glu Ser Pro Pro Tyr His Thr Gly Cys Gly 450 455 460
His Asp Gin 465
(2) INFORMATION FOR SEQ ID NO: 366:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 152 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 366:
Met Ala Asp Glu Ala Thr Arg Arg Val Val Ser Glu He Pro Val Leu 1 5 10 15 Lys Thr Asn Ala Gly Pro Arg Asp Arg Glu Leu Trp Val Gin Arg Leu 20 25 30
Lys Glu Glu Tyr Gin Ser Leu He Arg Tyr Val Glu Asn Asn Lys Asn 35 40 45
Ala Asp Asn Asp Trp Phe Arg Leu Glu Ser Asn Lys Glu Gly Thr Arg 50 55 60
Trp Phe Gly Lys Cys Trp Tyr He His Asp Leu Leu Lys Tyr Glu Phe 65 70 75 80
Asp He Glu Phe Asp He Pro He Thr Tyr Pro Thr Thr Ala Pro Glu 85 90 95 He Ala Val Pro Glu Leu Asp Gly Lys Thr Ala Lys Met Tyr Arg Gly 100 105 110
Gly Lys He Cys Leu Thr Asp His Phe Lys Pro Leu Trp Gly Gin Glu 115 120 125
Cys Ala Gin He Trp Thr Ser Ser Ser His Gly Ser Gly Ala Gly Ser 130 135 140
Met Xaa Gly Ser Gly Asn Pro Xaa 145 150
(2) INFORMATION FOR SEQ ID NO: 367:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 373 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 367:
Met Tyr Asp Gly Thr Lys Glu Val Pro Met Asn Pro Val Lys He Tyr 1 5 10 15 Gin Val Cys Asp He Pro Gin Pro Gin Gly Ser He He Asn Pro Gly 20 25 30
Ser Thr Gly Ser Ala Pro Trp Asp Glu Lys Asp Asn Asp Val Asp Glu 35 40 45
Glu Asp Glu Glu Asp Glu Leu Asp Gin Ser Gin His His Val Pro He 50 55 60
Gin Asp Thr Phe Pro Phe Leu Asn He Asn Gly Ser Pro Met Ala Pro 65 70 75 80
Ala Ser Val Gly Asn Cys Ser Val Gly Asn Cys Ser Pro Glu Ala Val
85 90 95 Trp Pro Lys Thr Glu Pro Leu Glu Met Glu Val Pro Gin Ala Pro He
100 105 110
Gin Pro Phe Tyr Ser Ser Pro Glu Leu Trp He Ser Ser Leu Pro Met 115 120 125
Thr Asp Leu Asp He Lys Phe Gin Tyr Arg Gly Lys Glu Tyr Gly Gin 130 135 140
Thr Met Thr Val Ser Asn Pro Gin Gly Cys Arg Leu Phe Tyr Gly Asp 145 150 155 160
Leu Gly Pro Met Pro Asp Gin Glu Glu Leu Phe Gly Pro Val Xaa Leu 165 170 175 Glu Gin Val Lys Phe Pro Gly Pro Glu His He Thr Asn Glu Lys Gin 180 185 190
Lys Leu Phe Thr Ser Lys Leu Leu Asp Val Met Asp Arg Gly Leu He 195 200 205
Leu Glu Val Ser Gly His Ala He Tyr Ala He Arg Leu Cys Gin Cys
210 215 220
Lys Val Tyr Trp Ser Gly Pro Cys Ala Pro Ser Leu Val Ala Pro Asn 225 230 235 240
Leu He Glu Arg Gin Lys Lys Val Lys Leu Phe Cys Leu Glu Thr Phe 245 250 255
Leu Ser Asp Leu He Ala His Gin Lys Gly Gin He Glu Lys Gin Pro 260 265 270
Pro Phe Glu He Tyr Leu Cys Phe Gly Glu Glu Trp Pro Asp Gly Lys 275 280 285
Pro Leu Glu Arg Lys Leu He Leu Val Gin Val He Pro Val Val Ala 290 295 300
Arg Met He Tyr Glu Met Phe Ser Gly Asp Phe Thr Arg Ser Phe Asp 305 310 315 320
Ser Gly Ser Val Arg Leu Gin He Ser Thr Pro Asp He Lys Asp Asn 325 330 335 He Val Ala Gin Leu Lys Gin Leu Tyr Arg He Leu Gin Thr Gin Glu 340 345 350
Ser Trp Gin Pro Met Gin Pro Thr Pro Ser Met Gin Leu Pro Pro Ala 355 360 365
Leu Pro Pro Gin Xaa 370
(2) INFORMATION FOR SEQ ID NO: 368:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 83 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 368:
Met Gly Ser Ser Val Leu Pro Phe Cys Val Cys Val Thr Ser Pro Ser 1 5 10 15
Leu Gly Gly Arg Cys He Gin Gly Arg Phe Ala Ser His Ser Lys Phe 20 25 30 Trp Gly Phe Gly Arg Lys Thr Ala Ser Phe Gly Ala Val Gly Glu Thr 35 40 45
Pro Pro Asp Gin Glu Pro Gin Lys Glu Thr Glu Pro Ala Thr Ser Ser 50 55 60
His Ala Arg Pro Trp Ala Arg Val He Gly Leu Arg He Trp Pro Gin 65 70 75 80
Pro Asn Xaa
(2) INFORMATION FOR SEQ ID NO: 369:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 369:
Met Leu Leu Ser Val Ala He Phe He Leu Leu Thr Leu Val Tyr Ala 1 5 10 15 Tyr Trp Thr Met Xaa 20
(2) INFORMATION FOR SEQ ID NO: 370:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 227 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 370:
Met Gly Ala Ser Ala Arg Leu Leu Arg Ala Val He Met Gly Ala Pro 1 5 10 15
Gly Ser Gly Lys Gly Thr Val Ser Ser Arg He Thr Thr His Phe Glu 20 25 30
Leu Lys His Leu Ser Ser Gly Asp Leu Leu Arg Asp Asn Met Leu Arg 35 40 45
Gly Thr Glu He Gly Val Leu Ala Lys Ala Phe He Asp Gin Gly Lys 50 55 60 Leu He Pro Asp Asp Val Met Thr Arg Leu Ala Leu His Glu Leu Lys 65 70 75 80
Asn Leu Thr Gin Tyr Ser Trp Leu Leu Asp Gly Phe Pro Arg Thr Leu 85 90 95
Pro Gin Ala Glu Ala Leu Asp Arg Ala Tyr Gin He Asp Thr Val He 100 105 110
Asn Leu Asn Val Pro Phe Glu Val He Lys Gin Arg Leu Thr Ala Arg 115 120 125
Trp He His Pro Ala Ser Gly Arg Val Tyr Asn He Glu Phe Asn Pro 130 135 140 Pro Lys Thr Val Gly He Asp Asp Leu Thr Gly Glu Pro Leu He Gin 145 150 155 160
Arg Glu Asp Asp Lys Pro Glu Thr Val He Lys Arg Leu Lys Ala Tyr 165 170 175
Glu Asp Gin Thr Lys Pro Val Leu Glu Tyr Tyr Gin Lys Lys Gly Val 180 185 190
Leu Glu Thr Phe Ser Gly Thr Glu Thr Asn Lys He Trp Pro Tyr Val 195 200 205
Tyr Ala Phe Leu Gin Thr Lys Val Pro Gin Arg Ser Gin Lys Ala Ser 210 215 220
Val Thr Pro 225
(2) INFORMATION FOR SEQ ID NO: 371:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 79 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 371:
Met Phe Leu Asn Cys Glu He Leu Glu Tyr Cys Tyr Tyr Leu Thr Gin
1 5 10 15
Leu Lys He Ser Met Gly Lys Tyr Leu Ser He Pro Thr Val Leu Leu 20 25 30
Lys He He Arg Cys Ser He Thr Ala Val Ser Asp Ser Ser Thr Ser 35 40 45
Trp Ala He Lys Ala Gin Leu Lys He Glu Asn Lys Asp Leu Asp Asn 50 55 60 Lys Thr Ala Lys Gly Gly Gly Gin Glu Ala Leu Thr Cys Thr Xaa 65 70 75
(2) INFORMATION FOR SEQ ID NO: 372:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 51 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 372:
Met Arg Ala Val Phe Pro Cys Cys Pro Phe Leu Thr Leu Met Leu Pro 1 5 10 15
Leu Leu Glu Cys Leu Val Gly Met He Met Cys Tyr Leu Gly He Ser 20 25 30
Phe Thr Asp Thr Arg Lys Thr Ala Gly Leu Lys Lys Lys Lys Lys Lys 35 40 45
Lys Xaa Xaa 50
(2) INFORMATION FOR SEQ ID NO: 373:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 61 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 373: Met Phe Leu Met Arg Met His Leu Cys Phe Cys Lys Tyr Cys Cys Ser 1 5 10 15
Phe He Val Thr Pro Thr Ser Thr Ser Asn Thr Ala Ser Tyr Leu Trp 20 25 30
Pro Trp He Ser Ala Ser Met Ala Gly Arg Gly Ser Ser Trp Ala Cys 35 40 45
Thr Leu Asn Ala Val Thr Arg Glu Gly Leu Pro Glu Xaa 50 55 60
(2) INFORMATION FOR SEQ ID NO: 374:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 40 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 374:
Met Ser Leu Leu Asn Thr His Thr Leu Cys Phe Val Leu Phe Cys Phe 1 5 10 15 Thr Leu Ser He Asn Gin Glu Lys Leu Ala Asn His Leu Ala Phe Arg 20 25 30
He Leu Phe Phe He Val Phe Xaa 35 40
(2) INFORMATION FOR SEQ ID NO: 375: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 44 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 375:
Met Cys Ser Gly Gin Ser Gin Val Trp Lys Met Ala Leu Gin Ala Leu 1 5 10 15
Asp Ser Glu Thr Val Val He Leu Pro Asp Met His Leu He Leu Ser 20 25 30
Leu Arg Leu He His Asn Ala Arg Pro Cys Leu Xaa 35 40
(2) INFORMATION FOR SEQ ID NO: 376:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 203 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 376:
Met Leu He Ser Glu Glu Glu He Pro Phe Lys Asp Asp Pro Arg Asp 1 5 10 15
Glu Thr Tyr Lys Pro His Leu Glu Arg Glu Thr Pro Lys Pro Arg Arg 20 25 30
Lys Ser Gly Lys Val Lys Glu Glu Lys Glu Lys Lys Glu He Lys Val 35 40 45
Glu Val Glu Val Glu Val Lys Glu Glu Glu Asn Glu He Arg Glu Asp 50 55 60
Glu Glu Pro Pro Arg Lys Arg Gly Arg Arg Arg Lys Asp Asp Lys Ser 65 70 75 80 Pro Arg Leu Pro Lys Arg Arg Lys Lys Pro Pro He Gin Tyr Val Arg
85 90 95
Cys Glu Met Glu Gly Cys Gly Thr Val Leu Ala His Pro Arg Tyr Leu 100 105 110
Gin His His He Lys Tyr Gin His Leu Leu Lys Lys Lys Tyr Val Cys 115 120 125
Pro His Pro Ser Cys Gly Arg Leu Phe Arg Leu Gin Lys Gin Leu Leu 130 135 140
Arg His Ala Lys His His Thr Asp Gin Arg Asp Tyr He Cys Glu Tyr 145 150 155 160 Cys Ala Arg Ala Phe Lys Ser Ser His Asn Leu Ala Val His Arg Met
165 170 175
He His Thr Gly Glu Lys His Tyr Asn Val Arg Ser Val Asp Leu Leu 180 185 190
Val Asp Lys Arg His Leu Leu He Gly Thr Xaa 195 200
(2) INFORMATION FOR SEQ ID NO: 377:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 29 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 377:
Met Leu Pro Arg Arg Thr Phe Tyr Phe Tyr Phe He Phe He Phe Phe 1 5 10 15
Leu Ala Ser Phe Trp Gly Phe Thr Leu Arg Ala Ser Phe 20 25
(2) INFORMATION FOR SEQ ID NO: 378:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 136 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 378: Met Phe Asp Ser Leu Ser Tyr Phe Lys Gly Ser Ser Leu Leu Leu Met 1 5 10 15
Leu Lys Thr Tyr Leu Ser Glu Asp Val Phe Gin His Ala Val Val Leu 20 25 30
Tyr Leu His Asn His Ser Tyr Ala Ser He Gin Ser Asp Asp Leu Trp 35 40 45
Asp Ser Phe Asn Glu Val Thr Asn Gin Thr Leu Asp Val Lys Arg Met 50 55 60
Met Lys Thr Trp Thr Leu Gin Lys Gly Phe Pro Leu Val Thr Val Gin 65 70 75 80 Lys Lys Gly Lys Glu Leu Phe He Gin Gin Glu Arg Phe Phe Leu Asn
85 90 95
Met Lys Pro Glu He Gin Pro Ser Asp Thr Arg Tyr Met Pro Ser Phe 100 105 110
Phe Ser Cys His Leu Phe Cys Thr Leu Arg Trp Lys Tyr Phe Glu Val 115 120 125
Phe Tyr Asn His Lys Phe Leu Xaa 130 ' 135
(2) INFORMATION FOR SEQ ID NO: 379:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 41 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 379:
Met Ala Trp Arg Arg Arg Glu Pro Ala Ser Gly Leu Ala Ala Cys Trp 1 5 10 15 Leu Trp Arg Cys Ser Pro Trp Pro Cys Ala Cys Pro Gly Pro Gly Ala 20 25 30
Gly Leu Ser Ser Gly Ser Arg Pro Trp 35 40
(2) INFORMATION FOR SEQ ID NO: 380: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 468 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 380:
Met Glu Phe Leu Lys Val Ala Arg Arg Asn Lys Arg Glu Gin Leu Glu 1 5 10 15
Gin He Gin Lys Glu Leu Ser Val Leu Glu Glu Asp He Lys Arg Val 20 25 30
Glu Glu Met Ser Gly Leu Tyr Ser Pro Val Ser Glu Asp Ser Thr Val 35 40 45 Pro Gin Phe Glu Ala Pro Ser Pro Ser His Ser Ser He He Asp Ser 50 55 60
Thr Glu Tyr Ser Gin Pro Pro Gly Phe Ser Gly Ser Ser Gin Thr Lys 65 70 75 80
Lys Gin Pro Trp Tyr Asn Ser Thr Leu Ala Ser Arg Arg Lys Arg Leu 85 90 95
Thr Ala His Phe Glu Asp Leu Glu Gin Cys Tyr Phe Ser Thr Arg Met 100 105 110
Ser Arg He Ser Asp Asp Ser Arg Thr Ala Ser Gin Leu Asp Glu Phe 115 120 125 Gin Glu Cys Leu Ser Lys Phe Thr Arg Tyr Asn Ser Val Arg Pro Leu 130 135 140
Ala Thr Leu Ser Tyr Ala Ser Asp Leu Tyr Asn Gly Ser Ser He Val 145 150 155 160
Ser Ser He Glu Phe Asp Arg Asp Cys Asp Tyr Phe Ala He Ala Gly 165 170 175
Val Thr Lys Lys He Lys Val Tyr Glu Tyr Asp Thr Val He Gin Asp 180 185 190
Ala Val Asp He His Tyr Pro Glu Asn Glu Met Thr Cys Asn Ser Lys 195 200 205 He Ser Cys He Ser Trp Ser Ser Tyr His Lys Asn Leu Leu Ala Ser 210 215 220
Ser Asp Tyr Glu Gly Thr Val He Leu Trp Asp Gly Phe Thr Gly Gin
225 230 235 240
Arg Ser Lys Val Tyr Gin Glu His Glu Lys Arg Cys Trp Ser Val Asp
245 250 255
Phe Asn Leu Met Asp Pro Lys Leu Leu Ala Ser Gly Ser Asp Asp Ala 260 265 270
Lys Val Lys Leu Trp Ser Thr Asn Leu Asp Asn Ser Val Ala Ser He 275 280 285 Glu Ala Lys Ala Asn Val Cys Cys Val Lys Phe Ser Pro Ser Ser Arg
290 295 300
Tyr His Leu Ala Phe Gly Cys Ala Asp His Cys Val His Tyr Tyr Asp 305 310 315 320
Leu Arg Asn Thr Lys Gin Pro He Met Val Phe Lys Gly His Arg Lys 325 330 335
Ala Val Ser Tyr Ala Lys Phe Val Ser Gly Glu Glu He Val Ser Ala 340 345 350
Ser Thr Asp Ser Gin Leu Lys Leu Trp Asn Val Gly Lys Pro Tyr Cys 355 360 365 Leu Arg Ser Phe Lys Gly His He Asn Glu Lys Asn Phe Val Gly Leu 370 375 380
Ala Ser Asn Gly Asp Tyr He Ala Cys Gly Ser Glu Asn Asn Ser Leu 385 390 395 400
Tyr Leu Tyr Tyr Lys Gly Leu Ser Lys Thr Leu Leu Thr Phe Lys Phe 405 410 415
Asp Thr Val Lys Ser Val Leu Asp Lys Asp Arg Lys Glu Asp Asp Thr 420 425 430
Asn Glu Phe Val Ser Ala Val Cys Trp Arg Ala Leu Pro Asp Gly Glu 435 440 445 Ser Asn Val Leu He Ala Ala Asn Ser Gin Gly Thr He Lys Val Leu 450 455 460
Glu Leu Val Xaa 465
(2) INFORMATION FOR SEQ ID NO: 381: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 29 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 381:
Met Arg Lys Glu Asp Gly Phe Trp Phe Phe Phe Phe Leu Phe Phe Phe 1 5 10 15
Val Val Gly Ser Lys Phe Val Asn Gly Asn Lys Leu Val 20 25
(2) INFORMATION FOR SEQ ID NO: 382:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 29 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 382:
Met Pro Leu Ala Pro Tyr Cys Asp Leu Leu Val Ala Leu Ser Phe Ala 1 5 10 15
Leu Val Leu Glu Ser Pro Val Asp Ser Ser Asp Phe Thr 20 25
(2) INFORMATION FOR SEQ ID NO: 383:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 138 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 383:
Met Asn Ser Leu Val Ser Trp Gin Leu Leu Leu Phe Leu Cys Ala Thr 1 5 10 15
His Phe Gly Glu Pro Leu Glu Lys Val Ala Ser Val Gly Asn Ser Arg 20 25 30
Pro Thr Gly Gin Gin Leu Glu Ser Leu Gly Leu Leu Ala Pro Gly Glu 35 40 45
Gin Ser Leu Pro Cys Thr Glu Arg Lys Pro Ala Ala Thr Ala Arg Leu 50 55 60 Ser Arg Arg Gly Thr Ser Leu Ser Pro Pro Pro Glu Ser Ser Gly Ser 65 70 75 80
Pro Gin Gin Pro Gly Leu Ser Ala Pro His Ser Arg Gin He Pro Ala 85 90 95
Pro Gin Gly Ala Val Leu Val Gin Arg Glu Lys Asp Leu Pro Asn Tyr 100 105 110
Asn Trp Asn Ser Phe Gly Leu Arg Phe Gly Lys Arg Glu Ala Ala Pro 115 120 125
Gly Asn His Gly Arg Ser Ala Gly Arg Gly 130 135
(2) INFORMATION FOR SEQ ID NO: 384:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 74 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 384: Met Ser Cys Phe He Asp Ser Xaa Asp Ser Lys He Leu His Leu Leu 1 5 10 15
Val Val Ser Phe He Cys Xaa Leu Phe Leu Leu He Leu Thr His Gly 20 25 30
He Leu He Leu Arg Xaa Phe Phe Ser Val Xaa Xaa His Ser Leu Lys 35 40 45
Asn Asn Leu Glu Glu Tyr Leu He Leu Met Asn Lys Ala Leu Leu Thr 50 55 60
Arg Glu Asp Phe Phe Val Leu Pro Xaa Ala 65 t 70
(2) INFORMATION FOR SEQ ID NO: 385:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 521 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 385: Met Ser Ala Gly Glu Val Glu Arg Leu Val Ser Glu Leu Ser Gly Gly 1 5 10 15
Thr Gly Gly Asp Glu Glu Glu Glu Trp Leu Tyr Gly Asp Glu Asn Glu 20 25 30
Val Glu Arg Pro Glu Glu Glu Asn Ala Ser Ala Asn Pro Pro Ser Gly 35 40 45
He Glu Asp Glu Thr Ala Glu Asn Gly Val Pro Lys Pro Lys Val Thr 50 55 60
Glu Thr Glu Asp Asp Ser Asp Ser Asp Ser Asp Asp Asp Glu Asp Asp 65 70 75 80 Val His Val Thr He Gly Asp He Lys Thr Gly Ala Pro Gin Tyr Gly
85 90 95
Ser Tyr Gly Thr Ala Pro Val Asn Leu Asn He Lys Thr Gly Gly Arg 100 105 110
Val Tyr Gly Thr Thr Gly Thr Lys Val Lys Gly Val Asp Leu Asp Ala 115 120 125
Pro Gly Ser He Asn Gly Val Pro Leu Leu Glu Val Asp Leu Asp Ser 130 135 140
Phe Glu Asp Lys Pro Trp Arg Lys Pro Gly Ala Asp Leu Ser Asp Tyr 145 150 155 160 Phe Asn Tyr Gly Phe Asn Glu Asp Thr Trp Lys Ala Tyr Cys Glu Lys
165 170 175
Gin Lys Arg He Arg Met Gly Leu Glu Val He Pro Val Thr Ser Thr 180 185 190
Thr Asn Lys He Thr Val Gin Gin Gly Arg Thr Gly Asn Ser Glu Lys 195 200 205
Glu Thr Ala Leu Pro Ser Thr Lys Ala Glu Phe Thr Ser Pro Pro Ser 210 215 220
Leu Phe Lys Thr Gly Leu Pro Pro Ser Arg Arg Leu Pro Gly Ala He 225 230 235 240
Asp Val He Gly Gin Thr He Thr He Ser Arg Val Glu Gly Arg Arg 245 250 255
Arg Ala Asn Glu Asn Ser Asn He Gin Val Leu Ser Glu Arg Ser Ala 260 265 270
Thr Glu Val Asp Asn Asn Phe Ser Lys Pro Pro Pro Phe Phe Pro Pro 275 280 285
Gly Ala Pro Pro Thr His Leu Pro Pro Pro Pro Phe Leu Pro Pro Pro 290 295 300
Pro Thr Val Ser Thr Ala Pro Pro Leu He Pro Pro Pro Gly Phe Pro
305 310 315 320 Pro Pro Pro Gly Ala Pro Pro Pro Ser Leu He Pro Thr He Glu Ser
325 330 335
Gly His Ser Ser Gly Tyr Asp Ser Arg Ser Ala Arg Ala Phe Pro Tyr 340 345 350
Gly Asn Val Ala Phe Pro His Leu Pro Gly Ser Ala Pro Ser Trp Pro 355 360 365
Ser Leu Val Asp Thr Ser Lys Gin Trp Asp Tyr Tyr Ala Arg Arg Glu 370 375 380
Lys Asp Arg Asp Arg Glu Arg Asp Arg Asp Arg Glu Arg Asp Arg Asp 385 390 395 400 Arg Asp Arg Glu Arg Glu Arg Thr Arg Glu Arg Glu Arg Glu Arg Asp
405 410 415
His Ser Pro Thr Pro Ser Val Phe Asn Ser Asp Glu Glu Arg Tyr Arg 420 425 430
Tyr Arg Glu Tyr Ala Glu Arg Gly Tyr Glu Arg His Arg Ala Ser Arg 435 440 445
Glu Lys Glu Glu Arg His Arg Glu Arg Arg His Arg Glu Lys Glu Glu 450 455 460
Thr Arg His Lys Ser Ser Arg Ser Asn Ser Arg Arg Arg His Glu Ser
465 470 475 480 Glu Glu Gly Asp Ser His Arg Arg His Lys His Lys Lys Ser Lys Arg
485 490 495
Ser Lys Glu Gly Lys Glu Ala Gly Ser Glu Pro Ala Pro Glu Gin Glu 500 505 510
Ser Thr Glu Ala Thr Pro Ala Glu Xaa 515 520
(2) INFORMATION FOR SEQ ID NO: 386:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 137 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 386:
Met Asn Ser Arg Gly He Trp Leu Ala Tyr He He Leu Val Gly Leu 1 5 10 15
Leu His Met Val Leu Leu Ser He Pro Phe Phe Ser He Pro Val Val 20 25 30 Trp Thr Leu Thr Asn Val He His Asn Leu Ala Thr Tyr Val Phe Leu 35 40 45
His Thr Val Lys Gly Thr Pro Phe Glu Thr Pro Asp Gin Gly Lys Ala 50 55 60
Arg Leu Leu Thr His Trp Glu Gin Met Asp Tyr Gly Leu Gin Phe Thr 65 70 75 80
Ser Ser Arg Lys Phe Leu Ser He Ser Pro He Val Leu Tyr Leu Leu 85 90 95
Ala Ser Phe Tyr Thr Lys Tyr Asp Ala Ala His Phe Leu He Asn Thr 100 105 110 Ala Ser Leu Leu Ser Val Leu Leu Pro Lys Leu Pro Gin Phe His Gly 115 120 125
Val Arg Val Phe Gly He Asn Lys Tyr 130 135
(2) INFORMATION FOR SEQ ID NO: 387: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 186 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 387:
Met Ala Ala Gin Lys Asp Gin Gin Lys Asp Ala Glu Ala Glu Gly Leu 1 5 10 15
Ser Gly Thr Thr Leu Leu Pro Lys Leu He Pro Ser Gly Ala Gly Arg 20 25 30
Glu Trp Leu Glu Arg Arg Arg Ala Thr He Arg Pro Trp Ser Thr Phe 35 40 45 Val Asp Gin Gin Arg Phe Ser Arg Pro Arg Asn Leu Gly Glu Leu Cys 50 55 60
Gin Arg Leu Val Arg Asn Val Glu Tyr Tyr Gin Ser Asn Tyr Val Phe 65 70 75 80
Val Phe Leu Gly Leu He Leu Tyr Cys Val Val Thr Ser Pro Met Leu 85 90 95
Leu Val Ala Leu Ala Val Phe Phe Gly Ala Cys Tyr He Leu Tyr Leu 100 105 .110
Arg Thr Leu Glu Ser Lys Leu Val Leu Phe Gly Arg Glu Val Ser Pro 115 120 125 Ala His Gin Tyr Ala Leu Ala Gly Gly He Ser Phe Pro Phe Phe Trp 130 135 140
Leu Ala Gly Ala Gly Ser Ala Val Phe Trp Val Leu Gly Ala Thr Leu 145 150 155 160
Val Val He Gly Ser His Ala Ala Phe His Gin He Glu Ala Val Asp 165 170 175
Gly Glu Glu Leu Gin Met Glu Pro Val Xaa 180 185
(2) INFORMATION FOR SEQ ID NO: 3?
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 388:
Met 1
(2) INFORMATION FOR SEQ ID NO: 389:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 299 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 389: Met Leu Ser He Phe Tyr Phe Ala He Pro Val Gly Ser Gly Leu Gly 1 5 10 15
Tyr He Ala Gly Ser Lys Val Lys Asp Met Ala Gly Asp Trp His Trp 20 25 30
Ala Leu Arg Val Thr Pro Gly Leu Gly Val Val Ala Val Leu Leu Leu 35 40 45
Phe Leu Val Val Arg Glu Pro Pro Arg Gly Ala Val Glu Arg His Ser 50 55 60
Asp Leu Pro Pro Leu Asn Pro Thr Ser Trp Trp Ala Asp Leu Arg Ala 65 70 75 80 Leu Ala Arg Asn Pro Ser Phe Val Leu Ser Ser Leu Gly Phe Thr Ala
85 90 95
Val Ala Phe Val Thr Gly Ser Leu Ala Leu Trp Ala Pro Ala Phe Leu 100 105 110
Leu Arg Ser Arg Val Val Leu Gly Glu Thr Pro Pro Cys Leu Pro Gly 115 120 125
Asp Ser Cys Ser Ser Ser Asp Ser Leu He Phe Gly Leu He Thr Cys 130 135 140
Leu Thr Gly Val Leu Gly Val Gly Leu Gly Val Glu He Ser Arg Arg 145 150 155 160 Leu Arg His Ser Asn Pro Arg Ala Asp Pro Leu Val Cys Ala Thr Gly
165 170 175
Leu Leu Gly Ser Ala Pro Phe Leu Phe Leu Ser Leu Ala Cys Ala Arg 180 185 190
Gly Ser He Val Ala Thr Tyr He Phe He Phe He Gly Glu Thr Leu 195 200 205
Leu Ser Met Asn Trp Ala He Val Ala Asp He Leu Leu Tyr Val Val 210 215 220
He Pro Thr Arg Arg Ser Thr Ala Glu Ala Phe Gin He Val Leu Ser 225 230 235 240 His Leu Leu Gly Asp Ala Gly Ser Pro Tyr Leu He Gly Leu He Ser
245 250 255
Asp Arg Leu Arg Arg Asn Trp Pro Pro Ser Phe Leu Ser Glu Phe Arg 260 265 270
Ala Leu Gin Phe Ser Leu Met Leu Cys Ala Phe Val Gly Ala Leu Gly 275 280 285
Gly Ala Leu Pro Gly His Arg His Leu His Xaa 290 295
(2) INFORMATION FOR SEQ ID NO: 390:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 49 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 390:
Met Gly Pro Gin Gly Trp Val Arg Pro Leu Lys Thr Ala Pro Lys Leu 1 5 10 15 Gly Glu Ala He Arg Leu He Leu Phe Leu Asn Phe Val Lys Gin Cys 20 25 30
He Ala Ser Val Asn Leu Cys He Leu Arg Leu Asn He Thr Pro Leu 35 40 45
Leu
(2) INFORMATION FOR SEQ ID NO: 391:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 61 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 391:
Met Tyr Val Asn Tyr Gly Thr Arg Asn Tyr Ser Thr Glu Gly Pro Ala 1 5 10 15
Ala Leu Leu Asp Gin Ala Lys Leu Ser Leu Leu Val Trp Val Leu Cys 20 25 30 Phe Val Leu Leu Phe Val Cys Phe Cys Gly Leu Ser Tyr Val Val He 35 40 45
Ala Gin Val Pro Val Gly Leu Leu Cys He Thr Glu Xaa 50 55 60
(2) INFORMATION FOR SEQ ID NO: 392: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 79 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 392:
Met Leu Trp Phe Ala Asn Phe Phe Thr Tyr Leu Phe Leu Ser Gin Ser 1 5 10 15
Val Ala Phe Val His He Ser His He Gly Val Arg Gin Val Asn Thr 20 25 30
Asn Cys Tyr Phe Ser Arg Lys Ser Tyr Cys Tyr Gly He Leu Asn Pro 35 40 45 He Asn Cys He Lys Gly Lys Lys Lys Lys Lys Lys Lys Lys Lys Lys 50 55 60
Lys Lys Lys Lys He Pro Ala Gly Arg Xaa Leu Phe Pro Phe Gly 65 70 75
(2) INFORMATION FOR SEQ ID NO: 393: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 36 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 393:
Met Pro Gly Ala Phe Ser Glu Thr Val He Asn Asp Leu Leu Ser Leu 1 5 10 15
Phe Leu Val Leu Pro Ala Glu Leu Ser Tyr Ser Thr Leu Ser Gly Val 20 25 30
Tyr Arg Asn Ala 35
(2) INFORMATION FOR SEQ ID NO: 394:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 180 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 394: Met Ala Gin Ser Arg Asp Gly Gly Asn Pro Phe Ala Glu Pro Ser Glu 1 5 10 15
Leu Asp Asn Pro Phe Gin Asp Pro Ala Val He Gin His Arg Pro Ser 20 25 30
Arg Gin Tyr Ala Thr Leu Asp Val Tyr Asn Pro Phe Glu Thr Arg Glu 35 40 45
Pro Pro Pro Ala Tyr Glu Pro Pro Ala Pro Ala Pro Leu Pro Pro Pro 50 55 60
Ser Ala Pro Ser Leu Gin Pro Ser Arg Lys Leu Ser Pro Thr Glu Pro 65 70 75 80 Lys Asn Tyr Gly Ser Tyr Ser Thr Gin Ala Ser Ala Ala Ala Ala Thr
85 90 95
Ala Glu Leu Leu Lys Lys Gin Glu Glu Leu Asn Arg Lys Ala Glu Glu 100 105 110
Leu Asp Arg Arg Ser Glu Ser Cys Ser Met Leu Pro Trp Xaa Ala Gin 115 120 125
Leu Leu Asp Arg Thr He Gly Pro Leu Tyr Leu Leu Phe Val Gin Phe 130 135 140
Ser Pro Ala Phe Ser Arg Thr Ser Pro Trp Arg Ser Pro Lys Asn Phe 145 150 155 160 Arg Arg Leu Tyr Pro Pro Cys Thr Thr Ser Gly Cys Ala Ala Arg Trp
165 170 175
Xaa Phe Ser Xaa 180
(2) INFORMATION FOR SEQ ID NO: 395: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 395:
Met Pro Thr Pro Cys Thr Ser Leu Pro Ser Cys Cys Gin His Arg Ser 1 5 10 15
He Thr Met Thr Leu 20
(2) INFORMATION FOR SEQ ID NO: 396:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 60 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 396:
Met Pro Leu Phe He Pro Leu He Phe Phe Leu Ser Leu Leu His Cys 1 5 10 15 Gin Ser Lys His Pro He Gin Met Ser Leu Cys Met Cys Val Asn He 20 25 30
Ser Leu Val Trp Ser Pro Val Arg Trp He Phe Gly Ser Lys Gly Leu 35 40 45
Phe Ser Val His Leu Gin Ser Ser Gin Arg Pro Ser 50 55 60
(2) INFORMATION FOR SEQ ID NO: 397:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 152 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 397:
Met Ala Gly Pro Arg Pro Xaa Trp Arg Asp Gin Leu Leu Phe Met Ser 1 5 10 15
He He Val Leu Val He Val Val He Cys Leu Met Leu Tyr Ala Leu 20 25 30 Leu Trp Glu Ala Gly Asn Leu Thr Asp Leu Pro Asn Leu Arg He Gly 35 40 45
Phe Tyr Asn Phe Cys Leu Trp Asn Glu Asp Thr Ser Thr Leu Gin Cys 50 55 60
His Gin Phe Pro Glu Leu Glu Ala Leu Gly Val Pro Arg Val Gly Leu 65 70 75 80
Gly Leu Ala Arg Leu Gly Val Tyr Gly Ser Leu Val Leu Thr Leu Phe 85 90 95
Ala Pro Gin Pro Leu Leu Leu Ala Gin Cys Asn Xaa Asp Glu Arg Ala 100 105 110 Trp Arg Leu Ala Val Gly Phe Leu Ala Val Ser Ser Val Leu Leu Ala 115 120 125
Gly Gly Leu Gly Leu Phe Leu Ser Tyr Val Trp Asn Gly Ser Xaa Ser 130 135 140
Pro Ser Arg Gly Leu Gly Phe Xaa 145 150
(2) INFORMATION FOR SEQ ID NO: 398:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 480 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 398:
Met Ser Asp Gly Phe Asp Arg Ala Pro Gly Ala Gly Arg Gly Arg Xaa 1 5 10 15
Arg Gly Leu Gly Arg Gly Gly Gly Gly Pro Xaa Gly Gly Gly Phe Pro 20 25 30 Xaa Gly Xaa Xaa Pro Ala Glu Arg Xaa Arg His Gin Pro Pro Gin Pro 35 40 45
Lys Ala Pro Gly Phe Leu Gin Pro Xaa Pro Leu Arg Gin Pro Arg Thr 50 55 60
Thr Pro Pro Pro Gly Ala Gin Cys Glu Val Pro Ala Ser Pro Gin Arg 65 70 75 80
Pro Ser Arg Pro Gly Ala Leu Pro Glu Gin Thr Arg Pro Leu Arg Ala 85 90 95
Pro Pro Ser Ser Gin Asp Lys He Pro Gin Gin Asn Ser Glu Ser Ala 100 105 110 Met Ala Lys Pro Gin Val Val Val Ala Pro Val Leu Met Ser Lys Leu 115 120 125
Ser Val Asn Ala Pro Glu Phe Tyr Pro Ser Gly Tyr Ser Ser Ser Tyr 130 135 140
Thr Glu Ser Tyr Glu Asp Gly Cys Glu Asp Tyr Pro Thr Leu Ser Glu 145 150 155 160
Tyr Val Gin Asp Phe Leu Asn His Leu Thr Glu Gin Pro Gly Ser Phe 165 170 175
Glu Thr Glu He Glu Gin Phe Ala Glu Thr Leu Asn Gly Cys Val Thr 180 185 190 Thr Asp Asp Ala Leu Gin Glu Leu Val Glu Leu He Tyr Gin Gin Ala
195 200 205
Thr Ser He Pro Asn Phe Ser Tyr Met Gly Ala Arg Leu Cys Asn Tyr 210 215 220
Leu Ser His His Leu Thr He Ser Pro Gin Ser Gly Asn Phe Arg Gin 225 230 235 240
Leu Leu Leu Gin Arg Cys Arg Thr Glu Tyr Glu Val Lys Asp Gin Ala 245 250 255
Ala Lys Gly Asp Glu Val Thr Arg Lys Arg Phe His Ala Phe Val Leu 260 265 270 Phe Leu Gly Glu Leu Tyr Leu Asn Leu Glu He Lys Gly Thr Asn Gly 275 280 285
Gin Val Thr Arg Ala Asp He Leu Gin Val Gly Leu Arg Glu Leu Leu 290 295 300
Asn Ala Leu Phe Ser Asn Pro Met Asp Asp Asn Leu He Cys Ala Val 305 310 315 320
Lys Leu Leu Lys Leu Thr Gly Ser Val Leu Glu Asp Ala Trp Lys Glu 325 330 335
Lys Gly Lys Met Asp Met Glu Glu He He Gin Arg He Glu Asn Val 340 345 350 Val Leu Asp Ala Asn Cys Ser Arg Asp Val Lys Gin Met Leu Leu Lys 355 360 365
Leu Val Glu Leu Arg Ser Ser Asn Trp Gly Arg Val His Ala Thr Ser 370 375 380
Thr Tyr Arg Glu Ala Thr Pro Glu Asn Asp Pro Asn Tyr Phe Met Asn 385 390 395 400
Glu Pro Thr Phe Tyr Thr Ser Asp Gly Val Pro Phe Thr Ala Ala Asp 405 410 415
Pro Asp Tyr Gin Glu Lys Tyr Gin Glu Leu Leu Glu Arg Glu Asp Phe 420 425 430 Phe Pro Asp Tyr Glu Glu Asn Gly Thr Asp Leu Ser Gly Ala Gly Asp 435 440 445
Pro Tyr Leu Asp Asp He Asp Asp Glu Met Asp Pro Glu He Glu Glu 450 455 460
Ala Tyr Glu Lys Phe Cys Leu Glu Ser Glu Arg Lys Arg Lys Gin Xaa 465 470 475 480
(2) INFORMATION FOR SEQ ID NO: 399:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 423 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 399:
Met Glu Pro Lys Thr He Thr Asp Ala Leu Ala Ser Ser He He Lys 1 5 10 15 Ser Val Leu Pro Asn Phe Leu Pro Tyr Asn Val Met Leu Tyr Ser Asp 20 25 30
Ala Pro Val Ser Glu Leu Ser Leu Glu Leu Leu Leu Leu Gin Val Val 35 40 45
Leu Pro Ala Leu Leu Glu Gin Gly His Thr Arg Gin Trp Leu Lys Gly 50 55 60
Leu Val Arg Ala Trp Thr Val Thr Ala Gly Tyr Leu Leu Asp Leu His 65 70 75 80
Ser Tyr Leu Leu Gly Asp Gin Glu Glu Asn Glu Asn Ser Ala Asn Gin 85 90 95 Gin Val Asn Asn Asn Gin His Ala Arg Asn Asn Asn Ala He Pro Val 100 105 110
Val Gly Glu Gly Leu His Ala Ala His Gin Ala He Leu Gin Gin Gly 115 120 125
Gly Pro Val Gly Phe Gin Xaa Tyr Arg Arg Pro Leu Asn Phe Pro Leu 130 135 140
Arg He Phe Leu Leu He Val Phe Met Cys He Thr Leu Leu He Ala 145 150 155 160
Ser Leu He Cys Leu Thr Leu Pro Val Phe Ala Gly Arg Trp Leu Met 165 170 175 Ser Phe Trp Thr Gly Thr Ala Lys He His Glu Leu Tyr Thr Ala Ala 180 185 190
Cys Gly Leu Tyr Val Cys Trp Leu Thr He Arg Ala Val Thr Val Met 195 200 205
Val Ala Trp Met Pro Gin Gly Arg Arg Val He Phe Gin Lys Val Lys 210 215 220
Glu Trp Ser Leu Met He Met Lys Thr Leu He Val Ala Val Leu Leu 225 230 235 240
Ala Gly Val Val Pro Leu Leu Leu Gly Leu Leu Phe Glu Leu Val He
245 250 255 Val Ala Pro Leu Arg Val Pro Leu Asp Gin Thr Pro Leu Phe Tyr Pro
260 265 270
Trp Gin Asp Trp Ala Leu Gly Val Leu His Ala Lys He He Ala Ala 275 280 285
He Thr Leu Met Gly Pro Gin Trp Trp Leu Lys Thr Val He Glu Gin 290 295 300
Val Tyr Ala Asn Gly He Arg Asn He Asp Leu His Tyr He Val Arg 305 310 315 320
Lys Leu Ala Ala Pro Val lie Ser Val Leu Leu Leu Ser Leu Cys Val 325 330 335 Pro Tyr Val He Ala Ser Gly Val Val Pro Leu Leu Gly Val Thr Ala 340 345 350
Glu Met Gin Asn Leu Val His Arg Arg He Tyr Pro Phe Leu Leu Met 355 360 365
Val Val Val Leu Met Ala He Leu Ser Phe Gin Val Arg Gin Phe Lys 370 375 380
Arg Leu Tyr Glu His He Lys Asn Asp Lys Tyr Leu Val Gly Gin Arg 385 390 395 400
Leu Val Asn Tyr Glu Arg Lys Ser Gly Lys Gin Gly Ser Ser Pro Pro
405 410 415 Pro Pro Gin Ser Ser Gin Glu 420
(2) INFORMATION FOR SEQ ID NO: 400:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 78 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 400:
Met Leu Arg Leu Asp He He Asn Ser Leu Val Thr Thr Val Phe Met 1 5 10 15
Leu He Val Ser Val Leu Ala Leu He Pro Glu Thr Thr Thr Leu Thr 20 25 30
Val Gly Gly Gly Val Phe Ala Leu Val Thr Ala Val Cys Cys Leu Ala 35 40 45
Asp Gly Ala Leu He Tyr Arg Lys Leu Leu Phe Asn Pro Ser Gly Pro 50 55 60 Tyr Gin Lys Lys Pro Val His Glu Lys Lys Glu Val Leu Xaa 65 70 75
(2) INFORMATION FOR SEQ ID NO: 401:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 74 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 401:
Met Leu Lys Gin Val Met Phe Val Phe Ser Gly Met Gly Pro Arg Ser 1 5 10 15
His Cys Trp Gly Leu Pro Leu His Val Ala Pro Leu Cys Arg Gly His 20 25 30
Gin Ala Asp Ser Ser His Leu Leu Pro Leu Lys His Gin Gly Ala Trp 35 40 45
Asn Arg Asn Leu Ala Asn Gin Arg His Phe Phe Cys Pro Ser He Phe 50 55 60 His Thr Cys Pro Thr Val Leu Phe Phe Xaa 65 70
(2) INFORMATION FOR SEQ ID NO: 402:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 20 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 402:
Ala Arg Thr He Leu Val Leu Tyr Leu Ser Leu Gin Arg Leu Glu Asn 1 5 10 15
Leu Ala Tyr His 20
(2) INFORMATION FOR SEQ ID NO: 403:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 87 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 403:
Met Pro Leu Pro Ser Val Pro He Leu Gly He Phe Ser Phe Leu He 1 5 10 15
Pro Ser Ser Gin Gly Val Ser Tyr Thr Lys Leu Pro He Ser Ser Pro 20 25 30 Gin Tyr Ser Pro Phe Val Asn Asp His Phe Ser Phe Leu Asn Pro Phe 35 40 45
Pro Val Gin He His Thr Gly Phe Ala Arg Val Gly Ser Tyr Met Gin 50 55 60
Met Pro Leu Val His Leu Cys Leu Leu Gin Thr Ser Leu Met Lys Asn 65 70 75 80
Ser Gly Val Gin Gin Gly Ser 85
(2) INFORMATION FOR SEQ ID NO: 404:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 92 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 404:
Met Asn Ala Ala Met Val His He Asn Arg Ala Leu Lys Leu He He 1 5 10 15 Arg Leu Phe Leu Val Glu Asp Leu Val Asp Ser Leu Lys Leu Ala Val 20 25 30
Phe Met Trp Leu Met Thr Tyr Val Gly Ala Val Phe Asn Gly He Thr 35 40 45
Leu Leu He Leu Ala Glu Leu Leu He Phe Ser Val Pro He Val Tyr 50 55 60
Glu Lys Tyr Lys Thr Gin He Asp His Tyr Val Gly He Ala Arg Asp 65 70 75 80
Gin Thr Lys Ser He Val Glu Lys He Pro Ser Lys 85 90
(2) INFORMATION FOR SEQ ID NO: 405:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 21 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 405: Met Ala Cys Ser Cys Leu Met He Gin Ser Phe Ser Thr Ser Ala Leu 1 5 10 15
Val Leu Phe Tyr Gly 20
(2) INFORMATION FOR SEQ ID NO: 406: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 174 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 406:
Met Glu Glu Gly Gly Asn Leu Gly Gly Leu He Lys Met Val His Leu 1 5 10 15
Leu Val Leu Ser Gly Ala Trp Gly Met Gin Met Trp Val Thr Phe Val 20 25 30
Ser Gly Phe Pro Ala Phe Pro Lys Pro Ser Pro Thr Tyr Leu Arg Thr
35 40 45 Ser Ala Glu Gin Thr Leu Pro Leu Leu Leu Pro His Leu His Gly Leu
50 55 60
Cys Leu His Gin Pro Leu His Leu Gly Phe Thr Ala Cys Leu Gly Ser 65 70 75 80
Ala His He Leu Gly Gly Gin Pro Ala Leu Pro Ala Val Pro Glu Pro 85 90 95
Tyr Ala Gly His Cys Gin Arg Pro Leu Ala Gly Thr Pro His His Ser 100 105 110
Cys His Val Gly Pro Ala Asn Arg Gly Arg Arg Ser Glu Ala Trp Val 115 120 125 Gly Arg Tyr Gin Ala Ala Asn Arg Phe Pro He Leu Asn Ala Xaa Cys 130 135 140
Glu Arg Arg Thr Pro Ser Thr Val Leu Ser Ala Arg He Ser Ser Ala 145 150 155 160
Thr Met Gly Cys Pro Leu Phe Ala He Trp Ala Ala Ser Xaa 165 170
(2) INFORMATION FOR SEQ ID NO: 407:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 64 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 407:
Met Ala Phe He Leu Leu Phe Tyr Cys Leu Met Thr Phe Leu Ser Leu 1 5 10 15
Glu Gin Asn Ser Ala Thr Val Glu Pro Ser Ser His Glu He Leu His 20 25 30 Leu Leu Gin Asn Cys Phe Glu Leu Leu Arg Thr Ser Thr Ser Gin Cys 35 40 45
Thr Glu Gly He Pro Cys Gin Arg Tyr Gin Asn Gly Leu His He Xaa 50 55 60
(2) INFORMATION FOR SEQ ID NO: 408:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 280 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 408:
Met Glu Ala Val Val Asn Leu Tyr Gin Glu Val Met Lys His Ala Asp 1 5 10 15
Pro Arg He Gin Gly Tyr Pro Leu Met Gly Ser Pro Leu Leu Met Thr 20 25 30 Ser He Leu Leu Thr Tyr Val Tyr Phe Val Leu Ser Leu Gly Pro Arg 35 40 45
He Met Ala Asn Arg Lys Pro Phe Gin Leu Arg Gly Phe Met He Val 50 55 60
Tyr Asn Phe Ser Leu Val Ala Leu Ser Leu Tyr He Val Tyr Glu Phe 65 70 75 80
Leu Met Ser Gly Trp Leu Ser Thr Tyr Thr Trp Arg Cys Asp Pro Val 85 90 95
Asp Tyr Ser Asn Ser Pro Glu Ala Leu Arg Met Val Arg Val Ala Trp 100 105 110 Leu Phe Leu Phe Ser Lys Phe He Glu Leu Met Asp Thr Val He Phe 115 120 125
He Leu Arg Lys Lys Asp Gly Gin Val Thr Phe Leu His Val Phe His 130 135 140
His Ser Val Leu Pro Trp Ser Trp Trp Trp Gly Val Lys He Ala Pro 145 150 155 160
Gly Gly Met Gly Ser Phe His Ala Met He Asn Ser Ser Val His Val 165 170 175
He Met Tyr Leu Tyr Tyr Gly Leu Ser Ala Phe Gly Pro Val Ala Gin 180 185 190 Pro Tyr Leu Trp Trp Lys Lys His Met Thr Ala He Gin Leu He Gin 195 200 205
Phe Val Leu Val Ser Leu His He Ser Gin Tyr Tyr Phe Met Ser Ser 210 215 220
Cys Asn Tyr Gin Tyr Pro Val He He His Leu He Trp Met Tyr Gly 225 230 235 240
Thr He Phe Phe Met Leu Phe Ser Asn Phe Trp Tyr His Ser Tyr Thr 245 250 255
Lys Gly Lys Arg Leu Pro Arg Ala Leu Gin Gin Asn Gly Ala Pro Gly 260 265 270 He Ala Lys Val Lys Ala Asn Xaa 275 280
(2) INFORMATION FOR SEQ ID NO: 409:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 284 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 409:
Met Xaa Leu Trp Pro Gin Thr Cys Ser Gly Lys Phe Asp Gly Thr Leu 1 5 10 15
Ala Phe Ser He His Xaa Leu Ala Val He Leu Gly Asp Gin Leu Thr 20 25 30
Ala Ala Asp Leu Val Pro He Phe Asn Gly Phe Leu Lys Asp Leu Asp 35 40 45
Glu Val Arg He Gly Val Leu Lys His Leu His Asp Phe Leu Lys Leu 50 55 60 Leu His He Asp Lys Arg Arg Glu Tyr Leu Tyr Gin Leu Gin Glu Phe 65 70 75 80
Leu Val Thr Asp Asn Ser Arg Asn Trp Arg Phe Arg Ala Glu Leu Ala 85 90 95
Glu Gin Leu He Leu Leu Leu Glu Leu Tyr Ser Pro Arg Asp Val Tyr 100 105 110
Asp Tyr Leu Arg Pro He Ala Leu Asn Leu Cys Ala Asp Lys Val Ser 115 120 125
Ser Val Arg Trp He Ser Tyr Lys Leu Val Ser Glu Met Val Lys Lys 130 135 140 Leu His Ala Ala Thr Pro Pro Thr Phe Gly Val Asp Leu He Asn Glu 145 150 155 160
Leu Val Glu Asn Phe Gly Arg Cys Pro Lys Trp Ser Gly Arg Gin Ala 165 170 175
Phe Val Phe Val Cys Gin Thr Val He Glu Asp Asp Cys Leu Pro Met 180 185 190
Asp Gin Phe Ala Val His Leu Met Pro His Leu Leu Thr Leu Ala Asn 195 200 205
Asp Arg Val Pro Asn Val Arg Val Leu Leu Ala Lys Thr Leu Arg Gin 210 215 220 Thr Leu Leu Glu Lys Asp Tyr Phe Leu Ala Ser Ala Ser Cys His Gin 225 230 235 240
Glu Ala Val Glu Gin Thr He Met Ala Leu Gin Met Asp Arg Asp Ser 245 250 255
Asp Val Lys Tyr Phe Ala Ser He His Pro Ala Ser Thr Lys He Ser 260 265 270
Glu Asp Ala Met Ser Thr Ala Ser Ser Thr Tyr Xaa 275 280
(2) INFORMATION FOR SEQ ID NO: 410:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 187 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 410:
Met Leu Phe Leu Phe Phe Val He He Phe Leu Phe Val Phe Leu He 1 5 10 15 Leu He He Gin Phe Ser Lys Pro Leu Thr Asn Pro His Pro Pro Ala 20 25 30
Gly Xaa Ser Asp Arg Arg Arg Arg Tyr Ser Ser Tyr Arg Ser His Asp 35 40 45
His Tyr Gin Arg Gin Arg Val Leu Gin Lys Glu Arg Ala He Glu Glu 50 55 60
Arg Arg Val Val Phe He Gly Lys He Pro Gly Arg Met Thr Arg Ser 65 70 75 80
Glu Leu Lys Gin Arg Phe Ser Val Phe Gly Glu He Glu Glu Cys Thr
85 90 95 He His Phe Arg Val Gin Gly Asp Asn Tyr Gly Phe Val Thr Tyr Arg 100 105 110
Tyr Ala Glu Glu Ala Phe Ala Ala He Glu Ser Gly His Lys Leu Arg 115 120 125
Gin Ala Asp Glu Gin Pro Phe Asp Leu Cys Phe Gly Gly Arg Arg Xaa 130 135 140
Xaa Cys Lys Arg Ser Tyr Ser Asp Leu Asp Ser Asn Arg Glu Asp Phe 145 150 155 160
Asp Pro Ala Pro Val Lys Ser Lys Phe Asp Ser Leu Asp Phe Asp Thr 165 170 175 Leu Leu Lys Gin Ala Gin Lys Asn Leu Arg Arg 180 185
(2) INFORMATION FOR SEQ ID NO: 411:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 237 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 411:
Met Lys Leu Pro Gly Lys Phe Arg Arg Ala His Gin Gly Asn Leu Glu 1 5 10 15
Ser Gin Leu Thr Ser Glu Ser Tyr Tyr Lys Glu Thr Leu Ser Val Pro 20 25 30
Thr Val Glu His He He Gin Glu Leu Lys Asp He Phe Ser Glu Gin 35 40 45
His Leu Lys Ala Leu Lys Cys Leu Ser Leu Val Pro Ser Val Met Gly 50 55 60 Gin Leu Lys Phe Asn Thr Ser Glu Glu His His Ala Asp Met Tyr Arg 65 70 75 80
Ser Asp Leu Pro Asn Pro Asp Thr Leu Ser Ala Glu Leu His Cys Trp 85 90 95
Arg He Lys Trp Lys His Arg Gly Lys Asp He Glu Leu Pro Ser Thr 100 105 110
He Tyr Glu Ala Leu His Leu Pro Asp He Lys Phe Phe Pro Asn Val 115 120 125
Tyr Ala Leu Leu Lys Val Leu Cys He Leu Pro Val Met Lys Val Glu 130 135 140 Asn Glu Arg Tyr Glu Asn Gly Arg Lys Arg Leu Lys Ala Tyr Leu Arg 145 150 155 160
Asn Thr Leu Thr Asp Gin Arg Ser Ser Asn Leu Ala Leu Leu Asn He 165 170 175
Asn Phe Asp He Lys His Asp Leu Asp Leu Met Val Asp Thr Tyr He 180 185 190
Lys Leu Tyr Thr Xaa Xaa Ser Xaa Leu Xaa Thr Xaa Xaa Ser Xaa Xaa 195 200 205
Val Glu Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Xaa Gly Xaa Xaa Xaa Xaa 210 215 220 Asp Xaa Xaa Xaa Arg Glu Lys Ala Val Arg Cys Met Xaa 225 230 235
(2) INFORMATION FOR SEQ ID NO: 412:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 192 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 412:
Met Lys Pro Met Ala Val Val Ala Ser Thr Val Leu Gly Leu Val Gin 1 5 10 15
Asn Met Arg Ala Phe Gly Gly He Leu Val Val Val Tyr Tyr Val Phe 20 25 30
Ala He He Gly He Asn Leu Phe Arg Gly Val He Val Ala Leu Pro 35 40 45
Gly Asn Ser Ser Leu Ala Pro Ala Asn Gly Ser Ala Pro Cys Gly Ser 50 55 60
Phe Glu Gin Leu Glu Tyr Trp Ala Asn Asn Phe Asp Asp Phe Ala Ala 65 70 75 80
Ala Leu Val Thr Leu Trp Asn Leu Met Val Val Asn Asn Trp Gin Val 85 90 95
Phe Leu Asp Ala Tyr Arg Arg Tyr Ser Gly Pro Trp Ser Lys He Tyr 100 105 110
Phe Val Leu Trp Trp Leu Val Ser Ser Val He Trp Val Asn Leu Phe 115 120 125
Leu Ala Leu He Leu Glu Asn Phe Leu His Lys Trp Asp Pro Arg Ser 130 135 140 His Leu Gin Pro Leu Ala Gly Thr Pro Glu Ala Thr Tyr Gin Met Thr 145 150 155 160
Val Glu Leu Leu Phe Arg Asp He Leu Glu Glu Pro Gly Glu Asp Glu 165 170 175
Leu Thr Glu Arg Leu Ser Gin His Pro His Leu Trp Leu Cys Arg Xaa 180 185 190
(2) INFORMATION FOR SEQ ID NO: 413:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 413:
Asn Val Val Val Val Ala Phe Gly Leu He Leu He He Glu Ser Leu 1 5 10 15 Gly Glu Gin Cys Pro 20
(2) INFORMATION FOR SEQ ID NO: 414:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 51 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 414:
Met Asn Trp Gly Leu Ser He Trp Leu His Tyr Tyr Glu Lys Lys Lys 1 5 10 15
Glu Gin Val Phe Leu Val He Leu Ala His Val Val Arg Arg Cys Ala 20 25 30
Ser Asp Gly He Leu Gin Phe Glu Ser Ser Leu Leu Lys Met Arg Arg 35 40 45
Ala Pro Xaa 50
(2) INFORMATION FOR SEQ ID NO: 415:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 32 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 415: Met Leu He He Ser Leu Arg Pro Gin Phe Pro Ser Leu He Val Gin 1 5 10 15
Leu Glu Cys Ser Val Leu Phe Leu Pro He Ser Leu Asn Leu Leu Leu 20 25 30
(2) INFORMATION FOR SEQ ID NO: 416:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 163 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 416:
Met Val Lys Val Cys Asn Asp Ser Asp Arg Trp Ser Leu He Ser Leu 1 5 10 15
Ser Asn Asn Ser Gly Lys Asn Val Glu Leu Lys Phe Val Asp Ser Leu 20 25 30 Arg Arg Gin Phe Glu Phe Ser Val Asp Ser Phe Gin He Lys Leu Asp 35 40 45
Ser Leu Leu Leu Phe Tyr Glu Cys Ser Glu Asn Pro Met Thr Glu Thr 50 55 60
Phe His Pro Thr He He Gly Glu Ser Val Tyr Gly Asp Phe Gin Glu 65 70 75 80
Ala Phe Asp His Leu Cys Asn Lys He He Ala Thr Arg Asn Pro Glu 85 90 95
Glu He Arg Gly Gly Gly Leu Leu Lys Tyr Cys Asn Leu Leu Val Arg 100 105 110 Gly Phe Arg Pro Ala Ser Asp Glu He Lys Thr Leu Gin Arg Tyr Met
115 120 125
Cys Ser Arg Phe Phe lie Asp Phe Ser Asp He Gly Glu Gin Gin Arg 130 135 140
Lys Leu Glu Ser Tyr Leu Gin Asn His Phe Val Gly He Gly Arg Pro 145 150 155 160
Gin Val Xaa
(2) INFORMATION FOR SEQ ID NO: 417:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 174 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 417:
Met Ala Pro Lys Gly Lys Val Gly Thr Arg Gly Lys Lys Gin He Phe 1 5 10 15 Glu Glu Asn Arg Glu Thr Leu Lys Phe Tyr Leu Arg He He Leu Gly 20 25 30
Ala Asn Ala He Tyr Cys Leu Val Thr Leu Val Phe Phe Tyr Ser Ser 35 40 45
Ala Ser Phe Trp Ala Trp Leu Ala Leu Gly Phe Ser Leu Ala Val Tyr 50 55 60
Gly Ala Ser Tyr His Ser Met Ser Ser Met Ala Arg Ala Ala Phe Ser 65 70 75 80
Glu Asp Gly Ala Leu Met Asp Gly Gly Met Asp Leu Asn Met Glu Gin
85 90 95 Gly Met Ala Glu His Leu Lys Asp Val He Leu Leu Thr Ala He Val 100 105 110
Gin Val Leu Ser Cys Phe Ser Leu Tyr Val Trp Ser Phe Trp Leu Leu 115 120 125
Ala Pro Gly Arg Ala Leu Tyr Leu Leu Trp Val Asn Val Leu Gly Pro 130 135 140
Trp Phe Thr Ala Asp Ser Gly Thr Pro Ala Pro Glu His Asn Glu Lys 145 150 155 160
Arg Gin Arg Arg Gin Glu Arg Arg Gin Met Lys Arg Leu Xaa 165 170
(2) INFORMATION FOR SEQ ID NO: 418:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 50 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 418:
Met Glu Leu Pro Lys Gly Leu Gin Gly Val Gly Pro Val Ala Met Met 1 10 15
Arg Pro Phe Tyr Leu Leu Leu Pro Val Leu Cys Thr Gin Ala Leu Arg 20 25 30
Gin Ser Gin Gly Lys Ser Pro Leu Leu Trp Lys Arg Thr Cys Cys Leu 35 40 45
Ala Xaa 50
(2) INFORMATION FOR SEQ ID NO: 419:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 120 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 419:
Met Leu Gly Lys Gly Gly Gly Arg Ala Gly Leu Leu Arg Tyr Arg Leu 1 5 10 15 Leu Tyr Phe Thr Leu Val Val Gly Glu Gly Glu Pro Gly Glu Asn Lys 20 25 30
Val Thr He Pro Phe Phe Glu Thr Gly Lys Lys He He Phe Cys Ser 35 40 45
Val Lys Met Val Glu Asn Ser Asn Val Pro Ser His Lys Gly Pro Val 50 55 60
Pro Leu Arg Ser Glu Gin Trp Glu Leu Lys He Ser Glu Thr Leu Gly 65 70 75 80
Glu Gly Lys He Gly Phe Leu Leu He Gly Arg Cys Ser Ser Gly Xaa 85 90 95 Gly Gly Leu Cys Phe Cys Trp Asp Val Leu Cys Cys Met Tyr Ala Tyr 100 105 110
Met Asp Arg Ser Leu Leu Ser Leu 115 120
(2) INFORMATION FOR SEQ ID NO: 420: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 159 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 420:
Met Thr His Leu Leu Leu Thr Ala Thr Val Thr Pro Ser Glu Gin Asn 1 5 10 15
Ser Ser Arg Glu Pro Gly Trp Glu Thr Ala Met Ala Lys Asp He Leu 20 25 30
Gly Glu Ala Gly Leu His Phe Asp Glu Leu Asn Lys Leu Arg Val Leu 35 40 45 Asp Pro Glu Val Thr Gin Gin Thr He Glu Leu Lys Glu Glu Cys Lys 50 55 60
Asp Phe Val Asp Lys He Gly Gin Phe Gin Lys He Val Gly Gly Leu 65 70 75 80
He Glu Leu Val Asp Gin Leu Ala Lys Glu Ala Glu Asn Glu Lys Met 85 90 95
Lys Ala He Gly Ala Arg Asn Leu Leu Lys Ser He Ala Lys Gin Arg 100 105 110
Glu Ala Gin Gin Gin Gin Leu Gin Ala Leu He Ala Glu Lys Lys Met 115 120 125 Gin Leu Glu Arg Tyr Arg Val Glu Tyr Glu Ala Leu Cys Lys Val Glu 130 135 140
Ala Glu Gin Asn Glu Phe He Asp Gin Phe He Phe Gin Lys Xaa 145 150 155
(2) INFORMATION FOR SEQ ID NO: 421: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 154 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 421:
Met Asn Val Gly Val Ala His Ser Glu Val Asn Pro Asn Thr Arg Val 1 5 10 15
Met Asn Ser Arg Gly Met Trp Leu Thr Tyr Ala Leu Gly Val Gly Leu 20 25 30
Leu His He Val Leu Leu Ser He Pro Phe Phe Ser Val Pro Val Ala 35 40 45 Trp Thr Leu Thr Asn He He His Asn Leu Gly Met Tyr Val Phe Leu 50 55 60
His Ala Val Lys Gly Thr Pro Phe Glu Thr Pro Asp Gin Gly Lys Ala 65 70 75 80
Arg Leu Leu Thr His Trp Glu Gin Leu Asp Tyr Gly Val Gin Phe Thr 85 90 95
Ser Ser Arg Lys Phe Phe Thr He Ser Pro He He Leu Tyr Phe Leu 100 105 110
Ala Ser Phe Tyr Thr Lys Tyr Asp Pro Thr His Phe He Leu Asn Thr 115 120 125
Ala Ser Leu Leu Ser Val Leu He Pro Lys Met Pro Gin Leu His Gly 130 135 140
Val Arg He Phe Gly He Asn Lys Tyr Xaa 145 150
(2) INFORMATION FOR SEQ ID NO: 422: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 204 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 422:
Met Val Cys Gly Gly Phe Ala Cys Ser Lys Asn Cys Leu Cys Ala Leu 1 5 10 15
Asn Leu Leu Tyr Thr Leu Val Ser Leu Leu Leu He Gly He Ala Ala 20 25 30
Trp Gly He Gly Phe Gly Leu He Ser Ser Leu Arg Val Val Gly Val 35 40 45 Val He Ala Val Gly He Phe Leu Phe Leu He Ala Leu Val Gly Leu 50 55 60
He Gly Ala Val Lys His His Gin Val Leu Leu Phe Phe Tyr Met He 65 70 75 80
He Leu Leu Leu Val Phe He Val Gin Phe Ser Val Ser Cys Ala Cys 85 90 95
Leu Ala Leu Asn Gin Glu Gin Gin Gly Gin Leu Leu Glu Val Gly Trp 100 105 110
Asn Asn Thr Ala Ser Ala Arg Asn Asp He Gin Arg Asn Leu Asn Cys 115 120 125 Cys Gly Phe Arg Ser Val Asn Pro Asn Asp Thr Cys Leu Ala Ser Cys 130 135 140
Val Lys Ser Asp His Ser Cys Ser Pro Cys Ala Pro He He Gly Glu 145 150 155 160
Tyr Ala Gly Glu Val Leu Arg Phe Val Gly Gly He Gly Leu Phe Phe 165 170 175
Ser Phe Thr Glu He Leu Gly Val Trp Leu Thr Tyr Arg Tyr Arg Asn 180 185 190
Gin Lys Asp Pro Arg Ala Asn Pro Ser Ala Phe Leu 195 200
(2) INFORMATION FOR SEQ ID NO: 423:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 67 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 423: Met Leu Gin Ser He He Lys Asn He Trp He Pro Met Lys Pro Tyr 1 5 10 15
Tyr Thr Lys Val Tyr Gin Glu He Trp He Gly Met Gly Leu Met Gly 20 25 30
Phe He Val Tyr Lys He Arg Ala Ala Asp Lys Arg Ser Lys Ala Leu 35 40 45
Lys Ala Ser Ala Pro Ala Pro Gly His His Asn Gin He Tyr Leu Glu 50 55 60
Tyr Met Xaa 65
(2) INFORMATION FOR SEQ ID NO: 424:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 25 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 424: Met Leu Gly Val Ser Leu Phe Leu Leu Val Val Leu Tyr His Tyr Val 1 5 10 15
Ala Val Asn Asn Pro Lys Lys Gin Glu 20 25
(2) INFORMATION FOR SEQ ID NO: 425: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 299 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 425:
Met Ala Ala Xaa Glu Pro Ala Val Leu Ala Leu Pro Asn Ser Gly Ala 1 5 10 15
Gly Gly Ala Gly Ala Pro Ser Gly Thr Val Pro Val Leu Phe Cys Phe 20 25 30
Ser Val Phe Ala Arg Pro Ser Ser Val Pro His Gly Ala Gly Tyr Glu 35 40 45 Leu Leu He Gin Lys Phe Leu Ser Leu Tyr Gly Asp Gin He Asp Met
50 55 60
His Arg Lys Phe Val Val Gin Leu Phe Ala Glu Glu Trp Gly Gin Tyr 65 70 75 80
Val Asp Leu Pro Lys Gly Phe Ala Val Ser Glu Arg Cys Lys Val Arg 85 90 95
Leu Val Pro Leu Gin He Gin Leu Thr Thr Leu Gly Asn Leu Thr Pro 100 105 110
Ser Ser Thr Val Phe Phe Cys Cys Asp Met Gin Glu Arg Phe Arg Pro 115 120 125 Ala He Lys Tyr Phe Gly Asp He He Ser Val Gly Gin Arg Leu Leu 130 135 140
Gin Gly Ala Arg He Leu Gly He Pro Val He Val Thr Glu Gin Tyr 145 150 155 160
Pro Lys Gly Leu Gly Ser Thr Val Gin Glu He Asp Leu Thr Gly Val 165 170 175
Lys Leu Val Leu Pro Lys Thr Lys Phe Ser Met Val Leu Pro Glu Val 180 185 190
Glu Ala Ala Leu Ala Glu He Pro Gly Val Arg Ser Val Val Leu Phe 195 200 205 Gly Val Glu Thr His Val Cys He Gin Gin Thr Ala Leu Glu Leu Val 210 215 220
Gly Arg Gly Val Glu Val His He Val Ala Asp Ala Thr Ser Ser Arg 225 230 235 240
Ser Met Met Asp Arg Met Phe Ala Leu Glu Arg Leu Ala Xaa Xaa Gly 245 250 255
He He Val Thr Thr Ser Glu Ala Val Leu Leu Gin Leu Val Ala Asp 260 265 270
Lys Asp His Pro Lys Phe Lys Glu He Gin Asn Leu He Lys Ala Ser 275 280 285 Ala Pro Glu Ser Gly Leu Leu Ser Lys Val Xaa 290 295
(2) INFORMATION FOR SEQ ID NO: 426:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 13 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 426:
Met Arg Asp Leu Gly Thr Leu Leu Ser Pro Val Cys Ser 1 5 10
(2) INFORMATION FOR SEQ ID NO: 427:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 198 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 427:
Met Phe Gly Cys Leu Val Ala Gly Arg Leu Val Gin Thr Ala Ala Gin 1 5 10 15
Gin Val Ala Glu Asp Lys Phe Val Phe Asp Leu Pro Asp Tyr Glu Ser 20 25 30
He Asn His Val Val Val Phe Met Leu Gly Thr He Pro Phe Pro Glu
35 40 45 Gly Met Gly Gly Ser Val Tyr Phe Ser Tyr Pro Asp Ser Asn Gly Met
50 55 60
Pro Val Trp Gin Leu Leu Gly Phe Val Thr Asn Gly Lys Pro Ser Ala 65 70 75 80
He Phe Lys He Ser Gly Leu Lys Ser Gly Glu Gly Ser Gin His Pro 85 90 95
Phe Gly Ala Met Asn He Val Arg Thr Pro Ser Val Ala Gin He Gly 100 105 110
He Ser Val Glu Leu Leu Asp Ser Met Ala Gin Gin Thr Pro Val Gly 115 120 125 Asn Ala Ala Val Ser Ser Val Asp Ser Phe Thr Gin Phe Thr Gin Lys 130 135 140
Met Leu Asp Asn Phe Tyr Asn Phe Ala Ser Ser Phe Ala Val Ser Gin 145 150 155 160
Ala Gin Met Thr Pro Ser Pro Ser Glu Met Phe He Pro Ala Asn Val 165 170 175
Val Leu Lys Trp Tyr Glu Asn Phe Gin Arg Arg Leu Ala Gin Asn Pro 180 185 190
Xaa Phe Trp Xaa Thr Xaa 195
(2) INFORMATION FOR SEQ ID NO: 428:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 47 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 428: Met Gly Leu Pro Leu Met Ala Leu Met Trp Ser Thr Leu Pro Ala Ser
1 5 10 15
Ala Gly Val Asn Phe He Leu Ala Leu Pro Leu Leu Leu Leu Trp Lys 20 25 30
Asn Arg Gly Gly Val Gly Arg Ser Val Met Ser Ala Val Glu Xaa 35 40 45
(2) INFORMATION FOR SEQ ID NO: 429:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 370 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 429:
Met Lys Lys Val Glu Glu Lys Arg Val Asp Val Asn Ser Ala Val Ala 1 5 10 15
Met Gly Glu Val He Leu Ala Val Cys His Pro Asp Cys He Thr Thr 20 25 30 He Lys His Trp He Thr He He Arg Ala Arg Phe Glu Glu Val Leu 35 40 45
Thr Trp Ala Lys Gin His Gin Gin Arg Leu Glu Thr Ala Leu Ser Glu 50 55 60
Leu Val Ala Asn Ala Glu Leu Leu Glu Glu Leu Leu Ala Trp He Gin
65 70 75 80
Trp Ala Glu Thr Thr Leu He Gin Arg Asp Gin Glu Pro He Pro Gin 85 90 95
Asn He Asp Arg Val Lys Ala Leu He Ala Glu His Gin Thr Phe Met 100 105 110 Glu Glu Met Thr Arg Lys Gin Pro Asp Val Asp Arg Val Thr Lys Thr 115 120 125
Tyr Lys Arg Lys Asn He Glu Pro Thr His Ala Pro Phe He Glu Lys 130 135 140
Ser Arg Ser Gly Gly Arg Lys Ser Leu Ser Gin Pro Thr Pro Pro Pro 145 150 155 160
Met Pro He Leu Ser Gin Ser Glu Ala Lys Asn Pro Arg He Asn Gin 165 170 175
Leu Ser Ala Arg Trp Gin Gin Val Trp Leu Leu Ala Leu Glu Arg Gin 180 185 190 Arg Lys Leu Asn Asp Ala Leu Asp Arg Leu Glu Glu Leu Lys Glu Phe 195 200 205
Ala Asn Phe Asp Phe Asp Val Trp Arg Lys Lys Tyr Met Arg Trp Met 210 215 220
Asn His Lys Lys Ser Arg Val Met Asp Phe Phe Arg Arg He Asp Lys 225 230 235 240
Asp Gin Asp Gly Lys He Thr Arg Gin Glu Phe He Asp Gly He Leu 245 250 255
Ala Ser Lys Phe Pro Thr Thr Lys Leu Glu Met Thr Ala Val Ala Asp 260 265 270 He Phe Asp Arg Asp Gly Asp Gly Tyr He Asp Tyr Tyr Glu Phe Val 275 280 285
Ala Ala Leu His Pro Asn Lys Asp Ala Tyr Arg Pro Thr Thr Asp Ala 290 295 300
Asp Lys He Glu Asp Glu Val Thr Arg Gin Val Ala Gin Cys Lys Cys 305 310 315 320
Ala Lys Arg Phe Gin Val Glu Gin He Gly Glu Asn Lys Tyr Arg Phe 325 330 335
Phe Leu Gly Asn Gin Phe Gly Asp Ser Gin Gin Leu Arg Leu Val Arg 340 345 350 He Leu Arg Asn Arg Asp Gly Ser Arg Trp Trp Arg Met Asp Gly Leu 355 360 365
Gly Xaa 370
(2) INFORMATION FOR SEQ ID NO: 430: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 30 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 430:
Met Asn Val Lys Thr Phe Ser Xaa Asp His Met His Phe Leu Cys Cys 1 5 10 15
Leu Tyr Leu Arg Tyr Val Thr Phe Val Tyr Leu Asn Leu Phe 20 25 30
(2) INFORMATION FOR SEQ ID NO: 431:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 24 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 431:
Met Glu Pro His Leu Arg Cys Arg Val Thr Arg Val Arg Gly Ser Leu 1 5 10 15 Gly Asn Thr Gly Arg Trp Leu Leu
20
(2) INFORMATION FOR SEQ ID NO: 432:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 53 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 432:
Met His Tyr Leu Val Leu Gly Gly Leu Gly Val Phe Leu Phe Phe Ser 1 5 10 15
Cys Phe Val Phe Leu Phe Phe Xaa Phe Ser Phe Ala Phe Phe Pro Phe 20 25 30
Tyr Leu Glu Gly Met Gly Gly Ser Gly Asn Arg Glu Val Gly Gly Gly 35 40 45
Phe Cys Leu Phe Phe 50
(2) INFORMATION FOR SEQ ID NO: 433:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 176 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 433: Met Val Ser Lys Ala Leu Leu Arg Leu Val Ser Ala Val Asn Arg Arg 1 5 10 15
Arg Met Lys Leu Leu Leu Gly He Ala Leu Leu Ala Tyr Val Ala Ser 20 25 30
Val Trp Gly Asn Phe Val Asn Met Arg Ser He Gin Glu Asn Gly Glu 35 40 45
Leu Lys He Glu Ser Lys He Glu Glu Met Val Glu Pro Leu Arg Glu 50 55 60
Lys He Arg Asp Leu Glu Lys Ser Phe Thr Gin Lys Tyr Pro Pro Val 65 70 75 80 Lys Phe Leu Ser Glu Lys Asp Arg Lys Arg He Leu He Thr Gly Gly
85 90 95
Ala Gly Phe Val Gly Ser His Leu Thr Asp Lys Leu Met Met Asp Gly 100 105 110
His Glu Val Thr Val Val Asp Asn Phe Phe Thr Gly Arg Lys Arg Asn 115 120 125
Val Glu His Trp He Gly His Glu Asn Phe Glu Leu He Asn His Asp 130 135 140
Val Trp Ser Pro Ser Thr Ser Arg Leu Thr Arg Tyr Thr He Trp His 145 150 155 160
Leu Gin Pro Pro Leu Gin Thr Thr Cys He He Leu Ser Arg His Xaa 165 170 175
(2) INFORMATION FOR SEQ ID NO: 434: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 77 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 434:
Met Leu Arg Cys Trp Pro Leu Phe Trp Leu Pro Leu Val Ser Pro Phe
1 5 10 15
Cys Ser Leu Phe Trp Leu Leu Val Glu Trp Phe Gly Thr Asn He Asp 20 25 30
Arg Glu Ser Tyr Asp Ala He Gly Gly Pro Ser Trp Met Thr Ala Ser 35 40 45 Ser Phe Cys Leu Ser Asn Ser Asn He Trp Ser Leu Glu He Ser Ser 50 55 60
Gly Ser Thr Ser Val Val His Ser Gin Gin Ala Met Asp 65 70 75
(2) INFORMATION FOR SEQ ID NO: 435: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 32 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 435:
Met Arg Ser Cys Glu He Gin Leu Cys Val Trp Leu Leu Val Ser Ser 1 5 10 15
His Val Asp Met Val Leu Gly Gly Ser Pro Ser Thr Leu Tyr Met Met 20 25 30
(2) INFORMATION FOR SEQ ID NO: 436:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 30 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 436: Met Val Val Asn Ser Leu Cys Phe Leu Ser Leu Leu Leu Val He Leu 1 5 10 15
Glu Leu Ser Thr Asp Ser Ser Ala Arg Leu Leu Tyr His Glu 20 25 30
(2) INFORMATION FOR SEQ ID NO: 437: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 69 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 437:
Met Asp Lys Gin Lys His Leu Glu Val Arg Arg Ser Val Phe Lys He 1 5 10 15
Gin Gly Lys He Ala Phe Ser Leu Met Phe Val Leu Lys Asp Leu Ser 20 25 30
Pro Thr He Phe Ser His Ser He Leu Leu Leu Leu Pro His His Val
35 40 45 Leu Pro Cys Thr Pro Gin Met Val Arg Gly Val Thr Gin Val Leu Arg 50 55 60
Glu Phe Gly Asp Gin 65
(2) INFORMATION FOR SEQ ID NO: 438: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 19 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 438:
Met Pro Leu Cys Phe Phe Ser Phe Leu Cys Cys Trp Val Leu Val Phe 1 5 10 15
Lys Leu He
(2) INFORMATION FOR SEQ ID NO: 439:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 43 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 439:
Met Lys Phe Ser Leu Val Leu Leu He Lys He He Ser Phe Glu Arg 1 5 10 15 Leu Leu He Phe Leu Phe Pro Leu Ser Phe Leu Pro Asn He Trp Arg 20 25 30
Arg Val Met Val Asn Leu Asn He Leu Phe Xaa 35 40
(2) INFORMATION FOR SEQ ID NO: 440: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 33 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 440:
Met Leu Leu Phe Pro Ser Leu Leu Phe Ala Ala Thr Tyr Asn Val Ala 1 5 10 15
Asn Pro Ser Arg Leu He Leu Tyr Met He Ser Ala Gly Ala Asp Ser 20 25 30
Gin
(2) INFORMATION FOR SEQ ID NO: 441:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 53 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 441: Met Trp Gin Val Arg Gly Leu Pro Pro Val Pro Leu Leu Leu Thr Met 1 5 10 15
Ser Pro Pro Pro Cys Leu Ser Ser Pro Phe Pro Phe He Ser Val Pro 20 25 30
Leu Phe Glu Ala Val Pro He Ser Val Ser Asp Gin Pro Ser Pro Xaa 35 40 45
Leu Thr Thr Leu Leu so
(2) INFORMATION FOR SEQ ID NO: 442:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 64 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 442:
Met He Thr Ser Val Leu Val Phe Leu He Phe Phe Phe Pro Tyr Leu 1 5 10 15
Ser Leu Val Thr Leu Leu Gin Ala Arg Asn Leu Trp Val He His Arg 20 25 30
Ala Ala Leu Cys Glu Ser Gly Leu Phe His Trp Arg Lys Gly He Glu 35 40 45
Asn Gin Leu Glu Pro Met Tyr Phe Leu Pro His Gly Thr Leu Phe Leu 50 55 60
(2) INFORMATION FOR SEQ ID NO: 443:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 34 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 443:
Met Leu Tyr Ser Cys Glu Pro Tyr Leu He He Leu Asn He Tyr Ser 1 5 10 15 Gin Lys Ala Phe Tyr Phe Tyr Phe Phe Glu Gly Ser Phe Ser Val Cys 20 25 30
Thr Leu
(2) INFORMATION FOR SEQ ID NO: 444: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 89 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 444:
Met Arg Gin Arg Gin Ala Ala Cys Gin Pro Pro Pro Ser Arg Asn Gly 1 5 10 15
Leu Ala Gin Glu Cys Pro Pro His He Pro Ser Ser Phe Phe Leu Val 20 25 30
Lys Leu Leu Phe He Pro Trp Leu Ala Ser Leu Leu Ser Ser Pro Leu 35 40 45 Asn Leu Leu Leu Leu Val Ser He Ser Trp Asp Leu Gly Leu Lys Leu 50 55 60
Asn Leu Gin Gin Cys Arg Gin His Gin Val Leu Gin Glu Lys Asn Thr 65 70 75 80
Lys Lys Phe Asn Lys Lys Lys Lys Lys 85
(2) INFORMATION FOR SEQ ID NO: 445:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 350 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 445:
Met Asp Phe He Thr Ser Thr Ala He Leu Pro Leu Leu Phe Gly Cys 1 5 10 15
Leu Gly Val Phe Gly Leu Phe Arg Leu Leu Gin Trp Val Arg Gly Lys 20 25 30 Ala Tyr Leu Arg Asn Ala Val Val Val He Thr Gly Ala Thr Ser Gly 35 40 45
Leu Gly Lys Glu Cys Ala Lys Val Phe Tyr Ala Ala Gly Ala Lys Leu 50 55 60
Val Leu Cys Gly Arg Asn Gly Gly Ala Leu Glu Glu Leu He Arg Glu 65 70 75 80
Leu Thr Ala Ser His Ala Thr Lys Val Gin Thr His Lys Pro Tyr Leu 85 90 95
Val Thr Phe Asp Leu Thr Asp Ser Gly Ala He Val Ala Ala Ala Ala 100 105 110 Glu He Leu Gin Cys Phe Gly Tyr Val Asp He Leu Val Asn Asn Ala 115 120 125
Gly He Ser Tyr Arg Gly Thr He Met Asp Thr Thr Val Asp Val Asp 130 135 140
Lys Arg Val Met Glu Thr Asn Tyr Phe Gly Pro Val Ala Leu Thr Lys 145 150 155 160
Ala Leu Leu Pro Ser Met He Lys Arg Arg Gin Gly His He Val Ala 165 170 175
He Ser Ser He Gin Gly Lys Met Ser He Pro Phe Arg Ser Ala Tyr
180 185 190 Ala Ala Ser Lys His Ala Thr Gin Ala Phe Phe Asp Cys Leu Arg Ala 195 200 205
Glu Met Glu Gin Tyr Glu He Glu Val Thr Val He Ser Pro Gly Tyr 210 215 220
He His Thr Asn Leu Ser Val Asn Ala He Thr Ala Asp Gly Ser Arg 225 230 235 240
Tyr Gly Val Met Asp Thr Thr Thr Ala Gin Gly Arg Ser Pro Val Glu 245 250 255
Val Ala Gin Asp Val Leu Ala Ala Val Gly Lys Lys Lys Lys Asp Val 260 265 270
He Leu Ala Asp Leu Leu Pro Ser Leu Ala Val Tyr Leu Arg Thr Leu 275 280 285
Ala Pro Gly Leu Phe Phe Ser Leu Met Pro Pro Gly Pro Glu Lys Ser 290 295 300
Gly Asn Pro Arg Thr Pro Ser Thr Leu Thr Ser Gin Gly Gin Gly Arg 305 310 315 320
Glu Ala Ala Leu Leu Gly Leu Leu Thr Leu Gin Gly Thr Val Ala Phe 325 330 335
Val Glu Thr Leu Met Glu He Cys Leu Thr Ser Gly Lys Asp 340 345 350
(2) INFORMATION FOR SEQ ID NO: 446:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 49 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 446: Met Val Phe Leu Pro Arg Gly Val Val Val Ser Gly Gly Ala Ala Cys 1 5 10 15
Leu Trp Leu Thr Phe He Leu Glu Thr Glu Val Tyr Leu Asp Leu Ala 20 25 30
Thr Glu Ala Arg Ala His Ser Arg Met Gly Leu Gly Leu Trp Pro Pro 35 40 45
Asn
(2) INFORMATION FOR SEQ ID NO: 447:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 278 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 447:
Met Ala Ser Ala Glu Leu Asp Tyr Thr He Glu He Pro Asp Gin Pro 1 5 10 15 Cys Trp Ser Gin Lys Asn Ser Pro Ser Pro Gly Gly Lys Glu Ala Glu 20 25 30
Thr Arg Gin Pro Val Val He Leu Leu Gly Trp Gly Gly Cys Lys Asp 35 40 45
Lys Asn Leu Ala Lys Tyr Ser Ala He Tyr His Lys Arg Gly Cys He 50 55 60
Val He Arg Tyr Thr Ala Pro Trp His Met Val Phe Phe Ser Glu Ser 65 70 75 80
Leu Gly He Pro Ser Leu Arg Val Leu Ala Gin Lys Leu Leu Glu Leu 85 90 95 Leu Phe Asp Tyr Glu He Glu Lys Glu Pro Leu Leu Phe His Val Phe 100 105 110
Ser Asn Gly Gly Val Met Leu Tyr Arg Tyr Val Leu Glu Leu Leu Gin 115 120 125
Thr Arg Arg Phe Cys Arg Leu Arg Val Val Gly Thr He Phe Asp Ser 130 135 140
Ala Pro Gly Asp Ser Asn Leu Val Gly Ala Leu Arg Ala Leu Ala Ala 145 150 155 160
He Leu Glu Arg Arg Ala Ala Met Leu Arg Leu Leu Leu Leu Val Ala 165 170 175 Phe Ala Leu Val Val Val Leu Phe His Val Leu Leu Ala Pro He Thr 180 185 190
Ala Xaa Phe His Thr His Phe Tyr Asp Arg Leu Gin Asp Ala Gly Ser 195 200 205
Arg Trp Pro Glu Leu Tyr Leu Tyr Ser Arg Ala Asp Glu Val Val Leu 210 215 220
Ala Arg Asp He Glu Arg Met Val Glu Ala Arg Leu Ala Arg Arg Val 225 230 235 240
Leu Ala Arg Ser Val Asp Phe Val Ser Ser Ala His Val Ser His Leu 245 250 255 Arg Asp Tyr Pro Thr Tyr Tyr Thr Ser Leu Cys Val Asp Phe Met Arg 260 265 270
Asn Cys Val Arg Cys Xaa 275
(2) INFORMATION FOR SEQ ID NO: 448: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 199 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 448:
Met Ser Phe He Phe Asp Trp He Tyr Ser Gly Phe Ser Ser Val Leu 1 5 10 15
Gin Phe Leu Gly Leu Tyr Lys Lys Thr Gly Lys Leu Val Phe Leu Gly 20 25 30
Leu Asp Asn Ala Gly Lys Thr Thr Leu Leu His Met Leu Lys Asp Asp 35 40 45
Arg Leu Gly Gin His Val Pro Thr Leu His Pro Thr Ser Glu Glu Leu 50 55 60
Thr He Ala Gly Met Thr Phe Thr Thr Phe Asp Leu Gly Gly His Val 65 70 75 80
Gin Ala Arg Arg Val Trp Lys Asn Tyr Leu Pro Ala He Asn Gly He 85 90 95
Val Phe Leu Val Asp Cys Ala Asp His Glu Arg Leu Leu Glu Ser Lys 100 105 110
Glu Glu Leu Asp Ser Leu Met Thr Asp Glu Thr He Ala Asn Val Pro 115 120 125 He Leu He Leu Gly Asn Lys He Asp Arg Pro Glu Ala He Ser Glu 130 135 140
Glu Arg Leu Arg Glu Met Phe Gly Leu Tyr Gly Gin Thr Thr Gly Lys 145 150 155 160
Gly Ser He Ser Leu Lys Glu Leu Asn Ala Arg Pro Leu Glu Val Phe 165 170 175
Met Cys Ser Val Leu Lys Arg Gin Gly Tyr Gly Glu Gly Phe Arg Trp 180 185 190
Met Ala Gin Tyr He Asp Xaa 195
(2) INFORMATION FOR SEQ ID NO: 449:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 258 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 449: Met Thr Leu Ser Arg Phe Ala Tyr Asn Gly Lys Arg Cys Pro Ser Ser 1 5 10 15
Tyr Asn He Leu Asp Asn Ser Lys He He Ser Glu Glu Cys Arg Lys 20 25 30
Glu Leu Thr Ala Leu Leu His His Tyr Tyr Pro He Glu He Asp Pro 35 40 45
His Arg Thr Val Lys Glu Lys Leu Pro His Met Val Glu Trp Trp Thr 50 55 60
Lys Ala His Asn Leu Leu Cys Gin Gin Lys He Gin Lys Phe Gin He 65 70 75 80 Ala Gin Val Val Arg Glu Ser Asn Ala Met Leu Arg Glu Gly Tyr Lys
85 90 95
Thr Phe Phe Asn Thr Leu Tyr His Asn Asn He Pro Leu Phe He Phe 100 105 110
Ser Ala Gly He Gly Asp He Leu Glu Glu He He Arg Gin Met Lys 115 120 125
Val Phe His Pro Asn He His He Val Ser Asn Tyr Met Asp Phe Asn 130 135 140
Glu Asp Gly Phe Leu Gin Gly Phe Lys Gly Gin Leu He His Thr Tyr 145 150 155 160 Asn Lys Asn Ser Ser Val Cys Glu Asn Xaa Gly Tyr Phe Gin Gin Leu
165 170 175
Glu Gly Lys Thr Asn Val He Leu Leu Gly Asp Ser He Gly Asp Leu 180 185 190
Thr Met Ala Asp Gly Val Pro Gly Val Gin Asn He Leu Lys He Gly 195 200 205
Phe Leu Asn Asp Lys Val Glu Glu Arg Arg Xaa Arg Tyr Met Asp Ser 210 215 220
Tyr Asp He Val Leu Glu Lys Asp Glu Thr Leu Asp Val Val Asn Gly 225 230 235 240 Leu Leu Gin His He Leu Cys Gin Gly Val Gin Leu Glu Met Gin Gly
245 250 255
Pro Xaa
(2) INFORMATION FOR SEQ ID NO: 450: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 87 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 450:
Met Ser His Val Leu Leu Cys Pro Ser Leu Ser Cys Ser Asn Leu Leu 1 5 10 15
Pro Pro Ser His Ser Leu Gly Thr Met Gly Ser Leu Ser Pro His Leu 20 25 30
Cys Gly His Thr Met Cys Pro Val Asn Pro Glu Leu Pro Leu Ser Ser 35 40 45 Arg Leu Thr Thr Asp Gin Pro Gin Pro Asp Ala Cys Ser Pro Thr Leu 50 55 60
Leu Thr Leu Pro Leu Pro Ser Ser Phe Leu Pro His Ser Lys Pro Thr 65 70 75 80
Phe Xaa His Pro Cys Ser Pro 85
(2) INFORMATION FOR SEQ ID NO: 451:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 315 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 451:
Met Phe Ser He Asn Pro Leu Glu Asn Leu Lys Val Tyr He Ser Ser 1 5 10 15
Arg Pro Pro Leu Val Val Phe Met He Ser Val Xaa Pro Met Ala He 20 25 30 Ala Phe Leu Thr Leu Gly Tyr Phe Phe Lys He Lys Glu He Lys Ser 35 40 45
Pro Glu Met Ala Glu Asp Trp Asn Thr Phe Leu Leu Arg Phe Asn Asp 50 55 60
Leu Asp Leu Cys Val Ser Glu Asn Glu Thr Leu Lys His Leu Thr Asn 65 70 75 80
Asp Thr Thr Thr Pro Glu Ser Thr Met Thr Ser Gly Gin Ala Arg Ala 85 90 95
Ser Thr Gin Ser Pro Gin Ala Leu Glu Asp Ser Gly Pro Val Asn He 100 105 110 Ser Val Ser He Thr Leu Thr Leu Asp Pro Leu Lys Pro Phe Gly Gly 115 120 125
Tyr Ser Arg Asn Val Thr His Leu Tyr Ser Thr He Leu Gly His Gin 130 135 140
He Gly Leu Ser Gly Arg Glu Ala His Glu Glu He Asn He Thr Phe 145 150 155 160
Thr Leu Pro Thr Ala Trp Ser Ser Asp Asp Cys Ala Leu His Gly His 165 170 175
Cys Glu Gin Val Val Phe Thr Ala Cys Met Thr Leu Thr Ala Ser Pro 180 185 190 Gly Val Phe Pro Val Thr Val Gin Pro Pro His Cys Val Pro Asp Thr 195 200 205
Tyr Ser Asn Ala Thr Leu Trp Tyr Lys He Phe Thr Thr Ala Arg Asp 210 215 220
Ala Asn Thr Lys Tyr Ala Gin Asp Tyr Asn Pro Phe Trp Cys Tyr Lys 225 230 235 240
Gly Ala He Gly Lys Val Tyr His Ala Leu Asn Pro Lys Leu Thr Val 245 250 255
He Val Pro Asp Asp Asp Arg Ser Leu He Asn Leu His Leu Met His 260 265 270
Thr Ser Tyr Phe Leu Phe Val Met Val He Thr Met Phe Cys Tyr Ala 275 280 285
Val He Lys Gly Arg Pro Ser Lys Leu Arg Gin Ser Asn Pro Glu Phe 290 295 300
Cys Pro Glu Lys Val Ala Leu Ala Glu Ala Xaa 305 310 315
(2) INFORMATION FOR SEQ ID NO: 452:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 52 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 452:
Met Pro Gly Leu Ser Leu Ala Leu Leu Pro Phe Gly Pro Gly Cys Thr 1 5 10 15
Glu Ala Leu His Ala Gly Cys Phe Pro Ala Phe Ala Ser Ala Thr Arg 20 25 30 Val Asn Gly Glu Ala Ala Leu Ser Pro Gly Leu Cys Asp Pro He Ser 35 40 45
Val Pro Tyr Val 50
(2) INFORMATION FOR SEQ ID NO: 453: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 383 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 453:
Met Ala Val Gly Gin He Met Thr Phe Gly Ser Pro Val He Gly Cys 1 5 10 15
Gly Phe He Ser Gly Trp Asn Leu Val Ser Met Cys Val Glu Tyr Val 20 25 30
Leu Leu Trp Lys Val Tyr Gin Lys Thr Pro Ala Leu Ala Val Lys Ala 35 40 45 Gly Leu Lys Glu Glu Glu Thr Glu Leu Lys Gin Leu Asn Leu His Lys 50 55 60
Asp Thr Glu Pro Lys Pro Leu Glu Gly Thr His Leu Met Gly Val Lys 65 70 75 80
Asp Ser Asn He His Glu Leu Glu His Glu Gin Glu Pro Thr Cys Ala 85 90 95
Ser Gin Met Ala Glu Pro Phe Arg Thr Phe Arg Asp Gly Trp Val Ser 100 105 110
Tyr Tyr Asn Gin Pro Val Phe Leu Ala Gly Met Gly Leu Ala Phe Leu 115 120 125 Tyr Met Thr Val Leu Gly Phe Asp Cys He Thr Thr Gly Tyr Ala Tyr 130 135 140
Thr Gin Gly Leu Ser Gly Phe His Pro Gin Tyr Phe Asp Gly Ser He 145 150 155 160
Ser Tyr Asn Trp Asn Asn Gly Asn Cys Ser Phe Tyr Leu Ala Thr Ser 165 170 175
Lys Met Trp Phe Gly Ser Ala Gly Leu He Ser Gly Leu Ala Gin Leu 180 185 190
Ser Cys Leu He Leu Cys Val He Ser Val Phe Met Pro Gly Ser Pro 195 200 205 Leu Asp Leu Ser Val Ser Pro Phe Glu Asp He Arg Ser Arg Phe He 210 215 220
Gin Gly Glu Ser He Thr Pro Thr Lys He Pro Glu He Thr Thr Glu 225 230 235 240
He Tyr Met Ser Asn Gly Ser Asn Ser Ala Asn He Val Pro Glu Thr 245 250 255
Ser Pro Glu Ser Val Pro He He Ser Val Ser Leu Leu Phe Ala Gly 260 265 270
Val He Ala Ala Arg He Gly Leu Trp Ser Phe Asp Leu Thr Val Thr 275 280 285 Gin Leu Leu Gin Glu Asn Val He Glu Ser Glu Arg Gly He He Asn 290 295 300
Gly Val Gin Asn Ser Met Asn Tyr Leu Leu Asp Leu Leu His Phe He 305 310 315 320
Met Val He Leu Ala Pro Asn Pro Glu Ala Phe Gly Leu Leu Val Leu 325 330 335
He Ser Val Ser Phe Val Ala Met Gly His He Met Tyr Phe Arg Phe 340 345 350
Ala Gin Asn Thr Leu Gly Asn Lys Leu Phe Ala Cys Gly Pro Asp Ala 355 360 365 Lys Glu Val Arg Lys Glu Asn Gin Ala Asn Thr Ser Val Val Xaa 370 375 380
(2) INFORMATION FOR SEQ ID NO: 454:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 186 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 454:
Met Arg Ser He Gly Asn Lys Asn Thr He Leu Leu Gly Leu Gly Phe 1 5 10 15
Gin He Leu Gin Leu Ala Trp Tyr Gly Phe Gly Ser Glu Pro Trp Met 20 25 30
Met Trp Ala Ala Gly Ala Val Ala Ala Met Ser Ser He Thr Phe Pro 35 40 45
Ala Val Ser Ala Leu Val Ser Arg Thr Ala Asp Ala Asp Gin Gin Gly 50 55 60 Val Val Gin Gly Met He Thr Gly He Arg Gly Leu Cys Asn Gly Leu 65 70 75 80
Gly Pro Ala Leu Tyr Gly Phe He Phe Tyr He Phe His Val Glu Leu 85 90 95
Lys Glu Leu Pro He Thr Gly Thr Asp Leu Gly Thr Asn Thr Ser Pro 100 105 110
Gin His His Phe Glu Gin Asn Ser He He Pro Gly Pro Pro Phe Leu 115 120 125
Phe Gly Ala Cys Ser Val Leu Leu Ala Leu Leu Val Ala Leu Phe He 130 135 140 Pro Glu His Thr Asn Leu Ser Leu Arg Ser Ser Ser Trp Arg Lys His 145 150 155 160
Cys Gly Ser His Ser His Pro His Asn Thr Gin Ala Pro Gly Glu Ala 165 170 175
Lys Glu Pro Leu Leu Gin Asp Thr Asn Val 180 185
(2) INFORMATION FOR SEQ ID NO: 455:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 163 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 455:
Met Leu Gin Thr Ser Asn Tyr Ser Leu Val Leu Ser Leu Gin Phe Leu 1 5 10 15
Leu Leu Ser Tyr Asp Leu Phe Val Asn Ser Phe Ser Glu Leu Leu Gin 20 25 30 Lys Thr Pro Val He Gin Leu Val Leu Phe He He Gin Asp He Ala
35 40 45
Val Leu Phe Asn He He He He Phe Leu Met Phe Phe Asn Thr Phe 50 55 60
Val Phe Gin Ala Gly Leu Val Asn Leu Leu Phe His Lys Phe Lys Gly 65 70 75 80
Thr He He Leu Thr Ala Val Tyr Phe Ala Leu Ser He Ser Leu His 85 90 95
Val Trp Val Met Asn Leu Arg Trp Lys Asn Ser Asn Ser Phe He Trp 100 105 110 Thr Asp Gly Leu Gin Met Leu Phe Val Phe Gin Arg Leu Ala Ala Val 115 120 125
Leu Tyr Cys Tyr Phe Tyr Lys Arg Thr Ala Val Arg Leu Gly Asp Pro 130 135 140
His Phe Tyr Gin Asp Ser Leu Trp Leu Arg Lys Glu Phe Met Gin Val 145 150 155 160
Arg Arg Xaa
(2) INFORMATION FOR SEQ ID NO: 456:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 46 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 456:
Met Arg He Gin Val Phe He Leu Leu Leu Gly Ala Gly Gly Thr Ser 1 5 10 15 Gin Phe Thr Lys Pro Pro Ser Leu Pro Leu Glu Pro Glu Pro Ala Val 20 25 30
Glu Ser Ser Pro Thr Glu Thr Ser Glu Gin He Arg Glu Lys 35 40 45
(2) INFORMATION FOR SEQ ID NO: 457: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 105 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 457:
Met Ser Tyr Leu Ala Phe Leu Tyr Met Thr Phe Asp Phe Cys Cys Leu 1 5 10 15
Tyr Phe Ser Thr Val Tyr Ala Pro Ser Phe Lys Tyr He Cys Val His 20 25 30
Thr Asp Thr His He Cys Val Cys Val Cys He Tyr Leu Ser Ser Val 35 40 45 Val Ser Lys Ser Ser Ala Glu Ala Asp Gly Val Leu Gin Pro Arg Arg 50 55 60
His Pro Ala Ser Leu Leu He Val Phe Ala Thr Ser He Ser Glu Ser 65 70 75 80
Ser Leu Leu He Phe Ser Phe Gin Lys Thr Glu Ala Lys Leu He Val 85 90 95
Phe Ala Val Ser Leu Ala Ala Lys Xaa 100 105
(2) INFORMATION FOR SEQ ID NO: 458:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 70 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 458:
Met Leu Pro Pro Phe Ser Leu Val Tyr Thr His Phe Leu Val Ala Ser 1 5 10 15 Leu Leu Pro Val He Leu Ala Val Phe Pro Asp Ser Ala Gin He Val 20 25 30
Pro Leu Leu Lys Pro He Pro Arg Pro Gin Pro Glu Val He Phe Pro 35 40 45
Ser Ser Glu Leu Leu Glu Gin Leu Leu Ser Val Gin Phe Val Trp Gin 50 55 60
Ala His Thr Val Ala Xaa 65 70
(2) INFORMATION FOR SEQ ID NO: 459:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 155 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 459:
Met Ala Leu Leu Leu Ser Val Leu Arg Val Leu Leu Gly Gly Phe Phe 1 5 10 15 Ala Leu Val Gly Leu Ala Lys Leu Ser Glu Glu He Ser Ala Pro Val 20 25 30
Ser Glu Arg Met Asn Ala Leu Phe Val Gin Phe Ala Glu Val Phe Pro 35 40 45
Leu Lys Val Phe Gly Tyr Gin Pro Asp Pro Leu Asn Tyr Gin He Ala 50 55 60
Val Gly Phe Leu Glu Leu Leu Ala Gly Leu Leu Leu Val Met Gly Pro 65 70 75 80
Pro Met Leu Gin Glu He Ser Asn Leu Phe Leu He Leu Leu Met Met 85 90 95 Gly Ala He Phe Thr Leu Ala Ala Leu Lys Glu Ser Leu Ser Thr Cys 100 105 110
He Pro Ala He Val Cys Leu Gly Phe Leu Leu Leu Leu Asn Val Gly 115 120 125
Gin Leu Leu Ala Gin Thr Lys Lys Val Val Arg Pro Thr Arg Lys Lys 130 135 140
Thr Leu Ser Thr Phe Lys Glu Ser Trp Lys Xaa 145 150 155
(2) INFORMATION FOR SEQ ID NO: 460:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 332 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 460:
Met Lys Leu Gly Arg Ala Val Leu Gly Leu Leu Leu Leu Ala Pro Ser 1 5 10 15 Val Val Gin Ala Val Glu Pro He Ser Leu Gly Leu Ala Leu Ala Gly 20 25 30
Val Leu Thr Gly Tyr He Tyr Pro Arg Leu Tyr Cys Leu Phe Ala Glu 35 40 45
Cys Cys Gly Gin Lys Arg Ser Leu Ser Arg Glu Ala Leu Gin Lys Asp 50 55 60
Leu Asp Asp Asn Leu Phe Gly Gin His Leu Ala Lys Lys He He Leu 65 70 75 80
Asn Ala Val Phe Gly Phe He Asn Asn Pro Lys Pro Lys Lys Pro Leu 85 90 95 Thr Leu Ser Leu His Gly Trp Thr Gly Thr Gly Lys Asn Phe Val Ser 100 105 110
Lys He He Ala Glu Asn He Tyr Glu Gly Gly Leu Asn Ser Asp Tyr 115 120 125
Val His Leu Phe Val Ala Thr Leu His Phe Pro His Ala Ser Asn He 130 135 140
Thr Leu Tyr Lys Asp Gin Leu Gin Leu Trp He Arg Gly Asn Val Ser 145 150 155 160
Ala Cys Ala Arg Ser He Phe He Phe Asp Glu Met Asp Lys Met His 165 170 175 Ala Gly Leu He Asp Ala He Lys Pro Phe Leu Asp Tyr Tyr Asp Leu 180 185 190
Val Asp Gly Val Ser Tyr Gin Lys Ala Met Phe He Phe Leu Ser Asn 195 200 205
Ala Gly Ala Glu Arg He Thr Asp Val Ala Leu Asp Phe Trp Arg Ser 210 215 220
Gly Lys Gin Arg Glu Asp He Lys Leu Lys Asp He Glu His Ala Leu 225 230 235 240
Ser Val Ser Val Phe Asn Asn Lys Asn Ser Gly Phe Trp His Ser Ser 245 250 255 Leu He Asp Arg Asn Leu He Asp Tyr Phe Val Pro Phe Leu Pro Leu 260 265 270
Glu Tyr Lys His Leu Lys Met Cys He Arg Val Glu Met Gin Ser Arg 275 280 285
Gly Tyr Glu He Asp Glu Asp He Val Ser Arg Val Ala Glu Glu Met 290 295 300
Thr Phe Phe Pro Lys Glu Glu Arg Val Phe Ser Asp Lys Gly Cys Lys 305 310 315 320
Thr Val Phe Thr Lys Leu Asp Tyr Tyr Tyr Asp Asp 325 330
(2) INFORMATION FOR SEQ ID NO: 461:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 5 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 461: Met Leu Lys Cys He 1 5
(2) INFORMATION FOR SEQ ID NO: 462:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 14 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 462:
Met He Leu Thr Leu Leu Ser Val Val Ser Thr Met Ala Ser 1 5 10
(2) INFORMATION FOR SEQ ID NO: 463:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 285 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 463:
Met Lys Leu His Pro Pro Pro Pro Ser Pro Val Thr Gin Asp His Arg 1 5 10 15
Ser Lys Ser Ser His Ser Asn Trp Met Pro Arg Met Gly Ala Cys Ser 20 25 30
Met Ser Arg Thr Ser Ser Ser Gly Pro Pro Ser Leu Cys Lys Ser Thr 35 40 45 Ser Gly Arg Ser Cys Thr Arg Pro His Cys Trp Pro Ser Leu Pro Ala 50 55 60
Trp Val Ser Val Phe Thr Arg Thr Asn Thr Gly Ser Trp Cys Tyr Pro 65 70 75 80
Ala Trp Gly Gly Ala Phe Ser Arg Pro Trp Met Ser Ala Gin Ser Met 85 90 95
Cys Cys Ala Glu Arg Ser Val Leu Gin Val Ala Cys Arg Leu Leu Asp 100 105 110
Ala Leu Glu Phe Leu His Glu Asn Glu Tyr Val His Gly Asn Val Thr 115 120 125 Ala Glu Asn He Phe Val Asp Pro Glu Asp Gin Ser Gin Val Thr Leu 130 135 140
Ala Gly Tyr Gly Phe Ala Phe Arg Tyr Cys Pro Ser Gly Lys His Val 145 150 155 160
Ala Tyr Val Glu Gly Ser Arg Ser Pro His Glu Gly Asp Leu Glu Phe 165 170 175
He Ser Met Asp Leu His Lys Gly Cys Gly Pro Ser Arg Arg Xaa Asp 180 185 190
Leu Gin Ser Leu Gly Tyr Cys Met Leu Lys Trp Leu Tyr Gly Phe Leu 195 200 205 Pro Trp Thr Asn Cys Leu Pro Xaa Xaa Glu Asp He Met Lys Gin Lys 210 215 220
Gin Lys Phe Val Asp Lys Pro Gly Pro Phe Val Gly Pro Cys Gly His 225 230 235 240
Trp He Arg Pro Ser Glu Thr Leu Gin Lys Tyr Leu Lys Val Val Met 245 250 255
Ala Leu Thr Tyr Glu Glu Lys Pro Pro Tyr Ala Met Leu Arg Asn Asn 260 265 270
Leu Glu Ala Leu Leu Gin Asp Leu Arg Val Ser Pro Tyr 275 280 285
(2) INFORMATION FOR SEQ ID NO: 464:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 80 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 464: Met Thr Ser Pro Pro Pro His Gin Gly Trp Glu Gin Arg Gly Cys Gly 1 5 10 15
Glu Ser Gin Val Pro Leu Ala Leu Ser Arg Val Phe Ser Thr Ser His 20 25 30
Tyr Cys Leu Leu Leu Val Ala Asn Gin Ser He Phe Phe Pro Cys Leu 35 40 45
Trp Ala Val Glu Arg Leu Leu Gly Val Arg Cys Thr Cys Pro Leu Ser 50 55 60
Trp Gly Lys Arg He He Ser Glu His Cys Ser Ala Gin Ser Ser Xaa 65 70 75 80
(2) INFORMATION FOR SEQ ID NO: 465:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 47 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 465:
Met His Thr Trp Tyr Asn Asp Arg Arg Gin Asn Cys His Cys Leu Leu 1 5 10 15
Phe Phe Leu He Tyr Leu Arg Lys He Tyr Gin Val Val Pro His Val 20 25 30
Pro Leu Leu Val Lys Cys Arg Gly Arg Leu Lys Gly Val Asn He 35 40 45
(2) INFORMATION FOR SEQ ID NO: 466:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 96 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 466:
Met Glu Leu Val Leu Val Phe Leu Cys Ser Leu Leu Ala Pro Met Val 1 5 10 15
Leu Ala Ser Ala Ala Glu Lys Glu Lys Glu Met Asp Pro Phe His Tyr 20 25 30
Asp Tyr Gin Thr Leu Arg He Gly Gly Leu Val Phe Ala Val Val Leu 35 40 45
Phe Ser Val Gly He Leu Leu He Leu Ser Arg Arg Cys Lys Cys Ser 50 55 60
Phe Asn Gin Lys Pro Arg Ala Pro Gly Asp Glu Glu Ala Gin Val Glu 65 70 75 80
Asn Leu He Thr Ala Asn Ala Thr Glu Pro Gin Lys Ala Glu Asn Xaa 85 90 95
(2) INFORMATION FOR SEQ ID NO: 467:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 399 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 467:
Met Ala Ser Gly Ala Asp Ser Lys Gly Asp Asp Leu Ser Thr Ala He 1 5 10 15
Leu Lys Gin Lys Asn Arg Pro Asn Arg Leu He Val Asp Glu Ala He 20 25 30
Asn Glu Asp Asn Ser Val Val Ser Leu Ser Gin Pro Lys Met Asp Glu 35 40 45
Leu Gin Leu Phe Arg Gly Asp Thr Val Leu Leu Lys Gly Lys Lys Arg
50 55 60 Arg Glu Ala Val Cys He Val Leu Ser Asp Asp Thr Cys Ser Asp Glu
65 70 75 80
Lys He Arg Met Asn Arg Val Val Arg Asn Asn Leu Arg Val Arg Leu 85 90 95
Gly Asp Val He Ser He Gin Pro Cys Pro Asp Val Lys Tyr Gly Lys 100 105 110
Arg He His Val Leu Pro He Asp Asp Thr Val Glu Gly He Thr Gly 115 120 125
Asn Leu Phe Glu Val Tyr Leu Lys Pro Tyr Phe Leu Glu Ala Tyr Arg 130 135 140 Pro He Arg Lys Gly Asp He Phe Leu Val Arg Gly Gly Met Arg Ala
145 150 155 160
Val Glu Phe Lys Val Val Glu Thr Asp Pro Ser Pro Tyr Cys He Val 165 170 175
Ala Pro Asp Thr Val He His Cys Glu Gly Glu Pro He Lys Arg Glu 180 185 190
Asp Glu Glu Glu Ser Leu Asn Glu Val Gly Tyr Asp Asp He Gly Gly 195 200 205
Cys Arg Lys Gin Leu Ala Gin He Lys Glu Met Val Glu Leu Pro Leu 210 215 220 Arg His Pro Ala Leu Phe Lys Ala He Gly Val Lys Pro Pro Arg Gly 225 230 235 240
He Leu Leu Tyr Gly Pro Pro Gly Thr Gly Lys Thr Leu He Ala Arg 245 250 255
Ala Val Ala Asn Glu Thr Gly Ala Phe Phe Phe Leu He Asn Gly Pro 260 265 270
Glu He Met Ser Lys Leu Ala Gly Glu Ser Glu Ser Asn Leu Arg Lys 275 280 285
Ala Phe Glu Glu Ala Glu Lys Asn Ala Pro Ala He He Phe He Asp 290 295 300 Glu Leu Asp Ala He Ala Pro Lys Arg Glu Lys Thr His Gly Glu Val 305 310 315 320
Glu Arg Arg He Val Ser Gin Leu Leu Thr Leu Met Asp Gly Leu Lys 325 330 335
Gin Arg Ala His Val He Val Met Ala Ala Thr Asn Arg Pro Asn Ser 340 345 350
He Asp Pro Ala Leu Arg Arg Phe Gly Arg Phe Asp Arg Glu Val Asp 355 360 365
He Gly He Pro Asp Ala Thr Gly Arg Leu Glu He Leu Gin He His 370 375 380 Thr Lys Asn Met Lys Leu Ala Asp Asp Val Asp Leu Glu Gin Xaa 385 390 395
(2) INFORMATION FOR SEQ ID NO: 468:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 468:
Leu 1
(2) INFORMATION FOR SEQ ID NO: 469:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 273 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 469:
Met Ala Ala Pro Lys Gly Ser Leu Trp Val Arg Thr Gin Leu Gly Leu 1 5 10 15
Pro Pro Leu Leu Leu Leu Thr Met Ala Leu Ala Gly Gly Ser Gly Thr 20 25 30
Ala Ser Ala Glu Ala Phe Asp Ser Val Leu Gly Asp Thr Ala Ser Cys 35 40 45 His Arg Ala Cys Gin Leu Thr Tyr Pro Leu His Thr Tyr Pro Lys Glu 50 55 60
Glu Glu Leu Tyr Ala Cys Gin Arg Gly Cys Arg Leu Phe Ser He Cys 65 70 75 80
Gin Phe Val Asp Asp Gly He Asp Leu Asn Arg Thr Lys Leu Glu Cys 85 90 95
Glu Ser Ala Cys Thr Glu Ala Tyr Ser Gin Ser Asp Glu Gin Tyr Ala 100 105 110
Cys His Leu Gly Cys Gin Asn Gin Leu Pro Phe Ala Glu Leu Arg Gin 115 120 125 Glu Gin Leu Met Ser Leu Met Pro Lys Met His Leu Leu Phe Pro Leu 130 135 140
Thr Leu Val Arg Ser Phe Trp Ser Asp Met Met Asp Ser Ala Gin Ser 145 150 155 160
Phe He Thr Ser Ser Trp Thr Phe Tyr Leu Gin Ala Asp Asp Gly Lys 165 170 175
He Val He Phe Xaa Ser Lys Pro Arg Asn Pro Arg Tyr Ala Pro His 180 185 190
Leu Glu Pro Gly Ala Leu Pro Asn Leu Xaa Xaa Xaa Ser Leu Ser Lys 195 200 205 Met Ser Xaa Xaa Ser Xaa Met Arg Asn Ser Gin Ala His Arg Asn Phe 210 215 220
Leu Glu Asp Gly Glu Ser Asp Gly Phe Leu Arg Cys Leu Ser Leu Asn 225 230 235 240
Ser Gly Trp He Leu Thr Thr Thr Leu Val Leu Ser Val Met Val Leu 245 250 255
Leu Trp He Cys Cys Ala Thr Cys Cys Tyr Thr Leu Leu Asp Ala Val 260 265 270
Xaa
(2) INFORMATION FOR SEQ ID NO: 470:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 192 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 470: Met Met Val Leu Ser Leu Gly He He Leu Ala Ser Ala Ser Phe Ser
1 5 10 15
Pro Asn Phe Thr Gin Val Thr Ser Thr Leu Leu Asn Ser Ala Tyr Pro 20 25 30
Phe He Gly Pro Phe Phe Phe He He Ser Gly Ser Leu Ser He Ala 35 40 45
Thr Glu Lys Arg Leu Thr Lys Leu Leu Val His Ser Ser Leu Val Gly 50 55 60
Ser He Leu Ser Ala Leu Ser Ala Leu Val Gly Phe He He Leu Ser 65 70 75 80 Val Lys Gin Ala Thr Leu Asn Pro Ala Ser Leu Gin Cys Glu Leu Asp
85 90 95
Lys Asn Asn He Pro Thr Arg Ser Tyr Val Ser Tyr Phe Tyr His Asp 100 105 110
Ser Leu Tyr Thr Thr Asp Cys Tyr Thr Ala Lys Ala Ser Leu Ala Gly 115 120 125
Xaa Leu Ser Leu Met Leu He Cys Thr Leu Leu Glu Phe Cys Leu Ala 130 135 140
Val Leu Thr Ala Val Leu Arg Trp Lys Gin Ala Tyr Ser Asp Phe Pro 145 150 155 160 Gly Ser Val Leu Phe Leu Pro His Ser Tyr He Gly Asn Ser Gly Met
165 170 175
Ser Ser Lys Met Thr His Asp Cys Gly Tyr Glu Glu Leu Leu Thr Ser 180 185 190
(2) INFORMATION FOR SEQ ID NO: 471:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 234 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 471:
Met Arg Lys Thr Arg Leu Trp Gly Leu Leu Trp Met Leu Phe Val Ser 1 5 10 15
Glu Leu Arg Ala Ala Thr Lys Leu Thr Glu Glu Lys Tyr Glu Leu Lys 20 25 30 Glu Gly Gin Thr Leu Asp Val Lys Cys Asp Tyr Thr Leu Glu Lys Phe 35 40 45
Ala Ser Ser Gin Lys Ala Trp Gin He He Arg Asp Gly Glu Met Pro 50 55 60
Lys Thr Leu Ala Cys Thr Glu Arg Pro Ser Lys Asn Ser His Pro Val 65 70 75 80
Gin Val Gly Arg He He Leu Glu Asp Tyr His Asp His Gly Leu Leu 85 90 95
Arg Val Arg Met Val Asn Leu Gin Val Glu Asp Ser Gly Leu Tyr Gin 100 105 110 Cys Val He Tyr Gin Pro Pro Lys Glu Pro His Met Leu Phe Asp Arg 115 120 125
He Arg Leu Val Val Thr Lys Gly Phe Ser Gly Thr Pro Gly Ser Asn 130 135 140
Glu Asn Ser Thr Gin Asn Val Tyr Lys He Pro Pro Thr Thr Thr Lys 145 150 155 160
Ala Leu Cys Pro Leu Tyr Thr Ser Pro Arg Thr Val Thr Gin Ala Pro 165 170 175
Pro Lys Ser Thr Ala Asp Val Ser Thr Pro Asp Ser Glu He Asn Leu 180 185 190 Thr Asn Val Thr Asp He He Arg Val Pro Val Phe Asn He Val He 195 200 205
Leu Leu Ala Gly Gly Phe Leu Ser Lys Ser Leu Val Phe Ser Val Leu 210 215 220
Phe Ala Val Thr Leu Arg Ser Phe Val Pro 225 230
(2) INFORMATION FOR SEQ ID NO: 472:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 105 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 472:
Met Leu His He Leu Pro Leu Lys Ser Tyr Asp Phe Pro His Phe Ser 1 5 10 15
Leu Met Gly Arg Tyr Arg Cys Ala Ser Leu Leu Phe Cys Phe Leu Leu 20 25 30
Leu Phe Phe Phe Phe Cys Ser Val Leu Trp Thr Phe Ser Asp Met His 35 40 45
Arg Ser Gly Glu Asp Gly Pro Trp Thr Pro Cys Val His His Leu Ala 50 55 60
Ala Ser Leu He Ser Tyr Gly Gin Pro Gly Phe He Cys He Ser Leu 65 70 75 80
Phe Ser Pro Val Leu Phe He Glu Asn Pro Arg His Tyr Ala Asn Ala 85 90 95
Thr Val Thr Thr Leu Gly Asp Trp Xaa 100 105
(2) INFORMATION FOR SEQ ID NO: 473:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 32 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 473: Met Val Phe Leu Lys Tyr Arg Phe Leu Phe Phe Leu Val Phe Leu Ala 1 5 10 15
Asn Cys He Tyr Ser Leu His Tyr Lys Pro Ser Leu Met Tyr Pro Lys 20 25 30
(2) INFORMATION FOR SEQ ID NO: 474:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 571 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 474:
Met Ala Leu Ser Arg Gly Leu Pro Arg Glu Leu Ala Glu Ala Val Ala 1 5 10 15
Gly Gly Arg Val Leu Val Val Gly Ala Gly Gly He Gly Cys Glu Leu 20 25 30 Leu Lys Asn Leu Val Leu Thr Gly Phe Ser His He Asp Leu He Asp 35 40 45
Leu Asp Thr He Asp Val Ser Asn Leu Asn Arg Gin Phe Leu Phe Gin 50 55 60
Lys Lys His Val Gly Arg Ser Lys Ala Gin Val Ala Lys Glu Ser Val 65 70 75 80
Leu Gin Phe Tyr Pro Lys Ala Asn He Val Ala Tyr His Asp Ser He 85 90 95
Met Asn Pro Asp Tyr Asn Val Glu Phe Phe Arg Gin Phe He Leu Val 100 105 110 Met Asn Ala Leu Asp Asn Arg Ala Ala Arg Asn His Val Asn Arg Met 115 120 125
Cys Leu Ala Ala Asp Val Pro Leu He Glu Ser Gly Thr Ala Gly Tyr 130 135 140
Leu Gly Gin Val Thr Thr He Lys Lys Gly Val Thr Glu Cys Tyr Glu 145 150 155 160
Cys His Pro Lys Pro Thr Gin Arg Thr Phe Pro Gly Cys Thr He Arg 165 170 175
Asn Thr Pro Ser Glu Pro He His Cys He Val Trp Ala Lys Tyr Leu 180 185 190 Phe Asn Gin Leu Phe Gly Glu Glu Asp Ala Asp Gin Glu Val Ser Pro 195 200 205
Asp Arg Ala Asp Pro Glu Ala Ala Trp Glu Pro Thr Glu Ala Glu Ala 210 215 220
Arg Ala Arg Ala Ser Asn Glu Asp Gly Asp He Lys Arg He Ser Thr 225 230 235 240
Lys Glu Trp Ala Lys Ser Thr Gly Tyr Asp Pro Val Lys Leu Phe Thr 245 250 255
Lys Leu Phe Lys Asp Asp He Arg Tyr Leu Leu Thr Met Asp Lys Leu 260 265 270 Trp Arg Lys Arg Lys Pro Pro Val Pro Leu Asp Trp Ala Glu Val Gin 275 280 285
Ser Gin Gly Glu Glu Thr Asn Ala Ser Asp Gin Gin Asn Glu Pro Gin 290 295 300
Leu Gly Leu Lys Asp Gin Gin Val Leu Asp Val Lys Ser Tyr Ala Arg 305 310 315 320
Leu Phe Ser Lys Ser He Glu Thr Leu Arg Val His Leu Ala Glu Lys 325 330 335
Gly Asp Gly Ala Glu Leu He Trp Asp Lys Asp Asp Pro Ser Ala Met 340 345 350 Asp Phe Val Thr Ser Ala Ala Asn Leu Arg Met His He Phe Ser Met 355 360 365
Asn Met Lys Ser Arg Phe Asp He Lys Ser Met Ala Gly Asn He He 370 375 380
Pro Ala He Ala Thr Thr Asn Ala Val He Ala Gly Leu He Val Leu 385 390 395 400
Glu Gly Leu Lys He Leu Ser Gly Lys He Asp Gin Cys Arg Thr He 405 410 415
Phe Leu Asn Lys Gin Pro Asn Pro Arg Lys Lys Leu Leu Val Pro Cys 420 425 430 Ala Leu Asp Pro Pro Asn Pro Asn Cys Tyr Val Cys Ala Ser Lys Pro 435 440 445
Glu Val Thr Val Arg Leu Asn Val His Lys Val Thr Val Leu Thr Leu 450 455 460
Gin Asp Lys He Val Lys Glu Lys Phe Ala Met Val Ala Pro Asp Val 465 470 475 480
Gin He Glu Asp Gly Lys Gly Thr He Leu He Ser Ser Glu Glu Gly 485 490 495
Glu Thr Glu Ala Asn Asn His Lys Lys Leu Ser Glu Phe Gly He Arg 500 505 510 Asn Gly Ser Arg Leu Gin Ala Asp Asp Phe Leu Gin Asp Tyr Thr Leu 515 520 525
Leu He Asn He Leu His Ser Glu Asp Leu Gly Lys Asp Val Glu Phe 530 535 540
Glu Val Val Gly Asp Ala Pro Glu Lys Val Gly Xaa Lys Gin Ala Glu 545 550 555 560
Asp Ala Ala Lys Ser He Thr Asn Gly Gin Xaa 565 570
(2) INFORMATION FOR SEQ ID NO: 475:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 312 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 475:
Met Gin Val Val Thr Cys Leu Thr Arg Asp Ser Tyr Leu Thr His Cys 1 5 10 15 Phe Leu Gin His Leu Met Val Val Leu Ser Ser Leu Glu Arg Thr Pro 20 25 30
Ser Pro Glu Pro Val Asp Lys Asp Phe Tyr Ser Glu Phe Gly Asn Lys 35 40 45
Thr Thr Gly Lys Met Glu Asn Tyr Glu Leu He His Ser Ser Arg Val 50 55 60
Lys Phe Thr Tyr Pro Ser Glu Glu Glu He Gly Asp Leu Thr Phe Thr 65 70 75 80
Val Ala Gin Lys Met Ala Glu Pro Glu Lys Ala Pro Ala Leu Ser He 85 90 95
Leu Leu Tyr Val Gin Ala Phe Gin Val Gly Met Pro Pro Pro Gly Cys 100 105 110
Cys Arg Gly Pro Leu Arg Pro Lys Thr Leu Leu Leu Thr Ser Ser Glu 115 120 125
He Phe Leu Leu Asp Glu Asp Cys Val His Tyr Pro Leu Pro Glu Phe 130 135 140
Ala Lys Glu Pro Pro Gin Arg Asp Arg Tyr Arg Leu Asp Asp Gly Arg 145 150 155 160
Arg Val Arg Asp Leu Asp Arg Val Leu Met Gly Tyr Gin Thr Tyr Pro 165 170 175 Gin Pro Ser Pro Ser Ser Ser Met Thr Cys Lys Val Met Thr Ser Trp 180 185 190
Ala Val Ser Pro Trp Thr Thr Leu Gly Arg Cys Gin Val Ala Arg Leu 195 200 205
Glu Pro Ala Arg Ala Val Lys Ser Ser Gly Arg Cys Leu Ser Pro Val 210 215 220
Leu Arg Ala Glu Arg Ser Ser Ser Arg Cys Trp Leu Ala Ser Gly Arg 225 230 235 240
Pro Cys Val Ala Val Ser Cys Leu Ser Ser Ser Pro Ala Ser Pro Gly
245 250 255 His Ser Gin Pro Val Val Ser Ser Leu Thr Pro Thr Gly Ala Gly Gin 260 265 270
Gin Ala Phe Val Phe Ser Lys Asn Val Leu Ser Ser Leu Trp Tyr Leu 275 280 285
Asn Leu Thr Val Leu Ala Glu Asn Val Asn Met Cys Val Cys Cys Val 290 295 300
Asn Ser Phe Ser Cys Trp Glu Xaa 305 310
(2) INFORMATION FOR SEQ ID NO: 476:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 329 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 476:
Met Ala Gin His His Leu Trp He Leu Leu Leu Cys Leu Gin Thr Trp
1 5 10 15 Pro Glu Ala Ala Gly Lys Asp Ser Glu He Phe Thr Val Asn Gly He
20 25 30
Leu Gly Glu Ser Val Thr Phe Pro Val Asn He Gin Glu Pro Arg Gin 35 40 45
Val Lys He He Ala Trp Thr Ser Lys Thr Ser Val Ala Tyr Val Thr 50 55 60
Pro Gly Asp Ser Glu Thr Ala Pro Val Val Thr Val Thr His Arg Asn 65 70 75 80
Tyr Tyr Glu Arg He His Ala Leu Gly Pro Asn Tyr Asn Leu Val He 85 90 95 Ser Asp Leu Arg Met Glu Asp Ala Gly Asp Tyr Lys Ala Asp He Asn 100 105 110
Thr Gin Ala Asp Pro Tyr Thr Thr Thr Lys Arg Tyr Asn Leu Gin He 115 120 125
Tyr Arg Arg Leu Gly Lys Pro Lys He Thr Gin Ser Leu Met Ala Ser 130 135 140
Val Asn Ser Thr Cys Asn Val Thr Leu Thr Cys Ser Val Glu Lys Glu 145 150 155 160
Glu Lys Asn Val Thr Tyr Asn Trp Ser Pro Leu Gly Glu Glu Gly Asn
165 170 175 Val Leu Gin He Phe Gin Thr Pro Glu Asp Gin Glu Leu Thr Tyr Thr 180 185 190
Cys Thr Ala Gin Asn Pro Val Ser Asn Asn Ser Asp Ser He Ser Ala 195 200 205
Arg Gin Leu Cys Ala Asp He Ala Met Gly Phe Arg Thr His His Thr 210 215 220
Gly Leu Leu Ser Val Leu Ala Met Phe Phe Leu Leu Val Leu He Leu 225 230 235 240
Ser Ser Val Phe Leu Phe Arg Leu Phe Lys Arg Arg Gin Asp Ala Ala 245 250 255
Ser Lys Lys Thr He Tyr Thr Tyr He Met Ala Ser Arg Asn Thr Gin 260 265 270
Pro Ala Glu Ser Arg He Tyr Asp Glu He Leu Gin Ser Lys Val Leu 275 280 285
Pro Ser Lys Glu Glu Pro Val Asn Thr Val Tyr Ser Glu Val Gin Phe 290 295 300
Ala Asp Lys Met Gly Lys Ala Ser Thr Gin Asp Ser Lys Pro Pro Gly 305 310 315 320
Thr Ser Ser Tyr Glu He Val He Xaa 325
(2) INFORMATION FOR SEQ ID NO: 477:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 178 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 477: Met Lys Leu Gin Cys Val Ser Leu Trp Leu Leu Gly Thr He Leu He 1 5 10 15
Leu Cys Ser Val Asp Asn His Gly Leu Arg Arg Cys Leu He Ser Thr 20 25 30
Asp Met His His He Glu Glu Ser Phe Gin Glu He Lys Arg Ala He 35 40 45
Gin Ala Lys Asp Thr Phe Pro Asn Val Thr He Leu Ser Thr Leu Glu 50 55 60
Thr Leu Gin He He Lys Pro Leu Asp Val Cys Cys Val Thr Lys Asn 65 70 75 80 Leu Leu Ala Phe Tyr Val Asp Arg Val Phe Lys Asp His Gin Glu Pro
85 90 95
Asn Pro Lys He Leu Arg Lys He Ser Ser He Ala Asn Ser Phe Leu 100 105 110
Tyr Met Gin Lys Thr Leu Arg Gin Cys Gin Glu Gin Arg Gin Cys His 115 120 125
Cys Arg Gin Glu Ala Thr Asn Ala Thr Arg Val He His Asp Asn Tyr 130 135 140
Asp Gin Leu Glu Val His Ala Ala Ala He Lys Ser Leu Gly Glu Leu 145 150 155 160 Asp Val Phe Leu Ala Trp He Asn Lys Asn His Glu Val Met Ser Ser
165 170 175
Ala Xaa
(2) INFORMATION FOR SEQ ID NO: 478: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 52 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 478:
Asp Thr Ala He Arg Val Ala Leu Ala Val Ala Val Leu Lys Thr Val 1 5 10 15
He Leu Gly Leu Leu Cys Leu Leu Leu Cys Gly Gly Gly Glu Gly Lys 20 25 30
Val Ala Gly Arg Gin Ala Val Thr Ser Asp Gin Gin Ser Val Gly Arg 35 40 45
Arg Asp Val Tyr 50
(2) INFORMATION FOR SEQ ID NO: 479:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 62 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 479:
Met Gin Lys Lys Asn Ser Leu Phe Phe Phe Phe Ala Phe Tyr Tyr Glu 1 5 10 15
Asn Lys Thr Asn Ala Pro Gly Glu Gly Ser Met He Thr Arg Asn He 20 25 30
Lys Glu Tyr Phe Leu Pro Phe Leu Phe Cys Cys Val Glu Ala Ser He 35 40 45
Ala He Asn Lys Leu Asn Tyr Leu His Trp Thr His Phe Gin 50 55 60
(2) INFORMATION FOR SEQ ID NO: 480:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 27 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 480: Met Pro Gly Leu Ser Leu He Leu Thr Val Thr Leu Leu Ala Val Ser 1 5 10 15
Asp Ser Ala Ala Thr Cys He Val Ala Lys Gly 20 25
(2) INFORMATION FOR SEQ ID NO: 481: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 339 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 481:
Met Ser Gly Pro Asp Val Glu Thr Pro Ser Ala He Gin He Cys Arg 1 5 10 15
He Met Arg Pro Asp Asp Ala Asn Val Ala Gly Asn Val His Gly Gly 20 25 30
Thr He Leu Lys Met He Glu Glu Ala Gly Ala He He Ser Thr Arg 35 40 45
His Cys Asn Ser Gin Asn Gly Glu Arg Cys Val Ala Ala Leu Ala Arg 50 55 60
Val Glu Arg Thr Asp Phe Leu Ser Pro Met Cys He Gly Glu Val Ala 65 70 75 80
His Val Ser Ala Glu He Thr Tyr Thr Ser Lys His Ser Val Glu Val 85 90 95
Gin Val Asn Val Met Ser Glu Asn He Leu Thr Gly Ala Lys Lys Leu 100 105 110
Thr Asn Lys Ala Thr Leu Trp Tyr Val Pro Leu Ser Leu Lys Asn Val 115 120 125 Asp Lys Val Leu Glu Val Pro Pro Val Val Tyr Ser Arg Xaa Glu Gin 130 135 140
Glu Glu Glu Gly Arg Lys Arg Tyr Glu Ala Gin Lys Leu Glu Arg Met 145 150 155 160
Glu Thr Lys Trp Arg Asn Gly Asp He Val Gin Pro Val Leu Asn Pro 165 170 175
Glu Pro Asn Thr Val Ser Tyr Ser Gin Ser Ser Leu He His Leu Val 180 185 190
Gly Pro Ser Asp Cys Thr Leu His Gly Phe Val His Gly Gly Val Thr 195 200 205 Met Lys Leu Met Asp Glu Val Ala Gly He Val Ala Ala Arg His Cys 210 215 220
Lys Thr Asn He Val Thr Ala Ser Val Asp Ala He Asn Phe His Asp
225 230 235 240
Lys He Arg Lys Gly Cys Val He Thr He Ser Gly Arg Met Thr Phe
245 250 255
Thr Ser Asn Lys Ser Met Glu He Glu Val Leu Val Asp Ala Asp Pro 260 265 270
Val Val Asp Ser Ser Gin Lys Arg Tyr Arg Ala Ala Ser Ala Phe Phe 275 280 285 Thr Tyr Val Ser Leu Ser Gin Glu Gly Arg Ser Leu Pro Val Pro Gin 290 295 300
Leu Val Pro Glu Thr Glu Asp Glu Lys Lys Arg Phe Glu Glu Gly Lys 305 310 315 320
Gly Arg Tyr Leu Gin Met Lys Ala Lys Xaa Gin Gly His Ala Xaa Xaa 325 330 335
Gin Pro Xaa
(2) INFORMATION FOR SEQ ID NO: 482:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 32 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 482:
Met Leu Asn Ser Asn He Asn Asp Leu Leu Met Val Thr Tyr Leu Ala 1 5 10 15 Asn Leu Thr Gin Ser Gin He Ala Leu Asn Glu Lys Leu Val Asn Leu 20 25 30
(2) INFORMATION FOR SEQ ID NO: 483: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 48 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 483:
Met Arg Glu Thr Ser He Arg Val Leu Leu Met Leu Pro Ala Leu Glu 1 5 10 15
Ser Thr Ser Gly Leu Ser Ala Phe Met Gly Leu Gly Thr Arg He Gly 20 25 30
Cys Phe Lys Thr He Thr Cys Trp Pro Thr Ser Leu Thr Gin Arg Xaa 35 40 45
(2) INFORMATION FOR SEQ ID NO: 484:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 38 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 484:
Met Tyr Met Tyr Ser Leu Asn Val Phe Leu Ser Phe He Phe Leu Ala
1 5 10 15
Leu Val Phe Lys Cys Val His Val Cys Gin Gly Ala Asn Ala Phe Leu 20 25 30
Phe Leu Lys Leu Val Phe 35
(2) INFORMATION FOR SEQ ID NO: 485:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 61 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 485:
Met Gly Leu Arg Leu He Cys Leu Glu Leu Thr Met Val Lys Ala Leu 1 5 10 15 Val Cys Glu Met Phe Leu Phe Phe Leu Met Thr Gin Lys Leu He Trp 20 25 30
Gin Glu Cys Thr Glu Lys Phe Ala Lys Leu Leu Val Gin Leu He Ser 35 40 45
Leu Val Phe Ala Trp Glu Phe Phe Ser Glu Asp Thr Pro 50 55 60
(2) INFORMATION FOR SEQ ID NO: 486:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 346 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 486:
Met Leu Ala Ala Arg Leu Val Cys Leu Arg Thr Leu Pro Ser Arg Val 1 5 10 15
Phe His Pro Ala Phe Thr Lys Ala Ser Pro Val Val Lys Asn Ser He 20 25 30 Thr Lys Asn Gin Trp Leu Leu Thr Pro Ser Arg Glu Tyr Ala Thr Lys 35 40 45
Thr Arg He Gly He Arg Arg Gly Arg Thr Gly Gin Glu Leu Lys Glu 50 55 60
Ala Ala Leu Glu Pro Ser Met Glu Lys He Phe Lys He Asp Gin Met 65 70 75 80
Gly Arg Trp Phe Val Ala Gly Gly Ala Ala Val Gly Leu Gly Ala Leu 85 90 95
Cys Tyr Tyr Gly Leu Gly Leu Ser Asn Glu He Gly Ala He Glu Lys 100 105 110 Ala Val He Trp Pro Gin Tyr Val Lys Asp Arg He His Ser Thr Tyr 115 120 125
Met Tyr Leu Ala Gly Ser He Gly Leu Thr Ala Leu Ser Ala He Ala 130 135 140
He Ser Arg Thr Pro Val Leu Met Asn Phe Met Met Arg Gly Ser Trp 145 150 155 160
Val Thr He Gly Val Thr Phe Ala Ala Met Val Gly Ala Gly Met Leu 165 170 175
Val Arg Ser He Pro Tyr Asp Gin Ser Pro Gly Pro Lys His Leu Ala 180 185 190 Trp Leu Leu His Ser Gly Val Met Gly Ala Val Val Ala Pro Leu Thr 195 200 205
He Leu Gly Gly Pro Leu Leu He Arg Ala Ala Trp Tyr Thr Ala Gly 210 215 220
He Val Gly Gly Leu Ser Thr Val Ala Met Cys Ala Pro Ser Glu Lys 225 230 235 240
Phe Leu Asn Met Gly Ala Pro Leu Gly Val Gly Leu Gly Leu Val Phe 245 250 255
Val Ser Ser Leu Gly Ser Met Phe Leu Pro Pro Thr Thr Val Ala Gly 260 265 270 Ala Thr Leu Tyr Ser Val Ala Met Tyr Gly Gly Leu Val Leu Phe Ser 275 280 285
Met Phe Leu Leu Tyr Asp Thr Gin Lys Val He Lys Arg Ala Glu Val 290 295 300
Ser Pro Met Tyr Gly Val Gin Lys Tyr Asp Pro He Asn Ser Met Leu 305 310 315 320
Ser He Tyr Met Asp Thr Leu Asn He Phe Met Arg Val Ala Thr Met 325 330 335
Leu Ala Thr Gly Gly Asn Arg Lys Lys Xaa 340 345
(2) INFORMATION FOR SEQ ID NO: 487:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 237 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 487: Met Glu Glu Val Leu Leu Leu Gly Leu Lys Asp Arg Glu Gly Tyr Thr 1 5 10 15
Ser Phe Trp Asn Asp Cys He Ser Ser Gly Leu Arg Gly Cys Met Leu 20 25 30
He Glu Leu Ala Leu Arg Gly Arg Leu Gin Leu Glu Ala Cys Gly Met 35 40 45
Arg Arg Lys Ser Leu Leu Thr Arg Lys Val He Cys Lys Ser Asp Ala 50 55 60
Pro Thr Gly Asp Val Leu Leu Asp Glu Ala Leu Lys His Val Lys Glu 65 70 75 80
Thr Gin Pro Pro Glu Thr Val Gin Asn Trp He Glu Leu Leu Ser Gly 85 90 95
Glu Thr Trp Asn Pro Leu Lys Leu His Tyr Gin Leu Arg Asn Val Arg 100 105 110
Glu Arg Leu Ala Lys Asn Leu Val Glu Lys Gly Val Leu Thr Thr Glu 115 120 125
Lys Gin Asn Phe Leu Leu Phe Asp Met Thr Thr His Pro Leu Thr Asn 130 135 140
Asn Asn He Lys Gin Arg Leu He Lys Lys Val Gin Glu Ala Val Leu 145 150 155 160 Asp Lys Trp Val Asn Asp Pro His Arg Met Asp Arg Arg Leu Leu Ala
165 170 175
Leu He Tyr Leu Ala His Ala Ser Asp Val Leu Glu Asn Ala Phe Ala 180 185 190
Pro Leu Leu Asp Glu Gin Tyr Asp Leu Ala Thr Lys Arg Val Arg Gin 195 200 205
Leu Leu Asp Leu Asp Pro Glu Val Glu Cys Leu Lys Ala Asn Thr Asn 210 215 220
Glu Val Leu Trp Ala Val Val Ala Ala Phe Thr Lys Xaa 225 230 235
(2) INFORMATION FOR SEQ ID NO: 488:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 200 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 488: Met Ala Gin Arg Met Val Trp Val Asp Leu Glu Met Thr Gly Leu Asp 1 5 10 15
He Glu Lys Asp Gin He He Glu Met Ala Cys Leu He Thr Asp Ser 20 25 30
Asp Leu Asn He Leu Ala Glu Gly Pro Asn Leu He He Lys Gin Pro 35 40 45
Asp Glu Leu Leu Asp Ser Met Ser Asp Trp Cys Lys Glu His His Gly 50 55 60
Lys Ser Gly Leu Thr Lys Ala Val Lys Glu Ser Thr He Thr Leu Gin 65 70 75 80 Gin Ala Glu Tyr Glu Phe Leu Ser Phe Val Arg Gin Gin Thr Pro Pro
85 90 95
Gly Leu Cys Pro Leu Ala Gly Asn Ser Val His Glu Asp Lys Lys Phe 100 105 110
Leu Asp Lys Tyr Met Pro Gin Phe Met Lys His Leu His Tyr Arg He 115 120 125
He Asp Val Ser Thr Val Lys Glu Leu Cys Arg Arg Trp Tyr Pro Glu 130 135 140
Glu Tyr Glu Phe Ala Pro Lys Lys Ala Ala Ser His Arg Ala Leu Asp 145 150 155 160 Asp He Ser Glu Ser He Lys Glu Leu Gin Phe Tyr Arg Asn Asn He
165 170 175
Phe Lys Lys Lys He Asp Glu Lys Lys Arg Lys He He Glu Asn Gly 180 185 190
Glu Asn Glu Lys Thr Val Ser Xaa 195 200
(2) INFORMATION FOR SEQ ID NO: 489:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 351 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 489:
Met Ala Thr Thr Ala Ala Pro Ala Gly Gly Ala Arg Asn Gly Ala Gly 1 5 10 15
Pro Glu Trp Gly Gly Phe Glu Glu Asn He Gin Gly Gly Gly Ser Ala 20 25 30 Val He Asp Met Glu Asn Met Asp Asp Thr Ser Gly Ser Ser Phe Glu 35 40 45
Asp Met Gly Glu Leu His Gin Arg Leu Arg Glu Glu Glu Val Asp Ala 50 55 60
Asp Ala Ala Asp Ala Ala Ala Ala Glu Glu Glu Asp Gly Glu Phe Leu 65 70 75 80
Gly Met Lys Gly Phe Lys Gly Gin Leu Ser Arg Gin Val Ala Asp Gin 85 90 95
Met Trp Gin Ala Gly Lys Arg Gin Ala Ser Arg Ala Phe Ser Leu Tyr 100 105 110 Ala Asn He Asp He Leu Arg Pro Tyr Phe Asp Val Glu Pro Ala Gin 115 120 125
Val Arg Thr Gly Leu Leu Glu Ser Met He Pro He Lys Met Val Asn 130 135 140
Phe Pro Gin Lys He Ala Gly Glu Leu Tyr Gly Pro Leu Met Leu Val 145 150 155 160
Phe Thr Leu Val Ala He Leu Leu His Gly Met Lys Thr Ser Asp Thr 165 170 175
He He Arg Glu Gly Thr Leu Met Gly Thr Ala He Gly Thr Cys Phe 180 185 190 Gly Tyr Trp Leu Gly Val Ser Ser Phe He Tyr Phe Leu Ala Tyr Leu 195 200 205
Cys Asn Ala Gin He Thr Met Leu Gin Met Leu Ala Leu Leu Gly Tyr 210 215 220
Gly Leu Phe Gly His Cys He Val Leu Phe He Thr Tyr Asn He His 225 230 235 240
Leu His Ala Leu Phe Tyr Leu Phe Trp Leu Leu Val Gly Gly Leu Ser 245 250 255
Thr Leu Arg Met Val Ala Val Leu Val Ser Arg Thr Val Gly Pro Thr 260 265 270 Gin Arg Leu Leu Leu Cys Gly Thr Leu Ala Ala Leu His Met Leu Phe 275 280 285
Leu Leu Tyr Leu His Phe Ala Tyr His Lys Val Val Glu Gly He Leu 290 295 300
Asp Thr Leu Glu Gly Pro Asn He Pro Pro He Gin Arg Val Pro Arg 305 310 315 320
Asp He Pro Ala Met Leu Pro Ala Ala Arg Leu Pro Thr Thr Val Leu 325 330 335
Asn Ala Thr Ala Lys Ala Val Ala Val Thr Leu Gin Ser His Xaa 340 345 350
(2) INFORMATION FOR SEQ ID NO: 490:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 265 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 490: Met Arg Gly Ser Arg Gly Gly Trp Ala Gly Glu Met Ala Ala Ser Gly 1 5 10 15
Glu Ser Gly Thr Ser Gly Gly Gly Gly Ser Thr Glu Glu Ala Phe Met 20 25 30
Thr Phe Tyr Ser Glu Val Lys Gin He Glu Lys Arg Asp Ser Val Leu 35 40 45
Thr Ser Lys Asn Gin He Glu Arg Leu Thr Arg Pro Gly Ser Ser Tyr 50 55 60
Phe Asn Leu Asn Pro Phe Glu Val Leu Gin He Asp Pro Glu Val Thr
65 70 75 80 Asp Glu Glu He Lys Lys Arg Phe Arg Gin Leu Ser He Leu Val His
85 90 95
Pro Asp Lys Asn Gin Asp Asp Ala Asp Arg Ala Gin Lys Ala Phe Glu 100 105 110
Ala Val Asp Lys Ala Tyr Lys Leu Leu Leu Asp Gin Glu Gin Lys Lys 115 120 125
Arg Ala Leu Asp Val He Gin Ala Gly Lys Glu Tyr Val Glu His Thr 130 135 140
Val Lys Glu Arg Lys Lys Gin Leu Lys Lys Glu Gly Lys Pro Thr He 145 150 155 160 Val Glu Glu Asp Asp Pro Glu Leu Phe Lys Gin Ala Val Tyr Lys Gin
165 170 175
Thr Met Lys Leu Phe Ala Glu Leu Glu He Lys Arg Lys Glu Arg Glu 180 185 190
Ala Lys Glu Met His Glu Arg Lys Arg Gin Arg Glu Glu Glu He Glu 195 200 205
Ala Gin Glu Lys Ala Lys Arg Glu Arg Glu Trp Gin Lys Asn Phe Glu 210 215 220
Glu Ser Arg Asp Gly Arg Val Asp Ser Trp Arg Asn Phe Gin Ala Asn 225 230 235 240 Thr Lys Gly Lys Lys Glu Lys Lys Asn Arg Thr Phe Leu Arg Pro Pro
245 250 255
Lys Val Lys Met Glu Gin Arg Glu Xaa 260 265
(2) INFORMATION FOR SEQ ID NO: 491: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 25 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 491:
Asp Ser Met Pro Thr Cys Pro Leu Xaa Ala Ser Leu Glu Cys Gly Pro 1 5 10 15
Leu Leu Pro Val Arg Leu Cys Cys Leu 20 25
(2) INFORMATION FOR SEQ ID NO: 492:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 159 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 492:
Met Asn Glu Tyr Arg Val Pro Glu Leu Asn Val Gin Asn Gly Val Leu 1 5 10 15 Lys Ser Leu Ser Phe Leu Phe Glu Tyr He Gly Glu Met Gly Lys Asp 20 25 30
Tyr He Tyr Ala Val Thr Pro Leu Leu Glu Asp Ala Leu Met Asp Arg 35 40 45
Asp Leu Val His Arg Gin Thr Ala Ser Ala Val Val Gin His Met Ser 50 55 60
Leu Gly Val Tyr Gly Phe Gly Cys Glu Asp Ser Leu Asn His Leu Leu 65 70 75 80
Asn Tyr Val Trp Pro Asn Val Phe Glu Thr Ser Pro His Val He Gin 85 90 95 Ala Val Met Gly Ala Leu Glu Gly Leu Arg Val Ala He Gly Pro Cys 100 105 110
Arg Met Leu Gin Tyr Cys Leu Gin Gly Leu Phe His Pro Ala Arg Lys 115 120 125
Val Arg Asp Val Tyr Trp Lys He Tyr Asn Ser He Tyr He Gly Ser 130 135 140
Gin Asp Ala Leu He Ala His Tyr Pro Arg He Tyr Gin Arg Xaa 145 150 155
(2) INFORMATION FOR SEQ ID NO: 493:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 279 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 493:
Met He Ser Asp Asn Ser Ala Glu Asn He Ala Leu Val Thr Ser Met 1 5 10 15 Tyr Asp Gly Leu Leu Gin Ala Gly Ala Arg Leu Cys Pro Thr Val Gin 20 25 30
Leu Glu Asp He Arg Asn Leu Gin Asp Leu Thr Pro Leu Lys Leu Ala 35 40 45
Ala Lys Glu Gly Lys He Glu He Phe Arg His He Leu Gin Arg Glu 50 55 60
Phe Ser Gly Leu Ser His Leu Ser Arg Lys Phe Thr Glu Trp Cys Tyr 65 70 75 80
Gly Pro Val Arg Val Ser Leu Tyr Asp Leu Ala Ser Val Asp Ser Cys 85 90 95
Glu Glu Asn Ser Val Leu Glu He He Ala Phe His Cys Lys Ser Pro 100 105 110
His Arg His Arg Met Val Val Leu Glu Pro Leu Asn Lys Leu Leu Gin 115 120 125
Ala Lys Trp Asp Leu Leu He Pro Lys Phe Phe Leu Asn Phe Leu Cys 130 135 140
Asn Leu He Tyr Met Phe He Phe Thr Ala Val Ala Tyr His Gin Pro 145 150 155 160
Thr Leu Lys Lys Gin Ala Ala Pro His Leu Lys Ala Glu Val Gly Asn 165 170 175 Ser Met Leu Leu Thr Gly His He Leu He Leu Leu Gly Gly He Tyr 180 185 190
Leu Leu Val Gly Gin Leu Trp Tyr Phe Trp Arg Arg His Val Phe He 195 200 205
Trp He Ser Phe He Asp Ser Tyr Phe Glu He Leu Phe Leu Phe Gin 210 215 220
Ala Leu Leu Thr Val Val Ser Gin Val Leu Cys Phe Leu Xaa He Glu 225 230 235 240
Trp Tyr Leu Pro Leu Leu Val Ser Ala Leu Val Leu Gly Trp Leu Asn 245 250 255 Leu Leu Tyr Tyr Thr Arg Gly Phe Gin His Thr Gly He Tyr Ser Val 260 265 270
Met He Gin Lys Pro Trp Xaa 275
(2) INFORMATION FOR SEQ ID NO: 494: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 193 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 494:
Met He Arg Cys Gly Leu Ala Cys Glu Arg Cys Arg Trp He Leu Pro 1 5 10 15
Leu Leu Leu Leu Ser Ala He Ala Phe Asp He He Ala Leu Ala Gly 20 25 30
Arg Gly Trp Leu Gin Ser Ser Asp His Gly Gin Thr Ser Ser Leu Trp 35 40 45 Trp Lys Cys Ser Gin Glu Gly Gly Gly Ser Gly Ser Tyr Glu Glu Gly
50 55 60
Cys Gin Ser Leu Met Glu Tyr Ala Trp Gly Arg Ala Ala Ala Ala Met 65 70 75 80
Leu Phe Cys Gly Phe He He Leu Val He Cys Phe He Leu Ser Phe 85 90 95
Phe Ala Leu Cys Gly Pro Gin Met Leu Val Phe Leu Arg Val He Gly 100 105 110
Gly Leu Leu Ala Leu Ala Ala Val Phe Gin He He Ser Leu Val He 115 120 125 Tyr Pro Val Lys Tyr Thr Gin Thr Phe Thr Leu His Ala Asn Xaa Ala 130 135 140
Val Thr Tyr He Tyr Asn Trp Ala Tyr Gly Phe Gly Trp Ala Ala Thr 145 150 155 160
He He Leu He Gly Cys Ala Phe Phe Phe Cys Cys Leu Pro Asn Tyr 165 170 175
Glu Asp Asp Leu Leu Gly Asn Ala Lys Pro Arg Tyr Phe Tyr Thr Ser 180 185 190
Ala
(2) INFORMATION FOR SEQ ID NO: 495:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 205 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 495: Met Ala Ala Gly Asp Gin Val Phe Ser Gly Ala Gly His Val Xaa Glu 1 5 10 15
His Val Ala Gly Gly Arg His Ala Trp Leu Leu Thr Trp Gin Ser Ala 20 25 30
Cys Pro Ala Asn Arg Leu Ser Leu Val Pro Leu Val Pro Ser Ala Ser 35 40 45
Met Thr Arg Leu Met Arg Xaa Arg Thr Ala Ser Gly Ser Ser Val He 50 55 60
Leu Trp Met Ala Pro Ala Ala Ala Pro Thr Pro Ala Arg Ala Pro Glu
65 70 75 80 Ala Ala Pro Thr Pro Ala Arg Ala Pro Ala Ala Ala Arg Thr Pro Ala
85 90 95
Arg Gly Pro Thr Trp Thr Ser Pro Pro Thr Arg Val Leu Leu Gly Thr 100 105 110
Xaa Pro Gly Pro Ser Pro Trp Arg Ser Pro Ala Arg Arg Pro Ala Gin 115 120 125
Leu Pro Pro Pro Asp Ser Asp Leu Cys Ser Gly Pro Leu Leu Pro Gly 130 135 140
Pro Phe Ser Pro Pro Ala Cys His Thr Ala Pro Asn Ser Val Leu He 145 150 155 160 Gin Ser Leu Phe Cys Lys Ser Glu Leu Trp Trp Arg Gin Met Arg Ser
165 170 175
He Thr Trp Val Pro Ser Pro Lys Ala Gly Trp Arg Trp Thr Lys Gly 180 185 190
Arg Lys Gin Ala Ser Pro His Arg He Leu Phe His Xaa 195 200 205
(2) INFORMATION FOR SEQ ID NO: 496:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 147 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 496:
Met Ala Leu Thr Leu Leu Pro Ser Val Ser Arg Leu Pro Gly Glu Arg 1 5 10 15
Met Ala Ala Ser Gly Leu Pro Tyr Val Leu His His Lys Ser Ser Leu 20 25 30 Met Lys Val He Phe Phe Pro Tyr Pro Val Leu Pro Leu Pro Ala Pro 35 40 45
Asn Gly Thr Trp Val Pro Arg Leu Val Leu Gly Leu Gly Ser Gly Asp
50 55 60
Gin Val His Tyr Leu Pro He Ser Ser Ser He Val Asn Tyr Gly Thr
65 70 75 80
Ser Val Ser Gly Lys Ser Trp Val Phe Leu Val Tyr Pro Leu His Pro 85 90 95
Thr Pro Thr Trp Ser Thr Arg Cys Phe Gin Val Trp Asp Leu Leu Ser 100 105 110 Val Glu Leu Pro Asp Lys Gly Glu Gly Asn Thr Arg Arg Ala Ser Gly 115 120 125
Val Pro Gly Leu Ser Gin Leu Pro Thr Ser His Lys Pro He Lys Gin 130 135 140
Glu Tyr Xaa 145
(2) INFORMATION FOR SEQ ID NO: 497:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 64 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 497:
Met Val Trp Val Leu Trp Ser Ala Pro Ser Leu Ala Pro Pro Trp Val 1 5 10 15
Gly Pro Cys Trp Pro Ser Thr Gly Asn Cys Cys Leu Cys Glu Val Gly 20 25 30 Ala Ala Leu Pro Pro Arg Gly Pro Ser Leu Ser Asp Cys Leu Gly Leu 35 40 45
Pro Pro Trp Thr Pro Trp Gly Pro Ala Trp Thr Leu Ala Gin Ser Xaa 50 55 60
(2) INFORMATION FOR SEQ ID NO: 498:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 94 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 498:
Met Ser Thr Gly Ala Leu Asn Thr Ser Pro Pro Ala Ser Asn Arg Leu 1 5 10 15
Glu Ser Thr Leu Asn Glu Tyr Leu He Gin Pro Gin Leu His Cys Ser 20 25 30 Ser Val Gin Arg Leu Thr Leu Lys Trp Gly Cys Ser Ser Leu Gin Arg 35 40 45
Asp Gly Gin Ala Val Pro Trp Gly Leu Trp Gin Arg Ala Tyr Pro Ser 50 55 60
Leu Leu Pro Thr Leu Pro Ser Asp Leu Leu Arg Pro His Ala Val Thr 65 70 75 80
Pro Ser Val Ser Val Ser Val His Thr Cys Glu Ser Ser Xaa 85 90
(2) INFORMATION FOR SEQ ID NO: 499:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 22 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 499:
Met Phe Leu He Phe Val Tyr Phe Leu Lys Xaa Leu Phe Ser Ser Ser 1 5 10 15
Leu Pro Phe Leu Trp Leu 20
(2) INFORMATION FOR SEQ ID NO: 500:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 33 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 500:
Arg Gly Gly Leu Cys Pro Leu Leu Val Pro Gly Pro Leu Ala Arg Gin 1 5 10 15
Glu Pro Ser Pro Ser Leu Gin Gly Cys Ser Glu Ser Pro Val Gly Met 20 25 30
Asp
(2) INFORMATION FOR SEQ ID NO: 501:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 28 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 501:
Met Gin Phe Leu Leu Thr Ala Phe Leu Leu Val Pro Leu Leu Ala Leu 1 5 10 15 Cys Asp Val Pro He Ser Leu Gly Phe Ser Pro Ser 20 25
(2) INFORMATION FOR SEQ ID NO: 502:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 15 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 502:
Pro Gly Lys Pro Gin Ala Cys Pro Glu Leu Thr Ser Val Leu Pro 1 5 10 15
(2) INFORMATION FOR SEQ ID NO: 503: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 19 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 503:
Asn Lys Ser Leu Xaa Ser Cys Leu Phe Val Leu His Phe Val Leu His 1 5 10 15
Cys Xaa Phe
(2) INFORMATION FOR SEQ ID NO: 504:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 29 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 504:
Met Glu Lys Thr His Arg Leu Arg He Arg Asn Pro Cys Leu Gin Phe 1 5 10 15 Ser He Leu Asn Leu Phe Leu Leu Lys Met He Val Ser 20 25
(2) INFORMATION FOR SEQ ID NO: 505:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 75 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 505:
Met Val Asp He Ser Lys Met His Met He Leu Tyr Asp Leu Gin Gin 1 5 10 15
Asn Leu Ser Ser Ser His Arg Ala Leu Glu Lys Gin He Asp Thr Leu 20 25 30
Ala Gly Lys Leu Asp Ala Leu Thr Glu Leu Leu Ser Thr Ala Leu Gly 35 40 45
Pro Ser Ser Phe Gin Asn Pro Ala Ser Ser Pro Ser Ser Trp Thr His 50 55 60 Glu Glu Glu Pro Gly Tyr Phe Pro Gin Tyr Xaa 65 70 75
(2) INFORMATION FOR SEQ ID NO: 506:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 10 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 506:
Leu Pro Leu Ala Glu Leu Lys Asn Trp Val 1 5 10
(2) INFORMATION FOR SEQ ID NO: 507: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 207 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 507:
Met Leu Trp Phe Gly Gly Cys Ser Ala Val Asn Ala Thr Gly His Leu 1 5 10 15
Ser Asp Thr Leu Trp Leu He Pro He Thr Phe Leu Thr He Gly Tyr 20 25 30
Gly Asp Val Val Pro Gly Thr Met Trp Gly Lys He Val Cys Leu Cys 35 40 45 Thr Gly Val Met Gly Val Cys Cys Thr Ala Leu Leu Val Ala Val Val 50 55 60
Ala Arg Lys Leu Glu Phe Asn Lys Ala Glu Lys His Val His Asn Phe 65 70 75 80
Met Met Asp He Gin Tyr Thr Lys Glu Met Lys Glu Ser Ala Ala Arg 85 90 95
Val Leu Gin Glu Ala Trp Met Phe Tyr Lys His Thr Arg Arg Lys Glu 100 105 110
Ser His Ala Ala Arg Arg His Gin Arg Xaa Leu Leu Ala Ala He Asn 115 120 125 Ala Phe Arg Gin Val Arg Leu Lys His Arg Lys Leu Arg Glu Gin Val 130 135 140
Asn Ser Met Val Asp He Ser Lys Met His Met He Leu Tyr Asp Leu 145 150 155 160
Gin Gin Asn Leu Ser Ser Ser His Arg Ala Leu Glu Lys Gin He Asp 165 170 175
Thr Leu Ala Gly Lys Leu Asp Ala Leu Thr Glu Leu Leu Ser Thr Ala 180 185 190
Leu Gly Pro Arg Gin Leu Pro Glu Pro Ser Gin Gin Ser Lys Xaa 195 200 205
(2) INFORMATION FOR SEQ ID NO: 508:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 36 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 508:
Met Trp Arg Cys Arg Gly Lys Leu Ser Phe Pro Leu Phe Ala Val Val 1 5 10 15
He Val Ser Cys Arg Lys Asp Gly Pro Asp Ala Ala Ala Ala Pro Ala 20 25 30
Val Xaa Lys Lys 35
(2) INFORMATION FOR SEQ ID NO: 509:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 19 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 509:
Met Ala Leu Val Ala Leu Phe Thr Gin Leu Met Arg Xaa Leu Gly Arg 1 5 10 15
Cys Pro Gin
(2) INFORMATION FOR SEQ ID NO: 510:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 32 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 510: Met Thr Phe Pro Phe Glu Lys Glu Asn Ser Cys Phe Gin Cys Leu Leu 1 5 10 15
Phe Asp Ser Trp Arg Glu Gin Thr Arg Thr Asn He Gin Pro Gin Arg 20 25 30
(2) INFORMATION FOR SEQ ID NO: 511:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 28 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 511:
Met His Leu Leu Asp Phe Phe Arg Asp Leu Val Leu Leu Val Leu Leu 1 5 10 15
Ala Leu Leu Asp Ser Phe Trp Leu Glu Val Gin Lys 20 25
(2) INFORMATION FOR SEQ ID NO: 512:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 26 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 512: Met Cys Leu He His Phe He Lys He He Leu Val Phe He Leu Lys 1 5 10 15
Leu Trp Leu Tyr Ser Gin Lys Cys Pro Lys 20 25
(2) INFORMATION FOR SEQ ID NO: 513: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 33 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 513:
Met He His Val His Glu Trp Asn Asp Gin Met Leu Met Val Tyr He 1 5 10 15
Phe Leu Tyr Pro Val Ser He Thr Phe Leu Asn Leu Cys Ser Leu Thr 20 25 30
Cys
(2) INFORMATION FOR SEQ ID NO: 514:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 47 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 514: Leu Asn Glu Ser Tyr Val Ser Arg Ala Gly Gly Trp Phe Ser Met Phe 1 5 10 15
Xaa Leu He Phe Phe Leu Leu Ala Leu Gly Ser Xaa Leu Cys Leu Leu 20 25 30
Leu Cys Leu Pro Ser Phe Asn Lys Thr Arg Arg Lys Gin Lys Pro 35 40 45
(2) INFORMATION FOR SEQ ID NO: 515:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 43 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 515:
Ser Ser Lys Thr Pro Leu Pro Ser Glu Arg Arg Trp He Ser Gly Ser 1 5 10 15
Ser Leu Met Ala Pro Arg Pro Trp Leu Leu Gly He Ala Leu Leu Gly 20 25 30 Leu Trp Ala Leu Glu Pro Ala Leu Gly His Trp 35 40
(2) INFORMATION FOR SEQ ID NO: 516:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 3 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 516:
Leu Asn Trp 1
(2) INFORMATION FOR SEQ ID NO: 517: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 174 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 517:
Phe Ala Phe Cys Ala Glu Leu Met He Gin Asn Trp Thr Leu Gly Ala 1 5 10 15
Val Asp Ser Gin Met Asp Asp Met Asp Met Asp Leu Asp Lys Glu Phe 20 25 30
Leu Gin Asp Leu Lys Glu Leu Lys Val Leu Val Ala Asp Lys Asp Leu 35 40 45 Leu Asp Leu His Lys Ser Leu Val Cys Thr Ala Leu Arg Gly Lys Leu 50 55 60
Gly Val Phe Ser Glu Met Glu Ala Asn Phe Lys Asn Leu Ser Arg Gly 65 70 75 80
Leu Val Asn Val Ala Ala Lys Leu Thr His Asn Lys Asp Val Arg Asp 85 90 95
Leu Phe Val Asp Leu Val Glu Lys Phe Val Glu Pro Cys Arg Ser Asp 100 105 110
His Trp Pro Leu Ser Asp Val Arg Phe Phe Leu Asn Gin Tyr Ser Ala 115 120 125
Ser Val His Ser Leu Asp Gly Phe Arg His Gin Ala Ser Gly Thr Ala 130 135 140
Thr Trp Ala Pro Ser Ala Ala Ala Ser Cys Ala Cys He Met Thr Glu 145 150 155 160
Val Pro Pro Asn Ala Pro Pro Thr Leu Thr He Lys Leu Leu 165 170
(2) INFORMATION FOR SEQ ID NO: 518:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 43 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 518:
Met Trp Lys Asn Leu Gly Ser Gly Ser Val Phe Val Thr Trp Phe Ser 1 5 10 15
Leu Val Met He Leu Ser Gly He Gly Pro Leu Gly Asp Ala Glu Asp 20 25 30 Ser He Ser Asp Val Ser His Arg Leu Arg Pro 35 40
(2) INFORMATION FOR SEQ ID NO: 519:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 13 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 519:
Phe Gin Phe Pro Leu Leu Thr He Ala Leu Gin Phe Leu 1 5 10
(2) INFORMATION FOR SEQ ID NO: 520: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 30 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 520:
Met His Tyr Val He Val Leu Ser Leu Phe Val Val Leu Glu Lys Lys 1 5 10 15
Asn Lys Met Gly Ser Asp Gly Cys Leu Arg Lys Asn Gly Ser 20 25 30
(2) INFORMATION FOR SEQ ID NO: 521:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 47 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 521:
Met Ser Arg Ser He Val Leu Arg Gly Ser Leu Phe Leu Phe Phe Ser
1 5 10 15 His Tyr Thr Leu Lys Leu Leu Ser Val He Lys Gin Thr Asn Arg Lys 20 25 30
He Val Trp Glu Lys Pro Cys He Arg Leu Phe Tyr Xaa Val Leu 35 40 45
(2) INFORMATION FOR SEQ ID NO: 522: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 26 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 522:
Met Pro Leu Pro Val Leu Leu Cys Leu Thr Leu Pro Met Pro Leu Pro 1 5 10 15
Ser Ala Thr Ala Arg Gly Gly Asn Arg Thr 20 25
(2) INFORMATION FOR SEQ ID NO: 523:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 58 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 523:
Ser Ser He Pro Val Ser He Leu He Gly Met Lys Leu He Leu Tyr 1 5 10 15 Leu Leu He Thr Glu Ser Gly Ser His Glu Lys Lys Ser Phe Tyr Pro 20 25 30
Ser Phe Lys Tyr Met Phe Lys He He He Tyr Val Ser Ala Tyr Cys 35 40 45
Arg Thr Ala Leu Arg Ala Thr Val Ser His 50 55
(2) INFORMATION FOR SEQ ID NO: 524:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 19 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 524:
Asn Arg Thr Leu Leu Phe Leu He Leu Phe Val Leu Phe Gly Leu Gly 1 5 10 15
Tyr Gly Phe
(2) INFORMATION FOR SEQ ID NO: 525:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 40 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 525: Met Phe Leu Leu Val Leu Ser Val Phe Cys Asp Phe Met Cys Ser He 1 5 10 15
Ala Pro Arg Cys His Ala Leu Ser Leu Val Ser Leu Arg Ala Gin His 20 25 30
Leu Ser Leu Phe He Thr Cys His 35 40
(2) INFORMATION FOR SEQ ID NO: 526:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 57 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 526:
Met Leu Leu Phe He Leu Leu Thr Leu Ser Ser Gly Cys Arg Leu Leu 1 5 10 15
Val Ser Ser Trp Lys Thr Phe Leu Pro His Phe Ser Leu Pro Gly Pro 20 25 30 Arg Glu His Pro Glu Gly Ser Arg Thr Trp Phe Phe Arg Tyr Trp Glu 35 40 45
Pro Gly Ala His Cys Leu His Cys Ala 50 55
(2) INFORMATION FOR SEQ ID NO: 527: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 527:
Ala Arg Leu Leu Leu Phe Leu Ser Ser Val His Pro Ser He Met Pro 1 5 10 15
Ser Cys Asn Gin Leu 20
(2) INFORMATION FOR SEQ ID NO: 528:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 528:
Met Ser Leu Thr Ser Ser Leu Thr Phe Leu Ser His He Leu Leu Leu 1 5 10 15 Pro Gin Lys Leu Gin Phe Leu Ser Trp Met Glu Arg Gin Gin Arg Cys 20 25 30
Thr Gly Val Ala Lys Tyr Ala 35
(2) INFORMATION FOR SEQ ID NO: 529: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 128 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 529:
Met Val Leu Arg Leu He Gin Leu He Phe Leu He Phe Phe He His 1 5 10 15
He He He Leu Leu He Pro Gly Ser Arg Pro Cys Gly Ser Trp Val 20 25 30
Asn Asp Arg Xaa Leu Gly Leu Arg Asp Val Thr His Leu He Tyr Leu 35 40 45 His Trp Val His Gly His Leu Pro Trp Cys His Pro Tyr He Gin Val 50 55 60
Glu Phe Ser Ala Leu He Glu Ser Thr Ala Gin Leu Gly Leu Pro Phe 65 70 75 80
Ser Trp Val Arg Val He His Pro Phe Leu Val Leu Pro Cys Leu Tyr 85 90 95
Ser Pro Gly Leu Lys Asn Gly He Phe Leu Phe Leu Leu Arg Ala Met 100 105 110
Pro Gly Gly Met Phe Pro Gly Asn Leu Glu Ala Phe Arg Val Pro Val 115 120 125
(2) INFORMATION FOR SEQ ID NO: 530:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 82 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 530:
Met Gly Ser Ser Val Leu Pro Phe Cys Val Cys Val Thr Ser Pro Ser 1 5 10 15
Leu Gly Gly Arg Cys He Gin Gly Arg Phe Ala Ser His Ser Lys Phe 20 25 30
Trp Gly Phe Gly Xaa Lys Thr Ala Ser Phe Gly Ala Val Gly Glu Thr 35 40 45
Pro Pro Asp Gin Glu Pro Gin Lys Glu Thr Glu Pro Ala Thr Ser Ser 50 55 60 His Ala Arg Pro Trp Ala Arg Val He Gly Leu Arg He Trp Pro Gin 65 70 75 80
Pro Asn
(2) INFORMATION FOR SEQ ID NO: 531: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 20 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 531:
Met Leu Leu Ser Val Ala He Phe He Leu Leu Thr Leu Val Tyr Ala 1 5 10 15
Tyr Trp Thr Met 20
(2) INFORMATION FOR SEQ ID NO: 532:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 75 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 532:
Asn Cys Glu He Leu Glu Tyr Cys Tyr Tyr Leu Thr Gin Leu Lys He 1 5 10 15
Ser Met Gly Lys Tyr Leu Ser He Pro Thr Val Leu Leu Lys lie He 20 25 30
Arg Cys Ser He Thr Ala Val Ser Asp Ser Ser Thr Ser Trp Ala He 35 40 45
Lys Ala Gin Leu Lys He Glu Asn Lys Asp Leu Asp Asn Lys Thr Ala 50 55 60
Lys Gly Gly Gly Gin Glu Ala Leu Thr Cys Thr 65 70 75
(2) INFORMATION FOR SEQ ID NO: 533:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 60 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 533:
Met Phe Leu Met Arg Met His Leu Cys Phe Cys Lys Tyr Cys Cys Ser 1 5 10 15 Phe He Val Thr Pro Thr Ser Thr Ser Asn Thr Xaa Ser Tyr Leu Trp 20 25 30
Pro Trp He Ser Ala Ser Met Ala Gly Arg Gly Ser Xaa Trp Ala Cys 35 40 45
Thr Leu Asn Ala Val Thr Arg Glu Gly Leu Pro Glu 50 55 60
(2) INFORMATION FOR SEQ ID NO: 534:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 534:
Met Ser Leu Leu Asn Thr His Thr Leu Cys Phe Val Leu Phe Cys Phe 1 5 10 15
Thr Leu Ser He Asn Gin Glu Lys Leu Ala Asn His Leu Ala Phe Arg 20 25 30 He Leu Phe Phe He Val Phe 35
(2) INFORMATION FOR SEQ ID NO: 535:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 2 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 535:
Met Leu 1
(2) INFORMATION FOR SEQ ID NO: 536: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 36 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 536:
Met Asp Gin Phe Lys He Phe Tyr Phe Leu Lys Ala Phe Phe Ala Cys 1 5 10 15
Cys Asn Val Gin Asp Pro Ser Pro Phe Met Gly Glu Thr Gly Ser Tyr 20 25 30
Leu Asn He Gly 35
(2) INFORMATION FOR SEQ ID NO: 537:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 14 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 537: Met Phe Asp Phe Leu Ser Tyr Phe Lys Asp Leu Leu Ser Cys 1 5 10
(2) INFORMATION FOR SEQ ID NO: 538:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 18 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 53£
Met Gly Phe Gly Phe Val Leu Asn He Phe Ser Phe Phe Leu Xaa Pro 1 5 10 15
Pro Leu
(2) INFORMATION FOR SEQ ID NO: 539:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 11 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 539:
Leu Leu Leu Trp Thr Leu Leu Ala Xaa Tyr Xaa 1 5 10
(2) INFORMATION FOR SEQ ID NO: 540:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 108 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 540:
Met Ala Ala Gin Lys Asp Gin Gin Lys Asp Ala Glu Ala Glu Gly Leu 1 5 10 15 Ser Gly Thr Thr Leu Leu Pro Lys Leu He Pro Ser Gly Ala Gly Arg 20 25 30
Glu Trp Leu Glu Arg Arg Arg Ala Thr He Arg Pro Trp Ser Thr Phe 35 40 45
Val Asp Gin Gin Arg Phe Ser Arg Pro Arg Asn Leu Gly Glu Leu Cys 50 55 60
Gin Arg Leu Val Arg Asn Val Glu Tyr Tyr Gin Ser Asn Tyr Val Phe 65 70 75 80
Val Phe Leu Gly Leu He Leu Tyr Cys Val Val Thr Ser Pro Met Leu 85 90 95 Leu Val Ala Leu Ala Val Phe Phe Gly Ala Cys Xaa 100 105
(2) INFORMATION FOR SEQ ID NO: 541:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 106 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 541:
Phe Val Phe Leu Gly Leu He Leu Tyr Cys Val Val Thr Ser Pro Met 1 5 10 15
Leu Leu Val Ala Leu Ala Val Phe Phe Gly Ala Cys Tyr He Leu Tyr 20 25 30
Leu Arg Thr Leu Glu Ser Lys Leu Val Leu Phe Gly Arg Glu Val Ser 35 40 45
Pro Ala His Gin Tyr Ala Leu Ala Gly Gly He Ser Phe Pro Phe Phe 50 55 60
Trp Leu Ala Gly Ala Gly Ser Ala Val Phe Trp Val Leu Gly Ala Thr 65 70 75 80
Leu Val Val He Gly Ser His Ala Ala Phe His Gin He Glu Ala Val 85 90 95
Asp Gly Glu Glu Leu Gin Met Glu Pro Val 100 105
(2) INFORMATION FOR SEQ ID NO: 542:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 136 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 542:
Met Asp Arg Phe Thr Val Ala Gly Val Leu Pro Asp He Glu Gin Phe 1 5 10 15
Phe Asn He Gly Asp Ser Ser Ser Gly Leu He Gin Thr Val Phe He 20 25 30 Ser Ser Tyr Met Val Leu Ala Pro Val Phe Gly Tyr Leu Gly Asp Arg 35 40 45
Tyr Asn Arg Lys Tyr Leu Met Cys Gly Gly He Ala Phe Trp Ser Leu 50 55 60
Val Thr Leu Gly Ser Ser Phe He Pro Gly Glu His Phe Trp Leu Leu 65 70 75 80
Leu Leu Thr Arg Gly Leu Val Gly Val Gly Glu Ala Ser Tyr Ser Thr 85 90 95
He Ala Pro Thr Leu He Ala Asp Leu Phe Val Ala Asp Gin Arg Thr 100 105 110 Gly Cys Ser Ala Ser Ser Thr Leu Pro Phe Arg Trp Ala Val Val Trp 115 120 125
Ala Thr Leu Gin Ala Pro Lys Xaa 130 135
(2) INFORMATION FOR SEQ ID NO: 543: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 424 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 543:
Met Ala Gly Asp Trp His Trp Ala Leu Arg Val Thr Pro Gly Leu Gly 1 5 10 15
Val Val Ala Val Leu Leu Leu Phe Leu Val Val Arg Glu Pro Pro Arg 20 25 30
Gly Ala Val Glu Arg His Ser Asp Leu Pro Pro Leu Asn Pro Thr Ser 35 40 45 Trp Trp Ala Asp Leu Arg Ala Leu Ala Arg Asn Pro Ser Phe Val Leu 50 55 60
Ser Ser Leu Gly Phe Thr Ala Val Ala Phe Val Thr Gly Ser Leu Ala 65 70 75 80
Leu Trp Ala Pro Ala Phe Leu Leu Arg Ser Arg Val Val Leu Gly Glu 85 90 95
Thr Pro Pro Cys Leu Pro Gly Asp Ser Cys Ser Ser Ser Asp Ser Leu 100 105 110
He Phe Gly Leu He Thr Cys Leu Thr Gly Val Leu Gly Val Gly Leu 115 120 125 Gly Val Glu He Ser Arg Arg Xaa Arg His Ser Asn Pro Arg Ala Asp 130 135 140
Pro Leu Val Cys Ala Thr Gly Leu Leu Gly Ser Ala Pro Phe Leu Phe 145 150 155 160
Leu Ser Leu Ala Cys Ala Arg Gly Ser He Val Ala Thr Tyr He Phe 165 170 175
He Phe He Gly Glu Thr Leu Leu Ser Met Asn Trp Ala He Val Ala 180 185 190
Asp He Leu Leu Tyr Val Val He Pro Thr Arg Arg Ser Thr Ala Glu 195 200 205 Ala Phe Gin He Val Leu Ser His Leu Leu Gly Asp Ala Gly Ser Pro 210 215 220
Tyr Leu He Gly Leu He Ser Asp Arg Leu Arg Arg Asn Trp Pro Pro 225 230 235 240
Ser Phe Leu Ser Glu Phe Arg Ala Leu Gin Phe Ser Leu Met Leu Cys 245 250 255
Ala Phe Val Gly Ala Leu Gly Gly Ala Leu Ser Trp Ala Pro Xaa Ser 260 265 270
Ser Leu Arg Pro Thr Ala Gly Gly His Ser Cys Thr Cys Arg Ala Cys 275 280 285 Cys Thr Lys Gin Gly Pro Gin Thr Thr Gly Leu Trp Cys Pro Ser Gly 290 295 300
Ala Ala Pro Pro Ala Cys Pro Trp Pro Val Cys Ser Ser Glu Arg Leu 305 310 315 320
Pro Leu Thr Tyr Leu His He Cys His Ser Xaa Pro Trp Ala His Pro 325 330 335
Thr Lys Gly Leu Gly Leu Thr Pro Trp Pro Gly Pro Ala Ser Arg Gly 340 345 350
Thr Leu Gly Arg Val Pro Ala Pro Arg His Tyr Xaa Gly Ser Ser Gly 355 360 365 Glu Glu Val Gly Val Gin Glu Gly Asp Pro Ser Pro Gin Gly Xaa Pro 370 375 380
Gin Gly Leu Gly Ala He Cys Asn Gly He Lys Phe Val Ala Arg Pro 385 390 395 400
Gin Val Pro Ala Leu Val Phe Leu Trp Val Ala Ser Asp Leu Ala Pro 405 410 415
Arg Leu His Pro Arg Ala Pro Glu 420
(2) INFORMATION FOR SEQ ID NO: 544:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 544:
Met Phe Arg Phe Val He Cys Leu Phe Leu Trp Leu Val Leu Cys Arg 1 5 10 15 Asp Ser Thr Ser Ala Ser Arg He Ala Leu Tyr Tyr Arg He Val Phe 20 25 30
Leu He His Gin Cys Ser Ser 35
(2) INFORMATION FOR SEQ" ID NO: 545: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 58 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 545:
Met Leu Pro Trp Xaa Ala Gin Leu Leu Asp Arg Thr He Gly Pro Leu 1 5 10 15
Tyr Leu Leu Phe Val Gin Phe Ser Pro Ala Phe Ser Arg Thr Ser Pro 20 25 30
Trp Arg Ser Pro Lys Asn Phe Arg Arg Leu Tyr Pro Pro Cys Thr Thr 35 40 45 Ser Gly Cys Ala Ala Arg Trp Leu Phe Ser
50 55
(2) INFORMATION FOR SEQ ID NO: 546:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 33 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 546:
Met Gly Leu Ser Val Leu Leu Pro Leu Cys Leu Leu Gly Pro Gly Arg 1 5 10 15
Phe Thr Ser Gly Gin Lys Pro Leu Asp Thr Pro Gly Leu Gly Val Pro 20 25 30
Phe
(2) INFORMATION FOR SEQ ID NO: 547:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 367 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 547:
Met Ala Lys Pro Gin Val Val Val Ala Pro Val Leu Met Ser Lys Leu 1 5 10 15 Ser Val Asn Ala Pro Glu Phe Tyr Pro Ser Gly Tyr Ser Ser Ser Tyr 20 25 30
Thr Glu Ser Tyr Glu Asp Gly Cys Glu Asp Tyr Pro Thr Leu Ser Glu 35 40 45
Tyr Val Gin Asp Phe Leu Asn His Leu Thr Glu Gin Pro Gly Ser Phe 50 55 60
Glu Thr Glu He Glu Gin Phe Ala Glu Thr Leu Asn Gly Cys Val Thr 65 70 75 80
Thr Asp Asp Ala Leu Gin Glu Leu Val Glu Leu He Tyr Gin Gin Ala 85 90 95 Thr Ser He Pro Asn Phe Ser Tyr Met Gly Ala Arg Leu Cys Asn Tyr 100 105 110
Leu Ser His His Leu Thr He Ser Pro Gin Ser Gly Asn Phe Arg Gin 115 120 125
Leu Leu Leu Gin Arg Cys Arg Thr Glu Tyr Glu Val Lys Asp Gin Ala 130 135 140
Ala Lys Gly Asp Glu Val Thr Arg Lys Arg Phe His Ala Phe Val Leu 145 150 155 160
Phe Leu Gly Glu Leu Tyr Leu Asn Leu Glu He Lys Gly Thr Asn Gly 165 170 175
Gin Val Thr Arg Ala Asp He Leu Gin Val Gly Leu Arg Glu Leu Leu 180 185 190
Asn Ala Leu Phe Ser Asn Pro Met Asp Asp Asn Leu He Cys Ala Val 195 200 205
Lys Leu Leu Lys Leu Thr Gly Ser Val Leu Glu Asp Ala Trp Lys Glu 210 215 220
Lys Gly Lys Met Asp Met Glu Glu He He Gin Arg He Glu Asn Val 225 230 235 240
Val Leu Asp Ala Asn Cys Ser Arg Asp Val Lys Gin Met Leu Leu Lys 245 250 255 Leu Val Glu Leu Arg Ser Ser Asn Trp Gly Arg Val His Ala Thr Ser 260 265 270
Thr Tyr Arg Glu Ala Thr Pro Glu Asn Asp Pro Asn Tyr Phe Met Asn 275 280 285
Glu Pro Thr Phe Tyr Thr Ser Asp Gly Val Pro Phe Thr Ala Ala Asp 290 295 300
Pro Asp Tyr Gin Glu Lys Tyr Gin Glu Leu Leu Glu Arg Glu Asp Phe 305 310 315 320
Phe Pro Asp Tyr Glu Glu Asn Gly Thr Asp Leu Ser Gly Ala Gly Asp 325 330 335 Pro Tyr Leu Asp Asp He Asp Asp Glu Met Asp Pro Glu He Glu Glu 340 345 350
Ala Tyr Glu Lys Phe Cys Leu Glu Ser Glu Arg Lys Arg Lys Gin 355 360 365
(2) INFORMATION FOR SEQ ID NO: 548: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 77 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 548:
Met Leu Arg Leu Asp He He Asn Ser Leu Val Thr Thr Val Phe Met 1 5 10 15
Leu He Val Ser Val Leu Ala Leu He Pro Glu Thr Thr Thr Leu Thr 20 25 30
Val Gly Gly Gly Val Phe Ala Leu Val Thr Ala Val Cys Cys Leu Ala 35 40 45 Asp Gly Ala Leu He Tyr Arg Lys Leu Leu Phe Asn Pro Ser Gly Pro
50 55 60
Tyr Gin Lys Lys Pro Val His Glu Lys Lys Glu Val Leu 65 70 75
(2) INFORMATION FOR SEQ ID NO: 549: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 47 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 549:
Met Leu Lys Gin Val Met Phe Val Phe Ser Gly Met Gly Pro Arg Ser 1 5 10 15
His Cys Trp Gly Leu Pro Leu Ala Cys Gly Thr Phe Val Gin Gly His 20 25 30
Gin Ala Asp Ser Ser His Leu Leu Pro Leu Lys His Gin Gly Ala 35 40 45
(2) INFORMATION FOR SEQ ID NO: 550:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 168 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 550: Met Leu Leu Ser Leu Ala Ala Phe Ser Val He Ser Val Val Ser Tyr 1 5 10 15
Leu He Leu Ala Leu Leu Ser Val Thr He Ser Phe Arg He Tyr Lys 20 25 30
Ser Val He Gin Ala Val Gin Lys Ser Glu Glu Gly His Pro Phe Lys 35 40 45
Ala Tyr Leu Asp Val Asp He Thr Leu Ser Ser Glu Ala Phe His Asn 50 55 60
Tyr Met Asn Ala Ala Met Val His He Asn Arg Ala Leu Lys Leu He
65 70 75 80 He Arg Leu Phe Leu Val Glu Asp Leu Val Asp Ser Leu Lys Leu Ala
85 90 95
Val Phe Met Trp Leu Met Thr Tyr Val Gly Ala Val Phe Asn Gly He 100 105 110
Thr Leu Leu He Leu Ala Glu Leu Leu He Phe Ser Val Pro He Val 115 120 125
Tyr Glu Lys Tyr Lys Thr Gin He Asp His Tyr Val Gly He Ala Arg 130 135 140
Asp Gin Thr Lys Ser He Val Glu Lys He Gin Ala Lys Leu Pro Gly 145 150 155 160
He Ala Lys Lys Lys Ala Glu Xaa 165
(2) INFORMATION FOR SEQ ID NO: 551:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 124 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 551:
Ser Val Pro Phe His Leu Leu Val Val Leu Arg Ser Arg Ala Val Arg
1 5 10 15
Ala Arg Arg Arg Arg Glu Pro Arg Ser Leu Pro Arg Pro Gly Asp Glu 20 25 30
Glu Leu Gin Leu Leu Leu Cys Gly Ala Arg Ser Asp Phe Leu Glu Arg 35 40 45
Cys Glu Glu Asp Trp Val Cys Leu Trp His His Ala Asp His Ala Ala 50 55 60 Phe Pro Gly Ser Phe Gin Cys His Gin Cys Gly Phe Leu Pro His Pro 65 70 75 80
Gly Ser Ser Leu Cys His His Gin Leu Gin Asp Leu Gin Val Arg His 85 90 95
Pro Ser Cys Thr Glu Val Arg Arg Arg Pro Ser He Gin Ser Leu Pro 100 105 110
Gly Arg Arg His Tyr Ser Val Leu Arg Ser Phe Pro 115 120
(2) INFORMATION FOR SEQ ID NO: 552:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 177 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 552:
Met Val His Leu Leu Val Leu Ser Gly Ala Trp Gly Met Gin Met Trp 1 5 10 15 Val Thr Phe Val Ser Gly Phe Leu Leu Phe Arg Ser Leu Pro Arg His 20 25 30
Thr Phe Gly Leu Val Gin Ser Lys Leu Phe Pro Phe Tyr Phe His He 35 40 45
Ser Met Gly Cys Ala Phe He Asn Leu Cys He Leu Ala Ser Gin His 50 55 60
Ala Trp Ala Gin Leu Thr Phe Trp Glu Ala Ser Gin Leu Tyr Leu Leu 65 70 75 80
Phe Leu Ser Leu Thr Leu Ala Thr Val Asn Ala Arg Trp Leu Glu Pro 85 90 95 Arg Thr Thr Ala Ala Met Trp Ala Leu Gin Thr Val Glu Lys Glu Arg 100 105 110
Gly Leu Gly Gly Glu Val Pro Gly Ser His Gin Gly Pro Asp Pro Tyr 115 120 125
Arg Gin Leu Arg Glu Lys Asp Pro Lys Tyr Ser Ala Leu Arg Gin Asn 130 135 140
Phe Phe Arg Tyr His Gly Leu Ser Ser Leu Cys Asn Leu Gly Cys Val 145 150 155 160
Leu Ser Asn Gly Leu Cys Leu Ala Gly Leu Ala Leu Glu He Arg Ser 165 170 175
Leu
(2) INFORMATION FOR SEQ ID NO: 553:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 72 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 553:
Met Ala Phe He Leu Leu Phe Tyr Cys Leu Met Thr Phe Leu Ser Leu 1 5 10 15
Glu Gin Asn Ser Ala Thr Val Glu Pro Ser Ser His Glu He Leu His 20 25 30
Leu Leu Gin Asn Cys Phe Glu Leu Leu Arg Thr Ser Thr Ser Gin Cys 35 40 45
Thr Glu Gly He Pro Cys Ala Lys He Pro Glu Trp Val Thr His Leu 50 55 60 Thr Trp Gin Thr Leu Lys Asn Ser 65 70
(2) INFORMATION FOR SEQ ID NO: 554:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 45 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 554:
Val Leu Arg He He Cys Leu Trp Pro Cys Gly Thr Thr Leu Pro Leu 1 5 10 15
Val Glu Lys Ala His Asp Ser His Ser Ala Asp Pro Val Cys Pro Gly 20 25 30
Leu Thr Ala His Leu Pro Val Leu Leu Tyr Val Gin Leu 35 40 45
(2) INFORMATION FOR SEQ ID NO: 555:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 251 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 555:
Met Lys His Ala Asp Pro Arg He Gin Gly Tyr Pro Leu Met Gly Ser 1 5 10 15 Pro Leu Leu Met Thr Ser He Leu Leu Thr Tyr Val Tyr Phe Val Leu 20 25 30
Ser Leu Gly Pro Arg He Met Ala Asn Arg Lys Pro Phe Gin Leu Arg 35 40 45
Gly Phe Met He Val Tyr Asn Phe Ser Leu Val Ala Leu Ser Leu Tyr 50 55 60
He Val Tyr Glu Phe Leu Met Ser Gly Trp Leu Ser Thr Tyr Thr Trp 65 70 75 80
Arg Cys Asp Pro Gin Asp Cys Thr Leu Gly Gin Cys Pro Ser Val Pro 85 90 95 Ser Pro Xaa Thr Pro Val Thr Lys Ala Tyr Val Val Arg Thr Glu Gin 100 105 110
Gly Thr Gly Pro Pro Leu Pro Thr Ala Ala Leu Gin Gly Pro Arg Leu 115 120 125
Trp Phe Leu Thr His Phe Pro Arg Ala Ala Pro Gly Met Trp Pro His 130 135 140
Cys Cys Leu Pro Leu Gin Ser Trp Gly Leu Lys Gly Leu Tyr Ser Tyr 145 150 155 160
Phe Pro Leu Pro Ala Leu Lys Leu Gly Arg Gly Ala Leu Arg Ala Gly 165 170 175 Pro Thr Lys Gly Leu Val Ala Phe Phe Leu Thr Gin Lys Arg Ser Ala 180 185 190
He Met Ser Leu Trp Thr Gin Ser His Ser Ser Thr Pro His Thr Glu 195 200 205
Ala Val Ala Ser Gly Pro Lys Val Arg Val Gly Gly Gly Leu Gly He 210 215 220
Gin Pro Val Glu Ala Ala Tyr Ser Thr Cys Val Leu He Lys Ser Asp 225 230 235 240
Arg Gly Asn His Glu Lys Lys Lys Lys Lys Lys 245 250
(2) INFORMATION FOR SEQ ID NO: 556:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 19 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 556: Gly Leu Ala Gly Leu Cys Gly Gin Leu Ser Ser Pro Ala Leu Cys Val
1 5 10 15
Asn Arg Leu
(2) INFORMATION FOR SEQ ID NO: 557: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 217 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 557:
Met He Thr Glu Lys Trp Gly Leu Asn Met Glu Tyr Cys Arg Gly Gin 1 5 10 15
Ala Tyr He Xaa Ser Ser Gly Phe Ser Ser Lys Met Lys Val Val Ala 20 25 30
Ser Arg Leu Leu Glu Lys Tyr Pro Gin Ala He Tyr Thr Leu Cys Ser 35 40 45 Ser Cys Ala Leu Asn Met Trp Leu Ala Lys Ser Val Pro Val Met Gly 50 55 60
Val Ser Val Ala Leu Gly Thr He Glu Glu Val Cys Ser Phe Phe His 65 70 75 80
Arg Ser Pro Gin Leu Leu Leu Glu Leu Asp Asn Val He Ser Val Leu 85 90 95
Phe Gin Asn Ser Lys Glu Arg Gly Lys Glu Leu Lys Glu He Cys His 100 105 110
Ser Gin Trp Thr Gly Arg His Asp Ala Phe Glu He Leu Val Glu Leu 115 120 125 Leu Gin Ala Leu Val Leu Cys Leu Asp Gly He Asn Ser Asp Thr Asn
130 135 140
He Arg Trp Asn Asn Tyr He Ala Gly Arg Ala Phe Val Leu Cys Ser 145 150 155 160
Ala Val Ser Asp Phe Asp Phe He Val Thr He Val Val Leu Lys Asn 165 170 175
Val Leu Ser Phe Thr Arg Ala Phe Gly Lys Asn Leu Gin Gly Gin Thr 180 185 190
Ser Asp Val Phe Phe Ala Ala Gly Ser Leu Thr Ala Val Leu His Ser 195 200 205 Leu Asn Glu Val He Gly Lys Tyr Xaa 210 215
(2) INFORMATION FOR SEQ ID NO: 558:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 82 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 55£
Leu Leu Lys Val Leu Cys He Leu Pro Val Met Lys Val Glu Asn Glu 1 5 10 15
Arg Tyr Glu Asn Gly Arg Lys Arg Leu Lys Ala Tyr Leu Arg Asn Thr 20 25 30
Leu Thr Asp Gin Arg Ser Ser Asn Leu Ala Leu Leu Asn He Asn Phe 35 40 45
Asp He Lys His Asp Leu Asp Leu Met Val Asp Thr Tyr He Lys Leu 50 55 60 Tyr Thr Ser Lys Ser Glu Leu Pro Thr Asp Asn Ser Glu Thr Val Glu 65 70 75 80
Asn Thr
(2) INFORMATION FOR SEQ ID NO: 559: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 95 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 559:
Met Val Leu He Leu Leu Asn Leu Leu Leu Gly Gin Phe Ser Cys Met 1 5 10 15
Ser Pro Ala Ser His His Cys His Pro Leu Pro Thr Glu Met Pro Cys 20 25 30
Ser Ser Asp Trp Gly Phe Asp Ser His Thr Val Tyr Pro Ser Cys Val 35 40 45
Asp Ala Leu Leu Pro Lys Pro Ser Ala Asn Ser Phe Pro Asn Gly Ser 50 55 60
Cys His Cys Gin Gly Leu Tyr Asn Gin Gin Gin Gin Asn Leu His Ala 65 70 75 80
Ala Glu Gly Pro Ala Ser Leu Arg Cys Asn Lys Tyr Val Ser Thr 85 90 95
(2) INFORMATION FOR SEQ ID NO: 560:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 54 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 560:
Met He Pro Ala Tyr Ser Lys Asn Arg Ala Tyr Ala He Phe Phe He 1 5 10 15
Val Phe Thr Val He Gly Asp Ala Pro Gly Ala Val Leu Ser Cys Ala 20 25 30 Gly His Pro Cys Val Gly Phe Ala Ala Val Leu Val Ala Pro Leu Thr 35 40 45
Val Ala Val Ser Ser Xaa 50
(2) INFORMATION FOR SEQ ID NO: 561: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 108 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 561:
Met Glu Val Pro Pro Pro Ala Pro Arg Ser Phe Leu Cys Arg Ala Leu 1 5 10 15
Cys Leu Phe Pro Arg Val Phe Ala Ala Glu Ala Val Thr Ala Asp Ser 20 25 30
Glu Val Leu Glu Glu Arg Gin Lys Arg Leu Pro Tyr Val Pro Glu Pro 35 40 45 Tyr Tyr Pro Glu Ser Gly Trp Asp Arg Leu Arg Glu Leu Phe Gly Lys 50 55 60
Asp Thr Val Asn Thr Ser Leu Asn Val Tyr Arg Asn Lys Asp Ala Leu 65 70 75 80
Ser His Phe Val He Ala Gly Ala Val Thr Gly Ser Leu Phe Arg He 85 90 95
Asn Val Gly Leu Arg Gly Trp Trp Leu Val Ala Xaa 100 105
(2) INFORMATION FOR SEQ ID NO: 562:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 50 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 562:
Met Asn Trp Gly Leu Ser He Trp Leu His Tyr Tyr Glu Lys Lys Lys
1 5 10 15 Glu Gin Val Phe Leu Val He Leu Ala His Val Val Arg Arg Cys Ala 20 25 30
Ser Asp Gly He Leu Gin Phe Glu Ser Ser Leu Leu Lys Met Arg Arg 35 40 45
Ala Pro 50
(2) INFORMATION FOR SEQ ID NO: 563:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 253 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 563:
Met Val Lys Val Cys Asn Asp Ser Asp Arg Trp Ser Leu He Ser Leu 1 5 10 15
Ser Asn Asn Ser Gly Lys Asn Val Glu Leu Lys Phe Val Asp Ser Leu 20 25 30 Arg Arg Gin Phe Glu Phe Ser Val Asp Ser Phe Gin He Lys Leu Asp 35 40 45
Ser Leu Leu Leu Phe Tyr Glu Cys Ser Glu Asn Pro Met Thr Glu Thr 50 55 60
Phe His Pro Thr He He Gly Glu Ser Val Tyr Gly Asp Phe Gin Glu 65 70 75 80
Ala Phe Asp His Leu Cys Asn Lys He He Ala Thr Arg Asn Pro Glu 85 90 95
Glu He Arg Gly Gly Gly Leu Leu Lys Tyr Cys Asn Leu Leu Val Arg 100 105 110 Gly Phe Arg Pro Ala Ser Asp Glu He Lys Thr Leu Gin Arg Tyr Met
115 120 125
Cys Ser Arg Phe Phe He Asp Phe Ser Asp He Gly Glu Gin Gin Arg 130 135 140
Lys Leu Glu Ser Tyr Leu Gin Asn His Phe Val Gly Leu Glu Asp Arg 145 150 155 160
Lys Tyr Glu Tyr Leu Met Thr Leu His Gly Val Val Asn Glu Ser Thr 165 170 175
Val Cys Leu Met Gly His Glu Arg Arg Gin Thr Leu Asn Leu He Thr 180 185 190 Met Leu Ala He Arg Val Leu Ala Asp Gin Asn Val He Pro Asn Val 195 200 205
Ala Asn Val Thr Cys Tyr Tyr Gin Pro Ala Pro Tyr Val Ala Asp Ala 210 215 220
Asn Phe Ser Asn Tyr Tyr He Ala Gin Val Gin Pro Val Phe Thr Cys 225 230 235 240
Gin Gin Gin Thr Tyr Ser Thr Trp Leu Pro Cys Asn Xaa 245 250
(2) INFORMATION FOR SEQ ID NO: 564:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 18 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 564:
Met Ser Phe Leu Met Trp Leu Met Ser Leu Ala He Thr Ser Gin Pro 1 5 10 15 Pro Met
(2) INFORMATION FOR SEQ ID NO: 565:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 80 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 565:
Met Ala Pro Lys Gly Lys Val Gly Thr Arg Gly Lys Lys Gin He Phe 1 5 10 15
Glu Glu Asn Arg Glu Thr Leu Lys Phe Tyr Leu Arg He He Leu Gly 20 25 30
Ala Asn Ala He Tyr Cys Leu Val Thr Leu Val Phe Phe Tyr Ser Ser 35 40 45
Ala Ser Phe Trp Ala Trp Leu Ala Leu Gly Phe Ser Leu Ala Val Tyr 50 55 60
Gly Ala Ser Tyr His Ser Met Ser Ser Met Ala Arg Ala Ala Phe Phe 65 70 75 80
(2) INFORMATION FOR SEQ ID NO: 566: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 73 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 566:
His Leu Lys Asp Val He Leu Leu Thr Ala He Val Gin Val Leu Ser 1 5 10 15
Cys Phe Ser Leu Tyr Val Trp Ser Phe Trp Leu Leu Ala Pro Gly Arg 20 25 30
Ala Leu Tyr Leu Leu Trp Val Asn Val Leu Gly Pro Trp Phe Thr Ala 35 40 45 Asp Ser Gly Thr Pro Ala Pro Glu His Asn Glu Lys Arg Gin Arg Arg 50 55 60
Gin Glu Arg Arg Gin Met Lys Arg Leu 65 70
(2) INFORMATION FOR SEQ ID NO: 567: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 263 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 567:
Met Asp Cys Pro Ala Leu Pro Pro Gly Trp Lys Lys Glu Glu Val He 1 5 10 15
Arg Lys Ser Gly Leu Ser Ala Gly Lys Ser Asp Val Tyr Tyr Phe Ser 20 25 30
Pro Ser Gly Lys Lys Phe Arg Ser Lys Pro Gin Leu Ala Arg Tyr Leu 35 40 45 Gly Asn Thr Val Asp Leu Ser Ser Phe Asp Phe Arg Thr Gly Lys Met 50 55 60
Met Pro Ser Lys Leu Gin Lys Asn Lys Gin Arg Leu Arg Asn Asp Pro 65 70 75 80
Leu Asn Gin Asn Lys Gly Lys Pro Asp Leu Asn Thr Thr Leu Pro He 85 90 95
Arg Gin Thr Ala Ser He Phe Lys Gin Pro Val Thr Lys Val Thr Asn 100 105 110
His Pro Ser Asn Lys Val Lys Ser Asp Pro Gin Arg Met Asn Glu Gin 115 120 125 Pro Arg Gin Leu Phe Trp Glu Lys Arg Leu Gin Gly Leu Ser Ala Ser 130 135 140
Asp Val Thr Glu Gin He He Lys Thr Met Glu Leu Pro Lys Gly Leu 145 150 155 160
Gin Gly Val Gly Pro Gly Ser Asn Asp Glu Thr Leu Leu Ser Ala Val 165 170 175
Ala Ser Ala Leu His Thr Ser Ser Ala Pro He Thr Gly Gin Val Ser 180 185 190
Ala Ala Val Glu Lys Asn Pro Ala Val Trp Leu Asn Thr Ser Gin Pro 195 200 205 Leu Cys Lys Ala Phe He Val Thr Asp Glu Asp He Arg Lys Gin Glu 210 215 220
Glu Arg Val Gin Gin Val Arg Lys Lys Leu Glu Glu Ala Leu Met Ala 225 230 235 240
Asp He Leu Ser Arg Ala Ala Asp Thr Glu Glu Met Asp He Glu Met 245 250 255
Asp Ser Gly Asp Glu Ala Xaa 260
(2) INFORMATION FOR SEQ ID NO: 568:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 70 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 568:
Met Met Arg Pro Phe Tyr Leu Leu Leu Pro Val Leu Cys Thr Gin Ala 1 5 10 15 Leu Arg Gin Ser Gin Gly Lys Ser Pro Leu Leu Trp Lys Arg Thr Leu 20 25 30
Leu Phe Gly Leu Thr His Leu Asn Pro Ser Ala Lys Leu Leu Leu Ser 35 40 45
Gin Met Lys Thr Ser Gly Asn Arg Lys Ser Glu Tyr Ser Lys Tyr Ala 50 55 60
Arg Asn Trp Lys Lys His 65 70
(2) INFORMATION FOR SEQ ID NO: 569:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 34 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 569:
Met Pro Val Thr Ser Lys Arg Thr Leu Phe Phe Pro Asp Pro Cys Ser 1 5 10 15 Tyr Asp Thr Pro Pro Pro Asp Cys His Cys His Ser Phe Arg Ala Glu 20 25 30
Leu Leu
(2) INFORMATION FOR SEQ ID NO: 570: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 104 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 570:
Met Asn Ser Arg Gly Met Trp Leu Thr Tyr Ala Leu Gly Val Gly Leu 1 5 10 15
Leu His He Val Leu Leu Ser He Pro Phe Phe Ser Val Pro Val Ala 20 25 30
Trp Thr Leu Thr Asn He He His Asn Leu Gly Met Tyr Val Phe Leu 35 40 45 His Ala Val Lys Gly Thr Pro Phe Glu Thr Pro Asp Gin Gly Lys Ser 50 55 60
Lys Ala Pro Asn Ser Leu Gly Thr Thr Gly Leu Trp Ser Thr Val Tyr 65 70 75 80
He Phe Thr Glu Val Phe His Asn Phe Ser Asn Asn Ser He Phe Ser 85 90 95
Gly Lys Phe Leu Tyr Glu Val Xaa
(2) INFORMATION FOR SEQ ID NO: 571:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 132 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 571:
Met Trp Leu Thr Tyr Ala Leu Gly Val Gly Leu Leu His He Val Leu 1 5 10 15
Leu Ser He Pro Phe Phe Ser Val Pro Val Ala Trp Thr Leu Thr Asn 20 25 30
He He His Asn Leu Gly Met Tyr Val Phe Leu His Ala Val Lys Gly 35 40 45
Thr Pro Phe Glu Thr Pro Asp Gin Gly Lys Ala Arg Leu Leu Thr His 50 55 60
Trp Glu Gin Leu Asp Tyr Gly Val Gin Phe Thr Ser Ser Arg Lys Phe 65 70 75 80
Phe Thr He Ser Pro He He Leu Tyr Phe Leu Ala Ser Phe Tyr Thr 85 90 95 Lys Tyr Asp Pro Thr His Phe He Leu Asn Thr Ala Ser Leu Leu Ser 100 105 110
Val Leu He Pro Lys Met Pro Gin Leu His Gly Val Arg He Phe Gly 115 120 125
He Asn Lys Tyr 130
(2) INFORMATION FOR SEQ ID NO: 572:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 32 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 572:
Met Asn Lys Trp He Cys Glu Met His Cys Tyr Leu Val Leu Leu Ser 1 5 10 15
Val Cys Ser Pro Ser Ala Leu Arg Arg Val Arg His Thr Leu Ser Arg 20 25 30
(2) INFORMATION FOR SEQ ID NO: 573:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 28 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 573:
Met Pro Val Leu Ser Leu Leu Cys Thr Leu He Val Ser Phe Gin Ser 1 5 10 15
Ala Asp Ser Cys Glu Val Phe Leu Asn Cys Ser Leu 20 25
(2) INFORMATION FOR SEQ ID NO: 574:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 40 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 574:
Met Lys Val Ser Thr Met Leu Trp Phe Leu Cys Trp Glu Gin Ser His 1 5 10 15
Phe Leu Arg Glu Trp Glu Asp Leu Ser Thr Phe Leu He Leu He Gin 20 25 30 Met Glu Cys Gin Tyr Gly Asn Ser 35 40
(2) INFORMATION FOR SEQ ID NO: 575:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 30 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 575:
Met Gly Leu Pro Leu Met Ala Leu Met Trp Ser Thr Leu Pro Ala Ser 1 5 10 15
Ala Gly Val Asn Phe He Leu Ala Leu Pro Leu Leu Xaa Leu 20 25 30
(2) INFORMATION FOR SEQ ID NO: 576:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 29 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 576:
Met Lys Arg Gly Cys Leu Gly Leu Leu Phe Phe Ser Cys Cys Ser Ser 1 5 10 15
Ala Pro Thr Met Leu Leu Cys Asp Tyr Leu Asn Trp Phe 20 25
(2) INFORMATION FOR SEQ ID NO: 577:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 92 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 577:
Met Lys Leu Leu Leu Gly He Ala Leu Leu Ala Tyr Val Ala Ser Val 1 5 10 15
Trp Gly Asn Phe Val Asn Met Arg Ser He Gin Glu Asn Gly Glu Leu 20 25 30
Lys He Glu Ser Lys He Glu Glu Met Val Glu Pro Leu Arg Glu Lys 35 40 45
He Arg Asp Leu Glu Lys Ser Phe Thr Gin Lys Tyr Pro Pro Val Lys 50 55 60
Phe Leu Ser Glu Lys Asp Arg Lys Arg He Leu Xaa Asn Arg Arg Arg 65 70 75 80 Xaa Val Arg Gly Leu Pro Ser Xaa Leu Thr Asn Ser
85 90
(2) INFORMATION FOR SEQ ID NO: 578:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 42 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 578:
Met Lys Phe Ser Leu Val Leu Leu He Lys He He Ser Phe Glu Arg 1 5 10 15
Leu Leu He Phe Leu Phe Pro Leu Ser Phe Leu Pro Asn He Trp Arg 20 25 30
Arg Val Met Val Asn Leu Asn He Leu Phe 35 40
(2) INFORMATION FOR SEQ ID NO: 579:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 70 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 579:
Leu Ala Gin Glu Cys Pro Pro His He Pro Ser Ser Phe Phe Leu Val 1 5 10 15 Lys Leu Leu Phe He Pro Trp Leu Ala Ser Leu Leu Pro Pro Leu Ser 20 25 30
Thr Phe Thr Ser Asp Phe Tyr Phe Met Glu Phe Gly He Glu Val Lys 35 40 45
Leu Gin Gin Cys Arg Gin His Gin Val Leu Gin Glu Lys Asn Thr Lys 50 55 60
Lys Phe Asn Lys Lys Lys 65 70
(2) INFORMATION FOR SEQ ID NO: 580:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 110 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 580:
Met Leu Arg Leu Leu Leu Leu Val Ala Phe Ala Leu Val Val Val Leu 1 5 10 15 Phe His Val Leu Leu Ala Pro He Thr Ala Leu Phe His Thr His Phe 20 25 30
Tyr Asp Arg Leu Gin Asp Ala Gly Ser Arg Trp Pro Glu Leu Tyr Leu 35 40 45
Tyr Ser Arg Ala Asp Glu Val Val Leu Ala Arg Asp He Glu Arg Met 50 55 60
Val Glu Ala Arg Leu Ala Arg Arg Val Leu Ala Arg Ser Val Asp Phe 65 70 75 80
Val Ser Ser Ala His Val Ser His Leu Arg Asp Tyr Pro Thr Tyr Tyr 85 90 95 Thr Ser Leu Cys Val Asp Phe Met Arg Asn Cys Val Arg Cys 100 105 110
(2) INFORMATION FOR SEQ ID NO: 581:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 30 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 581:
Met Phe Lys Leu Glu Glu Cys Gly Lys Thr Thr Phe Leu Leu Ser Met 1 5 10 15
Ala Leu Tyr Phe Trp Trp He Val Gin Thr Thr Lys Gly Cys 20 25 30
(2) INFORMATION FOR SEQ ID NO: 582:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 71 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 582:
Met Glu Ser Asp Ala Leu Leu Leu Thr He Phe Trp He He Ala Arg 1 5 10 15
Ser Ser Val Arg Ser Val Gly Lys Ser Ser Gin Arg Ser Phe Thr Thr 20 25 30 He Thr Gin Leu Arg Ser Thr His Thr Gly Pro Ser Arg Arg Ser Tyr 35 40 45
Leu He Trp Trp Asn Gly Gly Pro Lys Arg Thr He Ser Tyr Val Ser 50 55 60
Arg Arg Phe Arg Ser Phe Arg 65 70
(2) INFORMATION FOR SEQ ID NO: 583:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 47 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 583:
Val Gly Leu Phe Gin Pro Lys Thr Phe Gin Val Pro Val Thr Asp Leu 1 5 10 15
Tyr He Phe He Lys He Tyr Ser Glu He Gly Pro He Met His Val 20 ' 25 30 Leu Cys Pro Gly Tyr Ser Gin Ser Pro Ser Thr Pro Pro Trp Thr 35 40 45
(2) INFORMATION FOR SEQ ID NO: 584:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 39 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 584:
Met Trp Phe Gly Ser Asp Arg Ser Asp Leu Arg He Gly Thr Ala Phe 1 5 10 15
Leu Phe Asp Leu Val Cys Asp Leu Cys He His Ala Trp Lys Pro Pro 20 25 30
Gly Leu Val Arg Phe Ser Phe 35
(2) INFORMATION FOR SEQ ID NO: 585:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 19 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 585:
Met Leu Asn Thr Ala Ser Leu Asn Leu Pro Trp Lys Val Gin Leu Phe 1 5 10 15
Ala His Ala
(2) INFORMATION FOR SEQ ID NO: 586:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 23 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 586:
Met Ser Ala Cys Leu Leu Leu Phe Leu Ala Phe Ser Trp Lys Arg Lys 1 5 10 15
Gly Leu Trp Ser Gly Pro Gly 20
(2) INFORMATION FOR SEQ ID NO: 587:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 69 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 587:
Met Leu Pro Pro Phe Ser Leu Val Tyr Thr His Phe Leu Val Ala Ser 1 5 10 15
Leu Leu Pro Val He Leu Ala Val Phe Pro Asp Ser Ala Gin He Val 20 25 30 Pro Leu Leu Lys Pro He Pro Arg Pro Gin Pro Glu Val He Phe Pro 35 40 45
Ser Ser Glu Leu Leu Glu Gin Leu Leu Ser Val Gin Phe Val Trp Gin 50 55 60
Ala His Thr Val Ala 65
(2) INFORMATION FOR SEQ ID NO: 588:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 77 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 588:
Met Gly Pro Pro Met Leu Gin Glu He Ser Asn Leu Phe Leu He Leu 1 5 10 15
Leu Met Met Gly Ala He Phe Thr Leu Ala Ala Leu Lys Glu Ser Leu 20 25 30 Ser Thr Cys He Pro Ala He Val Cys Leu Gly Phe Leu Leu Leu Leu 35 40 45
Asn Val Gly Gin Leu Leu Ala Gin Thr Lys Lys Val Val Arg Pro Thr 50 55 60
Arg Lys Lys Thr Leu Ser Thr Phe Lys Glu Ser Trp Lys 65 70 75
(2) INFORMATION FOR SEQ ID NO: 589:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 155 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 589:
Met Ala Leu Leu Leu Ser Val Leu Arg Val Leu Leu Gly Gly Phe Phe 1 5 10 15
Ala Leu Val Gly Leu Ala Lys Leu Ser Glu Glu He Ser Ala Pro Val 20 25 30 Ser Glu Arg Met Asn Ala Leu Phe Val Gin Phe Ala Glu Val Phe Pro 35 40 45
Leu Lys Val Phe Gly Tyr Gin Pro Asp Pro Leu Asn Tyr Gin He Ala 50 55 60
Val Gly Phe Leu Glu Leu Leu Ala Gly Leu Leu Leu Val Met Gly Pro 65 70 75 80
Pro Met Leu Gin Glu He Ser Asn Leu Phe Leu He Leu Leu Met Met 85 90 95
Gly Ala He Phe Thr Leu Ala Ala Leu Lys Glu Ser Leu Ser Thr Cys 100 105 110 He Pro Ala He Val Cys Leu Gly Phe Leu Leu Leu Leu Asn Val Gly 115 120 125
Gin Leu Leu Ala Gin Thr Lys Lys Val Val Arg Pro Thr Arg Lys Lys 130 135 140
Thr Leu Ser Thr Phe Lys Glu Ser Trp Lys Xaa 145 150 155
(2) INFORMATION FOR SEQ ID NO: 590:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 24 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 590:
Met Pro Glu Thr Arg Leu Gly His Arg Gin Gin Phe Ala Val Phe His 1 5 10 15
Leu Xaa Pro Val Pro Pro Cys Gly 20
(2) INFORMATION FOR SEQ ID NO: 591:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 38 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 591: Met Leu Thr Phe Leu Phe Ser Ala Cys Ala Thr Cys Leu Gly Lys Leu 1 5 10 15
Ala Ser Pro Leu Ala Pro Val Gly Pro Gin Gin Arg Gly Xaa Pro Pro 20 25 30
Gly Pro Pro Leu Leu Ser 35
(2) INFORMATION FOR SEQ ID NO: 592:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 69 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 592:
Met Asp Pro Phe His Tyr Asp Tyr Gin Thr Leu Arg He Gly Gly Leu 1 5 10 15
Val Phe Ala Val Val Leu Phe Ser Val Gly He Leu Leu He Leu Ser 20 25 30 Arg Arg Cys Lys Cys Ser Phe Asn Gin Lys Pro Arg Ala Pro Gly Asp 35 40 45
Glu Glu Ala Gin Val Glu Asn Leu He Thr Ala Asn Ala Thr Glu Pro 50 55 60
Gin Lys Ala Glu Asn 65
(2) INFORMATION FOR SEQ ID NO: 593:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 308 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 593:
Asn Leu Arg Val Arg Leu Gly Asp Val He Ser He Gin Pro Cys Pro 1 5 10 15
Asp Val Lys Tyr Gly Lys Arg He His Val Leu Pro He Asp Asp Thr 20 25 30 Val Glu Gly He Thr Gly Asn Leu Phe Glu Val Tyr Leu Lys Pro Tyr 35 40 45
Phe Leu Glu Ala Tyr Arg Pro He Arg Lys Gly Asp He Phe Leu Val 50 55 60
Arg Gly Gly Met Arg Ala Val Glu Phe Lys Val Val Glu Thr Asp Pro 65 70 75 80
Ser Pro Tyr Cys He Val Ala Pro Asp Thr Val He His Cys Glu Gly 85 90 95
Glu Pro He Lys Arg Glu Asp Glu Glu Glu Ser Leu Asn Glu Val Gly 100 105 110 Tyr Asp Asp He Gly Gly Cys Arg Lys Gin Leu Ala Gin He Lys Glu 115 120 125
Met Val Glu Leu Pro Leu Arg His Pro Ala Leu Phe Lys Ala He Gly 130 135 140
Val Lys Pro Pro Arg Gly He Leu Leu Tyr Gly Pro Pro Gly Thr Gly 145 150 155 160
Lys Thr Leu He Ala Arg Ala Val Ala Asn Glu Thr Gly Ala Phe Phe 165 170 175
Phe Leu He Asn Gly Pro Glu He Met Ser Lys Leu Ala Gly Glu Ser 180 185 190 Glu Ser Asn Leu Arg Lys Ala Phe Glu Glu Ala Glu Lys Asn Ala Pro 195 200 205
Ala He He Phe He Asp Glu Leu Asp Ala He Ala Pro Lys Arg Glu 210 215 220
Lys Thr His Gly Glu Val Glu Arg Arg He Val Ser Gin Leu Leu Thr 225 230 235 240
Leu Met Asp Gly Leu Lys Gin Arg Ala His Val He Val Met Ala Ala 245 250 255
Thr Asn Arg Pro Asn Ser He Asp Pro Ala Leu Arg Arg Phe Gly Arg 260 265 270 Phe Asp Arg Glu Val Asp He Gly He Pro Asp Ala Thr Gly Arg Leu
275 280 285
Glu He Leu Gin He His Thr Lys Asn Met Lys Leu Ala Asp Asp Val 290 295 300
Asp Leu Glu Gin 305
(2) INFORMATION FOR SEQ ID NO: 594:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 22 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 594:
Met Gin He Lys Leu Leu Lys Ser Val Lys Thr Val Phe Ala He Thr 1 5 10 15
Leu Leu Val Leu Phe Leu 20
(2) INFORMATION FOR SEQ ID NO: 595:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 24 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 595: Met Phe Pro Lys Phe Cys Pro He Leu Ser Leu Val Asp Phe He Ser 1 5 10 15
His Arg Asp Lys Pro Glu Thr Glu 20
(2) INFORMATION FOR SEQ ID NO: 596: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 24 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 596:
Met Leu He Glu Cys Ala Trp Gin Leu Met Phe Leu Leu Leu Lys Val 1 5 10 15
Glu Gin Leu Gly He Leu Asp Lys 20
(2) INFORMATION FOR SEQ ID NO: 597:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 1 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 597:
Met 1
(2) INFORMATION FOR SEQ ID NO: 598:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 8 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 598: Met Cys He Met Ser Ala Leu Val 1 5
(2) INFORMATION FOR SEQ ID NO: 599:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 25 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 599:
Met Phe Leu Val Trp Phe Phe Trp Gly Leu He Ser Ala Leu Ser Asn 1 5 10 15
Val His Thr Pro Ser Arg Leu Pro Ala 20 25
(2) INFORMATION FOR SEQ ID NO: 600:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 600:
Met Xaa Gly Leu Ser Leu He Leu Thr Val Thr Leu Leu Ala Val Ser 1 5 10 15
Asp Ser Ala Ala Thr Cys He Val Ala Lys Gly 20 25
(2) INFORMATION FOR SEQ ID NO: 601:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 61 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 601:
Met Trp Thr Arg Ser Ser Arg Cys Leu Leu Leu Cys He Pro Gly Xaa 1 5 10 15
Ser Arg Arg Arg Arg Ala Gly Ser Gly Met Lys Pro Arg Ser Trp Ser 20 25 30
Ala Trp Arg Pro Ser Gly Gly Thr Gly Thr Ser Ser Ser Gin Ser Ser 35 40 45
Thr Gin Ser Arg Thr Leu Ser Ala Thr Ala Ser Pro Ala 50 55 60
(2) INFORMATION FOR SEQ ID NO: 602:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 29 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 602:
Met Arg Glu Thr Ser He Arg Val Leu Leu Met Leu Pro Ala Leu Glu 1 5 10 15 Ser Thr Ser Gly Leu Ser Ala Phe Met Gly Leu Gly Thr 20 25
(2) INFORMATION FOR SEQ ID NO: 603:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 69 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 603:
Met Pro Pro Lys Gin Glu Leu Gly Ser Gly Val Gly Glu Leu Ala Lys 1 5 10 15
Asn Ser Lys Arg Gin His Trp Asn His Arg Trp Lys Lys Tyr Leu Lys 20 25 30
Leu He Arg Trp Glu Asp Gly Leu Leu Leu Glu Gly Leu Leu Leu Val 35 40 45
Leu Glu His Cys Ala Thr Met Ala Trp Asp Cys Leu Met Arg Leu Glu 50 55 60 Leu Leu Lys Arg Leu 65
(2) INFORMATION FOR SEQ ID NO: 604:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 24 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 604:
Lys He Val Tyr He Leu Gly Asn Pro Leu Lys Phe Asn Ser Arg Val 1 5 10 15
He His His Leu Val Leu Leu Gin 20
(2) INFORMATION FOR SEQ ID NO: 605:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 35 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 605:
Met Asn Leu His Gin Arg Arg Leu Leu Leu He Gly His Leu Met Thr 1 5 10 15
Leu Val Lys Ala Ser Lys Ser Phe Ser Phe Thr Glu He Thr Ser Ser 20 25 30 Arg Lys Lys 35
(2) INFORMATION FOR SEQ ID NO: 606:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 130 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 606:
Leu Leu Gly Tyr Gly Leu Phe Gly His Cys He Val Leu Phe He Thr 1 5 10 15
Tyr Asn He His Leu His Ala Leu Phe Tyr Leu Phe Trp Leu Leu Val 20 25 30
Gly Gly Leu Ser Thr Leu Arg Met Val Ala Val Leu Val Ser Arg Thr 35 40 45
Val Gly Pro Thr Gin Arg Leu Leu Leu Cys Gly Thr Leu Ala Ala Leu 50 55 60 His Met Leu Phe Leu Leu Tyr Leu His Phe Ala Tyr His Lys Val Xaa 65 70 75 80
Glu Gly He Leu Asp Thr Leu Glu Gly Pro Asn He Pro Pro He Gin 85 90 95
Arg Val Pro Arg Asp He Pro Ala Met Leu Pro Ala Ala Arg Leu Pro 100 105 110
Thr Thr Val Leu Asn Ala Thr Ala Lys Ala Val Ala Val Thr Leu Gin 115 120 125
Ser His 130
(2) INFORMATION FOR SEQ ID NO: 607:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 23 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 607: Met Leu Val He Phe Leu Phe Thr Ser Leu Leu Lys He Pro Ser Ser
1 5 10 15
Val Pro Gly Leu He Asn Val 20
(2) INFORMATION FOR SEQ ID NO: 608: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 6 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 608:
Glu Leu Asp Tyr He Leu 1 5
(2) INFORMATION FOR SEQ ID NO: 609:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 232 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 609:
Met Ala Pro Pro Gly Trp Gin Xaa Xaa Xaa Xaa Xaa Trp Leu Ala Cys 1 5 10 15
Pro Asp Arg Gly Glu Leu Ser Ser Arg Ser Pro Pro Cys Arg Leu Ala 20 25 30 Arg Trp Ala Glu Gly Asp Arg Glu Thr Arg Thr Cys Leu Leu Glu Leu 35 40 45
Ser Ala Gin Ser Trp Gly Gly Arg Phe Arg Arg Ser Ser Ala Val Ser 50 55 60
Ala Gly Ser Pro Ser Arg Leu His Phe Leu Pro Gin Pro Leu Leu Leu 65 70 75 80
Arg Ser Ser Gly He Pro Ala Ala Ala Thr Pro Trp Pro Gin Pro Ala 85 90 95
Gly Leu Pro Val Arg Pro Thr Pro Thr Arg Thr Gly Glu Glu Asp Arg 100 105 110 Thr Leu Asp He Ser He Cys Thr Glu Val Leu Ala Gly Thr Glu Gin 115 120 125
Pro Pro Pro Pro Arg Met Thr Ser Pro Ser Ser Ser Pro Val Phe Arg 130 135 140
Leu Glu Thr Leu Asp Gly Gly Gin Glu Asp Gly Ser Glu Ala Asp Arg 145 150 155 160
Gly Lys Leu Asp Phe Gly Ser Gly Leu Pro Pro Met Glu Ser Gin Phe 165 170 175
Gin Gly Glu Asp Arg Lys Phe Ala Pro Ser Asp Lys Ser Gin Pro Pro 180 185 190 Thr Thr Glu Arg Glu Gin Val Pro Val Ser Arg He Gin Thr Asp Leu 195 200 205
Thr Glu He Gly Ser Ser Met Arg Ser Pro Gly Val Ser Pro Arg He 210 215 220
Trp Leu Asp Phe Gin Ser Thr Xaa 225 230
(2) INFORMATION FOR SEQ ID NO: 610:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 34 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 610:
Met Val Leu Leu Leu Leu Leu Ala Tyr Val Leu Leu Thr Tyr He Leu 1 5 10 15
Leu Leu Asn Met Leu He Ala Leu Met Xaa Arg Asp Arg Gin Gin Cys 20 25 30 Arg His
(2) INFORMATION FOR SEQ ID NO: 611:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 611:
Met Val Phe Glu Gly Phe Ser Ser Ala Phe Cys Leu Ser Ser Thr Ala 1 5 10 15
Pro Thr Ser His Pro 20
(2) INFORMATION FOR SEQ ID NO: 612:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 9 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 612:
Gly Lys Lys Asn Gin Leu Leu Val He l 5
(2) INFORMATION FOR SEQ ID NO: 613:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 29 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 613:
Met Val Trp Val Leu Trp Ser Ala Pro Ser Leu Ala Pro Pro Trp Val 1 5 10 15 Gly Pro Cys Trp Pro Ser Thr Gly Asn Cys Cys Leu Cys 20 25
(2) INFORMATION FOR SEQ ID NO: 614:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 30 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 614:
Met Ala Lys Arg Ser Pro Gly Gly Cys Gly Ser Gly Leu He Leu Leu 1 5 10 15
Cys Cys Gin Pro Cys Arg Pro Thr Ser Ser Ala Pro Met Arg 20 25 30
(2) INFORMATION FOR SEQ ID NO: 615:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 113 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 615:
He Thr He Ala He Gin Met He Cys Leu Val Asn Xaa Glu Leu Tyr 1 5 10 15
Pro Thr Phe Val Arg Asn Xaa Gly Val Met Val Cys Ser Ser Leu Cys 20 25 30 Asp He Gly Gly He He Thr Pro Phe He Val Phe Arg Leu Arg Glu 35 40 45
Val Trp Gin Ala Leu Pro Leu He Leu Phe Ala Val Leu Gly Leu Leu 50 55 60
Ala Ala Gly Val Thr Leu Leu Leu Pro Glu Thr Lys Gly Val Ala Leu 65 70 75 80
Pro Glu Thr Met Lys Asp Ala Glu Asn Leu Gly Arg Lys Ala Lys Pro 85 90 95
Lys Glu Asn Thr He Tyr Leu Lys Val Gin Thr Ser Glu Pro Ser Gly 100 105 110 Thr
(2) INFORMATION FOR SEQ ID NO: 616:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 18 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 616:
Thr Met Lys Asp Ala Glu Asn Leu Gly Arg Lys Ala Lys Pro Lys Glu 1 5 10 15
Asn Thr
(2) INFORMATION FOR SEQ ID NO: 617:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 21 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 617:
Pro Arg Val Arg Asn Ser Pro Glu Asp Leu Gly Leu Ser Leu Thr Gly 1 5 10 15
Asp Ser Cys Lys Leu 20
(2) INFORMATION FOR SEQ ID NO: 618:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 52 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 618: Gin Ala Asp Asp Leu Gin Ala Thr Val Ala Ala Leu Cys Val Leu Arg 1 5 10 15
Gly Gly Gly Pro Trp Ala Gly Ser Trp Leu Ser Pro Lys Thr Pro Gly 20 25 30
Ala Met Gly Gly Asp Leu Val Leu Gly Leu Gly Ala Leu Arg Arg Arg 35 40 45
Lys Arg Leu Leu 50
(2) INFORMATION FOR SEQ ID NO: 619:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 232 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 619:
Glu Gin Glu Lys Ser Leu Ala Gly Trp Ala Leu Val Leu Ala Xaa Xaa 1 5 10 15 Gly He Gly Leu Met Val Leu His Ala Glu Met Leu Trp Phe Gly Gly 20 25 30
Cys Ser Ala Val Asn Ala Thr Gly His Leu Ser Asp Thr Leu Trp Leu 35 40 45
He Pro He Thr Phe Leu Thr He Gly Tyr Gly Asp Val Val Pro Gly 50 55 60
Thr Met Trp Gly Lys He Val Cys Leu Cys Thr Gly Val Met Gly Val 65 70 75 80
Cys Cys Thr Ala Leu Leu Val Ala Val Val Ala Arg Lys Leu Glu Phe 85 90 95 Asn Lys Ala Glu Lys His Val His Asn Phe Met Met Asp He Gin Tyr 100 105 110
Thr Lys Glu Met Lys Glu Ser Ala Ala Arg Val Leu Gin Glu Ala Trp 115 120 125
Met Phe Tyr Lys His Thr Arg Arg Lys Glu Ser His Ala Ala Arg Xaa 130 135 140
His Gin Arg Xaa Leu Leu Ala Ala He Asn Ala Phe Arg Gin Val Arg 145 150 155 160
Leu Lys His Arg Lys Leu Arg Glu Gin Val Asn Ser Met Val Asp He 165 170 175 Ser Lys Met His Met He Leu Tyr Asp Leu Gin Gin Asn Leu Ser Ser 180 185 190
Ser His Arg Ala Leu Glu Lys Gin He Asp Thr Leu Ala Gly Lys Leu 195 200 205
Asp Ala Leu Thr Glu Leu Leu Ser Thr Ala Leu Gly Pro Arg Gin Leu 210 215 220
Pro Glu Pro Ser Gin Gin Ser Lys 225 230
(2) INFORMATION FOR SEQ ID NO: 620:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 36 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 620:
Tyr Gin Ala His His Val Ser Arg Asn Lys Arg Gly Gin Val Val Gly 1 5 10 15 Thr Arg Gly Gly Phe Arg Gly Cys Thr Val Trp Leu Thr Gly Leu Ser 20 25 30
Gly Ala Gly Lys 35
(2) INFORMATION FOR SEQ ID NO: 621: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 57 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 621:
Leu Gin Cys Glu He Cys Gly Phe Thr Cys Arg Gin Lys Ala Ser Leu 1 5 10 15
Asn Trp His Met Lys Lys His Asp Ala Asp Ser Phe Tyr Gin Phe Ser 20 25 30
Cys Asn He Cys Gly Lys Lys Phe Glu Lys Lys Asp Ser Val Val Ala
35 40 45 His Lys Ala Lys Ser His Pro Glu Val
50 55
(2) INFORMATION FOR SEQ ID NO: 622:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 19 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 622:
He Thr Ser Thr Asp He Leu Gly Thr Asn Pro Glu Ser Leu Thr Gin 1 5 10 15
Pro Ser Asp
(2) INFORMATION FOR SEQ ID NO: 623:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 19 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 623:
Asn Ser Thr Ser Gly Glu Cys Leu Leu Leu Glu Ala Glu Gly Met Ser 1 5 10 15
Lys Ser Tyr
(2) INFORMATION FOR SEQ ID NO: 624:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 51 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 624: Cys Ser Gly Thr Glu Arg Val Ser Leu Met Ala Asp Gly Lys He Phe 1 5 10 15
Val Gly Ser Gly Ser Ser Gly Gly Thr Glu Gly Leu Val Met Asn Ser 20 25 30
Asp He Leu Gly Ala Thr Thr Glu Val Leu He Glu Asp Ser Asp Ser 35 40 45
Ala Gly Pro 50
(2) INFORMATION FOR SEQ ID NO: 625:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 60 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 625:
He Gin Tyr Val Arg Cys Glu Met Glu Gly Cys Gly Thr Val Leu Ala 1 5 10 15 His Pro Arg Tyr Leu Gin His His He Lys Tyr Gin His Leu Leu Lys 20 25 30
Lys Lys Tyr Val Cys Pro His Pro Ser Cys Gly Arg Leu Phe Arg Leu 35 40 45
Gin Lys Gin Leu Leu Arg His Ala Lys His His Thr 50 55 60
(2) INFORMATION FOR SEQ ID NO: 626:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 31 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 626:
Asp Gin Arg Asp Tyr He Cys Glu Tyr Cys Ala Arg Ala Phe Lys Ser 1 5 10 15
Ser His Asn Leu Ala Val His Arg Met He His Thr Gly Glu Lys 20 25 30
(2) INFORMATION FOR SEQ ID NO: 627:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 25 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 627: Arg Ser Ser Arg Ser Lys Thr Gly Ser Leu Gin Leu He Cys Lys Ser 1 5 10 15
Glu Pro Asn Thr Asp Gin Leu Asp Tyr 20 25
(2) INFORMATION FOR SEQ ID NO: 628: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 183 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 628:
Leu Gin Cys Glu He Cys Gly Phe Thr Cys Arg Gin Lys Ala Ser Leu 1 5 10 15
Asn Trp His Met Lys Lys His Asp Ala Asp Ser Phe Tyr Gin Phe Ser 20 25 30
Cys Asn He Cys Gly Lys Lys Phe Glu Lys Lys Asp Ser Val Val Ala 35 40 45
His Lys Ala Lys Ser His Pro Glu Val Xaa He Thr Ser Thr Asp He 50 55 60
Leu Gly Thr Asn Pro Glu Ser Leu Thr Gin Pro Ser Asp Xaa Asn Ser 65 70 75 80
Thr Ser Gly Glu Cys Leu Leu Leu Glu Ala Glu Gly Met Ser Lys Ser 85 90 95
Tyr Xaa Cys Ser Gly Thr Glu Arg Val Ser Leu Met Ala Asp Gly Lys 100 105 110
He Phe Val Gly Ser Gly Ser Ser Gly Gly Thr Glu Gly Leu Val Met 115 120 125 Asn Ser Asp He Leu Gly Ala Thr Thr Glu Val Leu He Glu Asp Ser 130 135 140
Asp Ser Ala Gly Pro Xaa Gin Arg Asp Tyr He Cys Glu Tyr Cys Ala 145 150 155 160
Arg Ala Phe Lys Ser Ser His Asn Leu Ala Val His Arg Met He His 165 170 175
Thr Gly Glu Lys His Tyr Xaa 180
(2) INFORMATION FOR SEQ ID NO: 629:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 60 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 629:
Gin Tyr Val Arg Cys Glu Met Glu Gly Cys Gly Thr Val Leu Ala His 1 5 10 15 Pro Arg Tyr Leu Gin His His He Lys Tyr Gin His Leu Leu Lys Lys 20 25 30
Lys Tyr Val Cys Pro His Pro Ser Cys Gly Arg Leu Phe Arg Leu Gin 35 40 45
Lys Gin Leu Leu Arg His Ala Lys His His Thr Asp 50 55 60
(2) INFORMATION FOR SEQ ID NO: 630:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 27 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 630:
Pro Phe Lys Asp Asp Pro Arg Asp Glu Thr Tyr Lys Pro His Leu Glu 1 5 10 15
Arg Glu Thr Pro Lys Pro Arg Arg Lys Ser Gly 20 25
(2) INFORMATION FOR SEQ ID NO: 631:
(i) SEQUENCE CHARACTERISTICS: (A) LENGTH: 110 amino acids
(B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 631: Glu Met Phe Asp Ser Leu Ser Tyr Phe Lys Gly Ser Ser Leu Leu Leu 1 5 10 15
Met Leu Lys Thr Tyr Leu Ser Glu Asp Val Phe Gin His Ala Val Val 20 25 30
Leu Tyr Leu His Asn His Ser Tyr Ala Ser He Gin Ser Asp Asp Leu 35 40 45
Trp Asp Ser Phe Asn Glu Val Thr Asn Gin Thr Leu Asp Val Lys Arg 50 55 60
Met Met Lys Thr Trp Thr Leu Gin Lys Gly Phe Pro Leu Val Thr Val 65 70 75 80 Gin Lys Lys Gly Lys Glu Leu Phe He Gin Gin Glu Arg Phe Phe Leu
85 90 95
Asn Met Lys Pro Glu He Gin Pro Ser Asp Thr Arg Tyr Met 100 105 110
(2) INFORMATION FOR SEQ ID NO: 632: (i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 24 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 632:
Leu Glu Lys Val Ala Ser Val Gly Asn Ser Arg Pro Thr Gly Gin Gin 1 5 10 15
Leu Glu Ser Leu Gly Leu Leu Ala 20
(2) INFORMATION FOR SEQ ID NO: 633:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 18 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 633:
Val His Arg Glu Glu Ala Ser Cys Tyr Cys Gin Ala Glu Pro Ser Gly 1 5 10 15
Asp Leu
(2) INFORMATION FOR SEQ ID NO: 634:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 22 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 634:
Arg Pro Ala Leu Arg Gin Ala Gly Gly Gly Thr Arg Glu Pro Arg Gin 1 5 10 15
Lys Arg Trp Ala Gly Leu 20
(2) INFORMATION FOR SEQ ID NO: 635:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 12 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 635:
Ala Val Asn Phe Arg Pro Gin Arg Ser Gin Ser Met 1 5 10
(2) INFORMATION FOR SEQ ID NO: 636:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 37 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 636:
Met He Thr Asp Val Gin Leu Ala He Phe Ala Asn Met Leu Gly Val 1 5 10 15 Ser Leu Phe Leu Leu Val Val Leu Tyr His Tyr Val Ala Val Asn Asn 20 25 30
Pro Lys Lys Gin Glu 35
(2) INFORMATION FOR SEQ ID NO: 637:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 342 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 637:
Glu Glu Met Ala Asp Ser Val Lys Thr Phe Leu Gin Asp Leu Ala Arg 1 5 10 15
Gly He Lys Asp Ser He Trp Gly He Cys Thr He Ser Lys Leu Asp 20 25 30
Ala Arg He Gin Gin Lys Arg Glu Glu Gin Arg Arg Arg Arg Ala Ser 35 40 45 Ser Val Leu Ala Gin Arg Arg Ala Gin Ser He Glu Arg Lys Gin Glu 50 55 60
Ser Glu Pro Arg He Val Ser Arg He Phe Gin Cys Cys Ala Trp Asn 65 70 75 80
Gly Gly Val Phe Trp Phe Ser Leu Leu Leu Phe Tyr Arg Val Phe He 85 90 95
Pro Val Leu Gin Ser Val Thr Ala Arg He He Gly Asp Pro Ser Leu 100 105 110
His Gly Asp Val Trp Ser Trp Leu Glu Phe Phe Leu Thr Ser He Phe
115 120 125 Ser Ala Leu Trp Val Leu Pro Leu Phe Val Leu Ser Lys Val Val Asn 130 135 140
Ala He Trp Phe Gin Asp He Ala Asp Leu Ala Phe Glu Val Ser Gly 145 150 155 160
Arg Lys Pro His Pro Phe Pro Ser Val Ser Lys He He Ala Asp Met 165 170 175
Leu Phe Asn Leu Leu Leu Gin Ala Leu Phe Leu He Gin Gly Met Phe 180 185 190
Val Ser Leu Phe Pro He His Leu Val Gly Gin Leu Val Ser Leu Leu 195 200 205 His Met Ser Leu Leu Tyr Ser Leu Tyr Cys Phe Glu Tyr Arg Trp Phe 210 215 220
Asn Lys Gly He Glu Met His Gin Arg Leu Ser Asn He Glu Arg Asn 225 230 235 240
Trp Pro Tyr Tyr Phe Gly Phe Gly Leu Pro Leu Ala Phe Leu Thr Ala 245 250 255
Met Gin Ser Ser Tyr He He Ser Gly Cys Leu Phe Ser He Leu Phe 260 265 270
Pro Leu Phe He He Ser Ala Asn Glu Ala Lys Thr Pro Gly Lys Ala 275 280 285 Tyr Leu Phe Gin Leu Arg Leu Phe Ser Leu Val Val Phe Leu Ser Asn 290 295 300
Arg Leu Phe His Lys Thr Val Tyr Leu Gin Ser Ala Leu Ser Ser Ser 305 310 315 320
Thr Ser Ala Glu Lys Phe Pro Ser Pro His Pro Ser Pro Ala Lys Leu 325 330 335
Lys Ala Thr Ala Gly His 340
(2) INFORMATION FOR SEQ ID NO: 638:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 529 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 638:
Met Ala Lys Phe Met Thr Pro Val He Gin Asp Asn Pro Ser Gly Trp 1 5 10 15 Gly Pro Cys Ala Val Pro Glu Gin Phe Arg Asp Met Pro Tyr Gin Pro 20 25 30
Phe Ser Lys Gly Asp Arg Leu Gly Lys Val Ala Asp Trp Thr Gly Ala 35 40 45
Thr Tyr Gin Asp Lys Arg Tyr Thr Asn Lys Tyr Ser Ser Gin Phe Gly 50 55 60
Gly Gly Ser Gin Tyr Ala Tyr Phe His Glu Glu Asp Glu Ser Ser Phe 65 70 75 80
Gin Leu Val Asp Thr Ala Arg Thr Gin Lys Thr Ala Tyr Gin Arg Asn 85 90 95 Arg Met Arg Phe Ala Gin Arg Asn Leu Arg Arg Asp Lys Asp Arg Arg 100 105 110
Asn Met Leu Gin Phe Asn Leu Gin He Leu Pro Lys Ser Ala Lys Gin 115 120 125
Lys Glu Arg Glu Arg He Arg Leu Gin Lys Lys Phe Gin Lys Gin Phe 130 135 140
Gly Val Arg Gin Lys Trp Asp Gin Lys Ser Gin Lys Pro Arg Asp Ser 145 150 155 160
Ser Val Glu Val Arg Ser Asp Trp Glu Val Lys Glu Glu Met Asp Phe 165 170 175 Pro Gin Leu Met Lys Met Arg Tyr Leu Glu Val Ser Glu Pro Gin Asp
180 185 190
He Glu Cys Cys Gly Ala Leu Glu Tyr Tyr Asp Lys Ala Phe Asp Arg 195 200 205
He Thr Thr Arg Ser Glu Lys Pro Leu Arg Xaa Xaa Lys Arg He Phe 210 215 220
His Thr Val Thr Thr Thr Asp Asp Pro Val He Arg Lys Leu Ala Lys 225 230 235 240
Thr Gin Gly Asn Val Phe Ala Thr Asp Ala He Leu Ala Thr Leu Met 245 250 255 Ser Cys Thr Arg Ser Val Tyr Ser Trp Asp He Val Val Gin Arg Val 260 265 270
Gly Ser Lys Leu Phe Phe Asp Lys Arg Asp Asn Ser Asp Phe Asp Leu 275 280 285
Leu Thr Val Ser Glu Thr Ala Asn Glu Pro Pro Gin Asp Glu Gly Asn 290 295 300
Ser Phe Asn Ser Pro Arg Asn Leu Ala Met Glu Ala Thr Tyr He Asn 305 310 315 320
His Asn.Phe Ser Gin Gin Cys Leu Arg Met Gly Lys Glu Arg Tyr Asn 325 330 335 Phe Pro Asn Pro Asn Pro Phe Val Glu Asp Asp Met Asp Lys Asn Glu 340 345 350
He Ala Ser Val Ala Tyr Arg Tyr Arg Ser Gly Lys Leu Gly Asp Asp 355 360 365
He Asp Leu He Val Arg Cys Glu His Asp Gly Val Met Thr Gly Ala 370 375 380
Asn Gly Glu Val Ser Phe He Asn He Lys Thr Leu Asn Glu Trp Asp 385 390 395 400
Ser Arg His Cys Asn Gly Val Asp Trp Arg Gin Lys Leu Asp Ser Gin 405 410 415 Arg Gly Ala Val He Ala Thr Glu Leu Lys Asn Asn Ser Tyr Lys Leu 420 425 430
Ala Arg Trp Thr Cys Cys Ala Leu Leu Ala Gly Ser Glu Tyr Leu Lys 435 440 445
Leu Gly Tyr Val Ser Arg Tyr His Val Lys Asp Ser Ser Arg His Val 450 455 460
He Leu Gly Thr Gin Gin Phe Lys Pro Asn Glu Phe Ala Ser Gin He 465 470 475 480
Asn Leu Ser Val Glu Asn Ala Trp Gly He Leu Arg Cys Val He Asp 485 490 495 He Cys Met Lys Leu Glu Glu Gly Lys Tyr Leu He Leu Lys Asp Pro
500 505 510
Asn Lys Gin Val He Arg Val Tyr Ser Leu Pro Asp Gly Thr Phe Ser 515 520 525
Ser
(2) INFORMATION FOR SEQ ID NO: 639:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 194 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 639:
Lys Lys Arg His Thr Asp Val Gin Phe Tyr Thr Glu Val Gly Glu He 1 5 10 15
Thr Thr Asp Leu Gly Lys His Gin His Met His Asp Arg Asp Asp Leu 20 25 30 Tyr Ala Glu Gin Met Glu Arg Glu Met Arg His Lys Leu Lys Thr Ala 35 40 45
Phe Lys Asn Phe He Glu Lys Val Glu Ala Leu Thr Lys Glu Glu Leu 50 55 60
Glu Phe Glu Val Pro Phe Arg Asp Leu Gly Phe Asn Gly Ala Pro Tyr 65 70 75 80
Arg Ser Thr Cys Leu Leu Gin Pro Thr Ser Ser Ala Leu Val Asn Ala 85 90 95
Thr Glu Trp Pro Pro Phe Val Val Thr Leu Asp Glu Val Glu Leu He 100 105 110 His Phe Xaa Arg Val Gin Phe His Leu Lys Asn Phe Asp Met Val He 115 120 125
Val Tyr Lys Asp Tyr Ser Lys Lys Val Thr Met He Asn Ala He Pro 130 135 140
Val Ala Ser Leu Asp Pro He Lys Glu Trp Leu Asn Ser Cys Asp Leu 145 150 155 160
Lys Tyr Thr Glu Gly Val Gin Ser Leu Asn Trp Thr Lys He Met Lys 165 170 175
Thr He Val Asp Asp Pro Glu Gly Phe Phe Glu Gin Gly Gly Trp Ser 180 185 190 Phe Leu
(2) INFORMATION FOR SEQ ID NO: 640:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 70 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 640:
Arg Ser Gly Leu Gly Leu Gly He Thr He Ala Phe Leu Ala Thr Leu 1 5 10 15
He Thr Gin Phe Leu Val Tyr Asn Gly Val Tyr Gin Tyr Thr Ser Pro 20 25 30
Asp Phe Leu Tyr He Arg Ser Trp Leu Pro Cys He Phe Phe Ser Gly 35 40 45
Gly Val Thr Val Gly Asn He Gly Arg Gin Leu Ala Met Gly Val Pro 50 55 60 Glu Lys Pro His Ser Asp 65 70
(2) INFORMATION FOR SEQ ID NO: 641:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 101 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 641:
Val Thr Gin Pro Lys His Leu Ser Ala Ser Met Gly Gly Ser Val Glu 1 5 10 15
He Pro Phe Ser Phe Tyr Tyr Pro Trp Glu Leu Ala Xaa Xaa Pro Xaa 20 25 30
Val Arg He Ser Trp Arg Arg Gly His Phe His Gly Gin Ser Phe Tyr 35 40 45
Ser Thr Arg Pro Pro Ser He His Lys Asp Tyr Val Asn Arg Leu Phe 50 55 60 Leu Asn Trp Thr Glu Gly Gin Glu Ser Gly Phe Leu Arg He Ser Asn 65 70 75 80
Leu Arg Lys Glu Asp Gin Ser Val Tyr Phe Cys Arg Val Glu Leu Asp 85 90 95
Thr Arg Arg Ser Gly 100
(2) INFORMATION FOR SEQ ID NO: 642:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 233 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 642:
Met Glu Ala Gin Gin Val Asn Glu Ala Glu Ser Ala Arg Glu Gin Leu 1 5 10 15
Gin Xaa Leu His Asp Gin He Ala Gly Gin Lys Ala Ser Lys Gin Glu 20 25 30 Leu Glu Thr Glu Leu Glu Arg Leu Lys Gin Glu Phe His Tyr He Glu 35 40 45
Glu Asp Leu Tyr Arg Thr Lys Asn Thr Leu Gin Ser Arg He Lys Asp 50 55 60
Arg Asp Glu Glu He Gin Lys Leu Arg Asn Gin Leu Thr Asn Lys Thr 65 70 75 80
Leu Ser Asn Ser Ser Gin Ser Glu Leu Glu Asn Arg Leu His Gin Leu 85 90 95
Thr Glu Thr Leu He Gin Lys Gin Thr Met Leu Glu Ser Leu Ser Thr 100 105 110 Glu Lys Asn Ser Leu Val Phe Gin Leu Glu Arg Leu Glu Gin Gin Met 115 120 125
Asn Ser Ala Ser Gly Ser Ser Ser Asn Gly Ser Ser He Asn Met Ser 130 135 140
Gly He Asp Asn Gly Glu Gly Thr Arg Leu Arg Asn Val Pro Val Leu 145 150 155 160
Phe Asn Asp Thr Glu Thr Asn Leu Ala Gly Met Tyr Gly Lys Val Arg 165 170 175
Lys Ala Ala Ser Ser He Asp Gin Phe Ser He Arg Leu Gly He Phe 180 185 190 Leu Arg Arg Tyr Pro He Ala Arg Val Phe Val He He Tyr Met Ala 195 200 205
Leu Leu His Leu Trp Val Met He Val Leu Leu Thr Tyr Thr Pro Glu 210 215 220
Met His His Asp Gin Pro Tyr Gly Lys 225 230
(2) INFORMATION FOR SEQ ID NO: 643:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 43 amino acids (B) TYPE: amino acid
(D) TOPOLOGY: linear (xi) SEQUENCE DESCRIPTION: SEQ ID NO: 643:
He Arg His Glu Gin His Pro Asn Phe Ser Leu Glu Met His Ser Lys 1 5 10 15
Gly Ser Ser Leu Leu Leu Phe Leu Pro Gin Leu He Leu He Leu Pro 20 25 30
Val Cys Ala His Leu His Glu Glu Leu Asn Cys 35 40
(2) INFORMATION FOR SEQ ID NO: 644:
(i) SEQUENCE CHARACTERISTICS:
(A) LENGTH: 63 amino acids
(B) TYPE: amino acid (D) TOPOLOGY: linear
(xi) SEQUENCE DESCRIPTION: SEQ ID NO: 644:
Ser Phe Phe He Ser Glu Glu Lys Gly His Leu Leu Leu Gin Ala Glu 1 5 10 15
Arg His Pro Trp Val Ala Gly Ala Leu Val Gly Val Ser Gly Gly Leu 20 25 30
Thr Leu Thr Thr Cys Ser Gly Pro Thr Glu Lys Pro Ala Thr Lys Asn 35 40 45
Tyr Phe Leu Lys Arg Leu Leu Gin Glu Met His He Arg Ala Asn 50 55 60
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule \3bis)
A. The indications made below relate to the microorganism referred to in the description on page 1 16 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet r~ι
Name of depositary institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parklawn Drive Rockville, Maryland 20852 United States of America
Date of deposit February 26, 1997 Accession Number 97897
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet [
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (if the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank if not applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications, e.g., "Accession Number of Deposit")
For receiving Office use only , ■ For International Bureau use only
MThis sheet was received with the international application D This sheet was received by the International Bureau on:
Authorized officer Authorized officer
Susan White
PCT Internattonal Division
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule \3bιs)
A. The indications made below relate to the microorganism referred to in the description on page 1 16 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet r I
Name of depositary institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parklawn Drive Rockville, Maryland 20852 United States of America
Date of deposit May 15 , 1997 Accession Number 209043
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet rj
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (if the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank if not applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications, e g , "Accession Number of Deposit")
For receiving Office use only . ■ For International Bureau use only
| fThιs sheet was received with the international application D This sheet was received by the International Bureau on
Authorized officer Authorized officer
Susan White PCTIntemβtlonal Division
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule Ϊ3bιs)
A. The indications made below relate to the microorganism referred to in the description on page 1 19 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet r
Name of depositary institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parklawn Drive Rockville, Maryland 20852 United States of America
Date of deposit September 4, 1997 Accession Number 209235
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet rj
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (if the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank if not applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications, e g , "Accession Number of Deposit")
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule \ 3bιs)
A. The indications made below relate to the microorganism referred to in the description on page 122 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet r
Name of depositary institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parklawn Drive Rockville, Maryland 20852 United States of America
Date of deposit February 26, 1997 Accession Number 97898
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet ]
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (if the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank if not applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications, e g , "Accession Number of Deposit")
For receiving Office use only , ■ For International Bureau use only
.This sheet was received with the international application
Sϋ D This sheet was received by the International Bureau on
Authorized officer Authorized officer
Susan White
PCT International Division
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule Ϊ3bιs)
A. The indications made below relate to the microorganism referred to in the description on page 122 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet r ]
Name of depositary institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parklawn Drive Rockville, Maryland 20852 United States of America
Date of deposit May 15, 1997 Accession Number 209044
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet [
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (if the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank if not applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications e g "Accession Number of Deposit")
For receiving Office use only , ■ For International Bureau use only
'This sheet was received with the international application was received by the International Bureau on
Q D This sheet
Authorized officer Authorized officer
Susan White
PCT International Division
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule 13* is)
A. The indications made below relate to the microorganism referred to in the description on page 126 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet r~]
Name of depositary institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parklawn Drive Rockville, Maryland 20852 United States of America
Date of deposit February 26, 1997 Accession Number 97899
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet ]
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (if the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank if not applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications, e g , "Accession Number of Deposit")
For receiving Office use only . ■ For International Bureau use only
IT T!his sheet was received with the international application D This sheet was received by the International Bureau on
Authorized officer Authorized officer
Susan White
PCT International Division
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule \ 3bts)
The indications made below relate to the microorganism referred to in the description on page 126 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet I I
Name of depositary institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parklawn Drive Rockville, Maryland 20852 United States of America
Date of deposit May 15, 1997 Accession Number 209045
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet rπ
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (if the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank if not applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications e g "Accession Number of Deposit")
For receiving Office use only , ■ For International Bureau use only
B? is sheet was received with the international application D This sheet was received by the International Bureau on
Authorized officer Authorized officer
Susan White
PCT international Division
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule Ϊ3bιs)
The indications made below relate to the microorganism referred to in the description on page 130 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet rπ
Name of depositary institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parklawn Drive Rockville, Maryland 20852 United States of America
Date of deposit Apπl 28, 1997 Accession Number 20901 1
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet Q
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (if the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank if not applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications e g "Accession Number of Deposit")
For receiving Office use only , ■ For International Bureau use only
This sheet was received with the international application D This sheet was received by the International Bureau on
Authorized officer Authorized officer
Susan White
PCT International Division
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule \3bts)
A. The indications made below relate to the microorganism referred to in the description on page 131 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet r i
Name of depositary institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parklawn Drive Rockville, Maryland 20852 United States of America
Date of deposit February 26, 1997 Accession Number 97900
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet ]
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (if the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank if not applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications e g "Accession Number of Deposit")
For receiving Office use only , ■ For International Bureau use only
B This sheet was received with the international application D This sheet was received by the International Bureau on
Authorized officer Authorized officer
Stβan White
PCT International Otvtøon
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule \ 3bis)
A. The indications made below relate to the microorganism referred to in the description on page 137 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet rπ
Name of depositary institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parklawn Drive Rockville, Maryland 20852 United States of America
Date of deposit February 26, 1997 Accession Number 97901
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (if the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank if not applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications, e g , "Accession Number of Deposit")
For receiving Office use only , ■ For International Bureau use only the international application
D This sheet was received by the International Bureau on
Authorized officer
Susan White Authorized officer
PCT International Division
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule \3bιs)
A. The indications made below relate to the microorganism referred to in the description on page 131 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet Q~j
Name of depositary institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parklawn Drive Rockville, Maryland 20852 United States of America
Date of deposit May 15, 1997 Accession Number 209046
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet j^j
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (if the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank if not applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications e g "Accession Number of Deposit")
For receiving Office use only , ■ For International Bureau use only
This sheet was received with the international application
D This sheet was received by the International Bureau on
Authorized officer
Susan White Authorized officer
PCT IπtemβfloπaJ Division
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule \ 3bts)
A. The indications made below relate to the microorganism referred to in the description on page 137 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet I |
Name of depositary institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parklawn Drive Rockville, Maryland 20852 United States of America
Date of deposit May 15, 1997 Accession Number 209047
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet r ]
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (f the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank if not applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications e g "Accession Number of Deposit")
For receiving Office use only , ■ For International Bureau use only
This sheet was received with the international application
M D This sheet was received by the International Bureau on
Authorized officer SUSβn White Authorized officer
PCT Iπtemβrtonal Division
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule \ 3bιs)
For receiving Office use only , ■ For International Bureau use only with the international application
D This sheet was received bv the International Bureau on
Authorized officer Susan White Authorized officer
PCT International DlvWon
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule \ 3bts)
A. The indications made below relate to the microorganism referred to in the description on page 140 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet I l
Name of depositary institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parklawn Drive Rockville, Maryland 20852 . United States of America
Date of deposit August 21 , 1997 Accession Number 209215
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet [ ]
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (if the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank if not applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications e g "Accession Number of Deposit")
For receiving Office use onl , ■ For International Bureau use only T" sheet was received with the international application D This sheet was received by the International Bureau on
Authorized officer Authorized officer
Susan White
PCT international Dlvlsiøy
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule \3bts)
A. The indications made below relate to the microorganism referred to in the description on page 160 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet ι — |
Name of depositary institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parklawn Drive Rockville, Maryland 20852 United States of America
Date of deposit February 26, 1997 Accession Number 97904
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet r
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (if the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank if not applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications e g "Accession Number of Deposit")
For receiving Office use only , i For International Bureau use only
W This sheet was received with the international application D This sheet was received by the International Bureau on
Authorized officer SUSβn White Authorized officer
PCT International DMsioø
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule \ 3bιs)
A. The indications made below relate to the microorganism referred to in the description on page 154 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet ι |
Name of depositary institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parkla n Drive Rockville, Maryland 20852 United States of America
Date of deposit July 3, 1997 Accession Number 209139
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet [~J
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (if the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank if not applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications, e g , "Accession Number of Deposit")
For receiving Office use only , ■ For International Bureau use only
I T Thhis sheet was received with the international application
D This sheet was received by the International Bureau on
Authorized officer Susan White Authorized officer
PCT International Division
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule \3bts)
The indications made below relate to the microorganism referred to in the description on page 153 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet I l
Name of depositary institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parklawn Drive Rockville, Maryland 20852 United States of America
Date of deposit May 15, 1997 Accession Number 209049
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet [J
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (if the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank if not applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications, e g "Accession Number of Deposit")
For receiving Office use only , ' For International Bureau use only
® This sheet was received with the international application D This sheet was received by the International Bureau on
Authorized officer Authorized officer
Susan White
PCT International Division
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule \ 3bιs)
A. The indications made below relate to the microorganism referred to in the description on page 153 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet | ι
Name of depositary institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parklawn Drive Rockville, Maryland 20852 United States of America
Date of deposit February 26, 1997 Accession Number 97903
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet r~]
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (If the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank , fnot applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications, e g "Accession Number of Deposit")
For receiving Office use only , i For International Bureau use only sheet was received with the international application
D This sheet was received by the International Bureau on
Authorized officer Authorized officer
Susan White
PCT International Division
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule \ 3bts)
A. The indications made below relate to the microorganism referred to in the description on page 142 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet l I
Name of depositary institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parklawn Drive Rockville, Maryland 20852 United States of America
Date of deposit June 12, 1997 Accession Number 2091 19
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet I 1
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (if the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank if not applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications, e g , "Accession Number of Deposit")
For receiving Office use only , For International Bureau use only iThis sheet was received with the international application D This sheet was received by the International Bureau on
Authorized officer Susan White Authorized officer
PCT International Division
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule \ 3bιs)
A. The indications made below relate to the microorganism referred to in the description on page 146 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet r ]
Name of depositary institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parklawn Drive Rockville, Maryland 20852 United States of America
Date of deposit February 26, 1997 Accession Number 97902
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (if the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank ifnot applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications e g "Accession Number of Deposit")
For receiving Office use only , ■ For International Bureau use only sheet was received with the international application
D This sheet was received by the International Bureau on
Authorized officer SUSβD Whttff Authorized officer
PCT International DMsion
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule \ 3bιs)
A. The indications made below relate to the microorganism referred to in the description on page 146 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet ■" l
Name of depositary institution
American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parklawn Drive Rockville, Maryland 20852 United States of America
Date of deposit May 15, 1997 Accession Number 209048
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet | ]
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (if the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank not applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications e g Accession Number of Deposit")
For receiving Office use only , ■ For International Bureau use only
VT This sheet was received with the international application D This sheet was received by the International Bureau on
Authorized officer Authorized officer
Susan White
PCT International Division
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule \ 3bts)
For receiving Office use only , ' For International Bureau use only
This sheet was received with the international application
D This sheet was received by the International Bureau on
Authorized officer SUSBn W ttβ Authorized officer
PCT International Division
INDICATIONS RELATING TO A DEPOSITED MICROORGANISM
(PCT Rule \ 3bιs)
The indications made below relate to the microorganism referred to in the description on page 142 , line N/A
B. IDENTIFICATION OF DEPOSIT Further deposits are identified on an additional sheet
Name of depositary institution American Type Culture Collection
Address of depositary institution (including postal code and country)
12301 Parklawn Drive Rockville, Maryland 20852 United States of America
Date of deposit February 12, 1998 Accession Number 209627
C. ADDITIONAL INDICATIONS (leave blank if not applicable) This information is continued on an additional sheet [~]
D. DESIGNATED STATES FOR WHICH INDICATIONS ARE MADE (if the indications are not for all designated States)
E. SEPARATE FURNISHING OF INDICATIONS (leave blank if not applicable)
The indications listed below will be submitted to the International Bureau later (specify the general nature of the indications, e g , "Accession Number of Deposit")
For receiving Office use only . ■ For International Bureau use only sheet was received with the international application
D This sheet was received by the International Bureau on
Authorized officer Susan White Authorized officer
PCT IntemaHonβf Division